Abstract
Mitochondria are highly dynamic organelles that mediate essential cell functions such as apoptosis and cell-cycle control in addition to their role as efficient ATP generators. Mitochondrial morphology changes are tightly regulated, and their shape can shift between small, fragmented units and larger networks of elongated mitochondria. We demonstrate that mitochondrial elements become significantly elongated and interconnected shortly after nutrient depletion. This mitochondrial morphological shift depends on the type of starvation, with an additive effect observed when multiple nutrients are depleted simultaneously. We further show that starvation-induced mitochondrial elongation is mediated by down-regulation of dynamin-related protein 1 (Drp1) through modulation of two Drp1 phosphorylation sites, leading to unopposed mitochondrial fusion. Finally, we establish that mitochondrial tubulation upon nutrient deprivation protects mitochondria from autophagosomal degradation, which could permit mitochondria to maximize energy production and supply autophagosomal membranes during starvation.
Keywords: autophagy, mitofusin
Mitochondria are dynamic organelles that mediate many essential cell functions. Depending on the cellular context, mitochondria shift between fragmented and tubular network-like morphologies by means of coordinated fission and fusion (1, 2). Proteins responsible for mitochondrial fusion include the outer mitochondrial membrane proteins mitofusin 1 (Mfn1) and mitofusin 2 (Mfn2) (3–5) and the inner membrane protein optic atrophy 1 (Opa1) (6). Fission is mediated by dynamin-related protein 1 (Drp1) (7, 8) and its interaction with binding partners Fis1 and/or mitochondrial fission factor (Mff) (9, 10). Multiple mechanisms, including phosphorylation, sumoylation, and ubiquitination, coordinate Drp1 fission capacity (11–14). Phosphorylation of Drp1 at S616 by Cdk1/cyclin B results in increased Drp1 fission activity (13). Conversely, phosphorylation of Drp1 at S637 by PKA decreases fission by causing Drp1 retention in the cytosol, whereas dephosphorylation of S637 by calcineurin causes Drp1 translocation to the mitochondria and increased mitochondrial fission (11, 15).
Mitochondrial morphological dynamics are linked to regulation of many specific cell functions. Changes in mitochondrial cristae and mitochondrial fragmentation, for example, play a vital role in apoptosis (16, 17). Ca2+ transfer (18), cell-cycle regulation (13, 19), and mitochondrial quality control (20, 21) are all closely tied to changes in mitochondrial morphology. Furthermore, stress conditions and changes in energy source can induce significant mitochondrial morphological changes (22, 23). Very recently, nutrient starvation was shown to induce mitochondrial elongation and to protect mitochondria from autophagic degradation (24). In addition to the above functions, mitochondria have recently been linked to autophagosome biogenesis during starvation conditions (25), and it is possible that mitochondrial morphological changes play a role in this process.
Autophagy is a catabolic process that proceeds via formation of a double-membrane vesicle, the autophagosome. Both cytosolic and organellar components can be engulfed by autophagosomes, which then fuse with lysosomes. Once delivered to the lysosome, autophagosomal contents are recycled, allowing prolonged cell survival when nutrients are scarce (26). Recent research shows that the autophagy machinery can target specific substrates for degradation depending on the cell's needs in specific contexts such as mitochondrial stress (27, 28). Autophagy induced by starvation, however, has traditionally been viewed as a nonspecific process in which cytosolic and organellar components are turned over en masse. Cells become highly dependent on autophagy when nutrients are severely limited because autophagy allows for replenishment of vital macromolecular precursors, including amino acids, sugars, and fatty acids. These breakdown products provide substrates for biosynthesis of core cell components and metabolic substrates for the ATP production required to drive macromolecule synthesis during starvation. Despite a presumed need under starvation conditions for efficient ATP production provided by mitochondria, it has been unclear whether changes in mitochondrial morphology occur to facilitate promotion of cell survival and how those dynamic changes in mitochondrial structure might be mediated.
Here, we address these questions by examining mitochondrial morphology under nutrient deprivation. Our results demonstrate that mitochondrial elongation is induced shortly after starvation. This mitochondrial tubulation is reversible and depends on the specific nutrients depleted. Mitochondrial elongation does not occur when autophagy is induced by serum depletion or glucose elimination. Rather, mitochondrial elongation is a specific response to deprivation of glutamine and/or amino acids, with faster, more extensive mitochondrial tubulation upon additional depletion of glucose and serum. We demonstrate that starvation-induced mitochondrial elongation is mediated by down-regulation of Drp1, leading to unopposed mitochondrial fusion. The mitochondrial elongation response is further shown to protect mitochondria from autophagic turnover under starvation conditions. This ability of autophagosomes to exclude mitochondria from autophagic sequestration during starvation may serve to provide efficient aerobic ATP production when energy sources are limited and also highlights the previously unappreciated substrate specificity of the autophagosomal pathway during nutrient deprivation.
Results
Mitochondria Elongate During Cell Starvation.
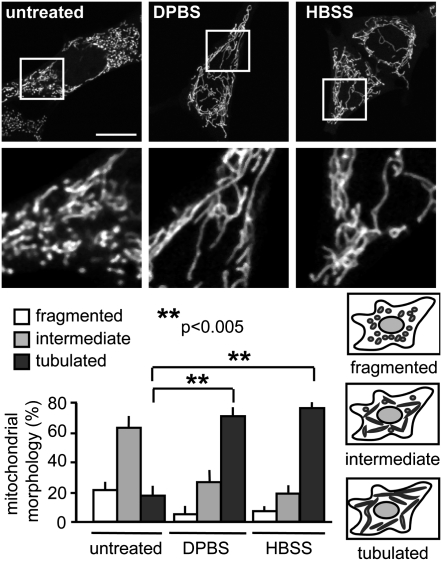
To determine whether mitochondria undergo morphological changes upon starvation, we imaged mouse embryonic fibroblasts (MEFs) expressing the mitochondrial matrix targeted YFP (mito-YFP). Starvation for 2 h with either Dulbecco's PBS (DPBS; see Fig. 2 for specific medium compositions) or HBSS resulted in a dramatic increase in the percentage of cells with tubulated mitochondria from 17% under normal growth conditions to 70% in DPBS and 76% in HBSS (Fig. 1), with significant tubulation observed within 1 h (Fig. S1A). MEF cells starved for 2 h and immunostained for the endogenous mitochondrial matrix protein Hsp60 also showed a significant degree of mitochondrial tubulation, demonstrating that mitochondrial fusion is not a result of mito-YFP overexpression (Fig. S2). Mitochondrial tubulation was also observed upon starvation in COS-7, HCT116, and HEK293T cells, suggesting that mitochondrial elongation is a common response to nutrient deprivation in many cell culture types (Fig. S1B). Replenishment of serum, glucose, and glutamine after starvation resulted in a return to less tubulated mitochondrial morphology within 2 h, demonstrating that starvation-induced elongation of mitochondria is reversible (Fig. S1C).
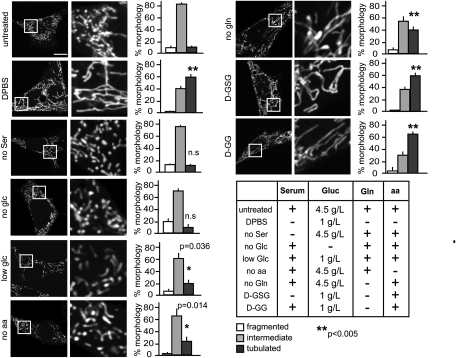
Fig. 2.
Mitochondrial tubulation is more efficiently induced by additive nutrient deprivation. MEFs were transfected with mito-YFP and starved for 2 h (DPBS, D-GSG, and no Ser) or 6 h (no glc, low glc, no aa, no gln, and D-GG). Media compositions are given in the table. Mitochondrial morphology was scored as described in Fig. 1 (n ≥ 3). Representative images of cells are shown in the first column. Images in the second column are magnified views of the boxed areas in the first column. glc, glucose; gln, glutamine; aa, amino acids. (Scale bar: 15 μm.)
Fig. 1.
Starvation induces mitochondrial tubulation. MEFs were transfected with mito-YFP. (Upper) Cells were starved 16 h after transfection for 2 h, and live images were acquired. (Images in the second row are magnified views of the boxed areas in first row.) (Lower) Mitochondrial morphology was scored as follows: fragmented, mainly small and round; intermediate, mixture of round and shorter tubulated; and tubulated, long and higher interconnectivity. The percentage of cells with indicated mitochondrial morphologies was determined as a percentage of the total number of transfected cells counted (≥100 cells per experiment). n = 6 independent experiments. (Scale bar: 15 μm.)
Because starvation is known to induce autophagy, we next investigated whether induction of the autophagy machinery is a prerequisite for mitochondrial elongation. To address this question, we used Atg5-deficient MEFs (Atg5KO), which lack the ability to elongate the isolation membrane (29). Starvation of Atg5KO cells led to increased mitochondrial tubulation relative to untreated cells (Fig. S1D). The retained ability of Atg5KO MEF cells to induce mitochondrial tubulation under nutrient depletion suggests that the early autophagosomal machinery is not required for starvation-induced mitochondrial elongation.
Starvation Conditions Differentially Induce Mitochondrial Tubulation.
To determine the minimal starvation conditions capable of inducing mitochondrial tubulation, we consecutively depleted single groups of nutrients. A summary of the nutrient-depletion conditions and their respective effects on mitochondrial morphology is presented in Fig. 2. Neither glucose nor serum elimination resulted in mitochondrial elongation. In fact, increased mitochondrial fragmentation was observed after 6–9 h of either serum or glucose depletion. Reduction of glucose from 4.5 g/L to 1 g/L prevented the increased mitochondrial fragmentation driven by complete glucose starvation and induced some mitochondrial elongation. Interestingly, elimination of either glutamine or amino acids from the growth medium resulted in increased mitochondrial tubulation after 6 h of starvation, indicating that a nitrogen-source deficiency can effect mitochondrial fusion.
Combinations of nutrient depletions proved the fastest and most effective way to induce mitochondrial elongation. Starvation in low-glucose (1 g/L) and no-glutamine conditions resulted in more cells containing fused mitochondria (69%) than in cells deprived of single nutrients after 6 h (23% after reduced glucose and 39% after glutamine elimination). Additional deprivation of growth factors by serum depletion promoted an equally substantial mitochondrial elongation and decreased the time needed for robust tubulation from 6 to 1.5 h. The additive effect of combined nutrient starvations on the extent and rate of mitochondrial elongation indicates that mitochondrial tubulation can be modulated according to the type and severity of starvation stress.
Starvation-Induced Mitochondrial Elongation Depends on Mfn1 and Opa1 and Is Mediated by Two Drp1 Posttranslational Modifications.
To identify the machinery involved in mitochondrial elongation upon starvation, we used MEF cell lines with deficiencies in the indicated mitochondrial fusion proteins: Mfn1 (Mfn1KO), Mfn2 (Mfn2KO), Mfn1 and 2 (Mfn1/2DKO), and Opa1 (Opa1KO) (5, 30). All of the knockout cell lines analyzed exhibited heavily fragmented mitochondria during normal growth conditions. After 2 h of incubation in medium with low glucose (1 g/L), no serum, and no glutamine (hereafter referred to as D-GSG), increased mitochondrial tubulation did not occur in Mfn1/2DKO and Opa1KO cells, in contrast to the elongation observed in WT MEFs (Fig. S2). D-GSG starvation of Mfn1KO MEFs resulted in slight mitochondrial elongation, in which a small increase in mitochondrial length was observed but no truly tubulated phenotype was apparent. The Mfn2KO cells' elongation capacity mirrored that of the WT cells when starved with D-GSG (Fig. S2). The high degree of elongation in nutrient-deprived Mfn2KO MEFs is particularly remarkable in light of their initial highly fragmented state. These results indicate that Mfn1 is essential for starvation-driven mitochondrial tubulation, whereas Mfn2 is dispensable for this activity.
To verify that starvation-induced mitochondrial fusion is independent of Mfn2, we analyzed cells deficient for Bax and Bak (Bax/BakDKO) (31), which have been implicated in the control of Mfn2-mediated mitochondrial fusion (32, 33). D-GSG starvation of Bax/BakDKO cells induced extensive mitochondrial fusion, similar to the effect observed in starved WT and Mfn2KO cells (Fig. S2). The ability of Mfn2KO and Bax/BakDKO MEFs to elongate mitochondria further suggests a minimal role for Mfn2 and indicates that starvation-induced mitochondrial fusion is mainly mediated through Opa1 and Mfn1. A prominent role for Mfn1 in effecting mitochondrial fusion during starvation is consistent with evidence implicating Mfn1 as the primary driver of mitochondrial fusion activity, whereas Mfn2 has a more diverse set of functions, including responsibility for endoplasmic reticulum–mitochondria tethering (34, 35).
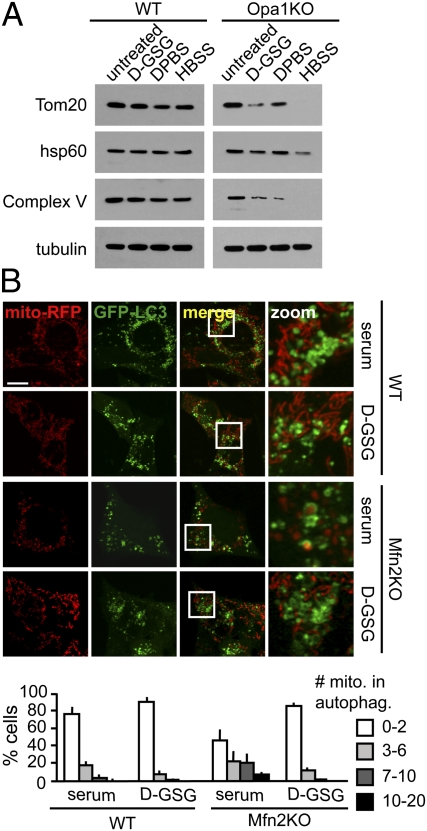
To better understand the mechanism underlying starvation-induced mitochondrial elongation, we conducted a biochemical analysis of the core mitochondrial fission and fusion machinery for changes elicited under starvation conditions. No change was observed in total Mfn2, Opa1, or Drp1 protein levels or migration rates by performing denaturing gel electrophoresis on D-GSG, DPBS, and HBSS starvation of MEF cells (Fig. 3A). We observed an apparent decrease in Mfn1 levels upon serum depletion, but the change was not statistically significant (Fig. S3A).
Fig. 3.
Unopposed fusion mediates mitochondrial tubulation during starvation. (A, C, and D) MEFs were treated with the media indicated for 2.5 h (A) or 5 h (C and D) and lysed in ice-cold lysis buffer A, and samples were subjected to immunoblotting against Mfn1, Mfn2, Opa1, Drp1, and pS616-Drp1 as indicated (whole-cell lysates were probed in A and C; isolated mitochondria were probed in D). (B) At 16 h after transfection with the indicated YFP-tagged Drp1 constructs, cells were treated with full medium or starved in D-GSG for 2 h. Mitochondrial morphology was scored as described in Fig. 1 (n = 3).
Because Drp1 is regulated by phosphorylation, we sought to assess the potential involvement of Drp1 phosphorylation sites in starvation-induced mitochondrial elongation. Drp1-S616 phosphorylation decreased upon starvation (Fig. 3C) as determined by immunoblotting with a phospho-S616–specific antibody. This decreased Drp1-S616 phosphorylation is consistent with an increase in mitochondrial elongation attributable to inhibited Drp1 fission activity. A phosphomimetic mutant (Drp1-S616E) overexpressed in MEFs led to less mitochondrial elongation than observed in cells transfected with WT Drp1 (wtDrp1) during starvation (Fig. 3B and Fig. S3B). Overexpression of a nonphosphorylatable Drp1-S616A mutant, on the other hand, did not affect mitochondrial elongation after starvation (Fig. S3C).
We next investigated the effect of a second posttranslational modification on Drp1-mediated mitochondrial fission during starvation. Overexpression of Drp1-S637A in MEFs resulted in significantly fewer cells with elongated mitochondria compared with wtDrp1-transfected cells after starvation (Fig. 3B and Fig. S3B). This finding is consistent with a report showing that phosphorylation of Drp1-S637 results in cytosolic retention of Drp1 and a decrease in mitochondrial fission (15). Overexpression of the double mutant Drp1-S616E/S637A during starvation resulted in a statistically significant decrease in mitochondrial tubulation compared with either Drp1-S616E– or Drp1-S637A–tranfected cells, indicating that a synergistic effect of the two posttranslational modifications is possible (Fig. 3B and Fig. S3D).
A comparison of Drp1 levels associated with mitochondria revealed a substantial decrease in mitochondrial localization of Drp1 upon starvation, whereas total Drp1 levels remained constant (Fig. 3 A and D), suggesting a reduction in the ability of Drp1 to effect fission after nutrient depletion. Together, these data suggest that starvation inhibits mitochondrial fission both by decreasing Drp1 fission activity and by preventing Drp1 localization to mitochondria, leading to unopposed Opa1/Mfn1-dependent mitochondrial fusion.
Elongation Protects Mitochondria from Mitophagy.
Mitochondrial fragmentation and dysfunction have been described as preconditions for autophagy of mitochondria (mitophagy) under different conditions (21, 27, 36), which led us to investigate whether starvation-induced mitochondrial fusion can prevent mitochondria from being targeted for autophagy. We did not observe a decrease in mitochondrial mass during a 5-h starvation in WT cells, consistent with our previously reported data (25). However, mitochondrial fusion-deficient Opa1KO cells showed a significant decrease in mitochondrial proteins Tom20, Hsp60, and Complex V (subunit α) under starvation conditions (Fig. 4A). Furthermore, autophagosome-engulfed mitochondria were observed in nutrient-deprived Opa1KO cells containing fragmented mitochondria but not in WT cells with elongated mitochondria (Fig. S4).
Fig. 4.
Mitochondrial tubulation ameliorates autophagosomal degradation of mitochondria during starvation. (A) WT and Opa1KO MEFs were treated with the indicated media for 5 h, and equal numbers of cells were lysed. Whole-cell lysates were subjected to immunoblotting against the proteins indicated. (B) Mfn2KO cells show increased mitophagy during serum starvation, which can be rescued through mitochondrial tubulation. WT and Mfn2KO MEF cells were transfected with GFP-LC3 and mito-RFP and starved for serum or D-GSG for 4 h in the presence of bafilomycin A1, and live images were acquired. Cells were grouped into categories according to the number of mitochondria in autophagosomes and are presented as percentage of the total number of transfected cells counted (n = 3). (Scale bar: 15 μm.)
To more clearly establish whether elongation can protect mitochondria from autophagy, we investigated whether high levels of mitophagy could be reversed by starvation-induced mitochondrial fusion. Here, we took advantage of the fact that serum starvation induces autophagy without inducing mitochondrial fusion (Fig. 2). First, we quantified mitophagy levels in WT and Mfn2KO MEFs transfected with GFP-LC3 and mitochondrially targeted red fluorescent protein (mito-RFP) using live-cell imaging. For better identification of mitochondrial material engulfed in autophagosomes, we blocked lysosomal turnover with bafilomycin A1. A lower incidence of mitophagy was observed in serum-starved WT cells compared with Mfn2KO cells (Fig. 4B). Additionally, there was a large subpopulation (29% of cells) in the starved Mfn2KO cells with >7 mitochondria-containing autophagosomes (approximately 5% in WT cells). The increased incidence of mitophagy could suggest that mitochondria from Mfn2-deficient cells are more prone to dysfunction because of the lack of endoplasmic reticulum–mitochondria tethering and altered calcium handling (34) and is consistent with evidence that serum starvation is associated with enhanced turnover of dysfunctional mitochondria (37). Next, we investigated whether the high levels of mitophagy in serum-starved Mfn2KO cells could be reduced if mitochondrial fusion was induced through glutamine depletion and glucose reduction. Indeed, although 29% of serum-starved Mfn2KO cells had >7 mitochondria in autophagosomes, only 1% of the D-GSG–starved cells contained >7 engulfed mitochondria (Fig. 4B). These findings support the hypothesis that mitochondrial elongation has a strong capacity to prevent mitochondrial turnover during starvation-induced autophagy.
Discussion
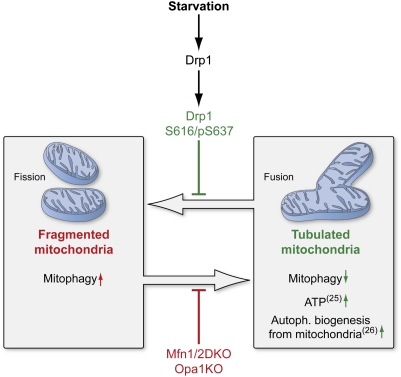
Mitochondrial morphological changes have been associated with many vital cellular functions. Mitochondrial fragmentation is important for progression of apoptosis (38, 39) and M phase (40) and for degradation of defective mitochondria (21). The number of processes involving mitochondrial fusion is equally diverse, underscoring the importance of understanding how changes in mitochondrial morphology are related to specific outcomes in the cell. Several lines of evidence, including the data presented here, indicate that mitochondrial fusion can serve a protective function, leading to the exchange of mitochondrial DNA, reorganization of mitochondrial cristae, and delay in apoptosis (22, 38, 39, 41). It is therefore reasonable that such mitochondrial morphological changes could also be beneficial for cells under conditions of low nutrient supply. Precedent for such energy source-induced mitochondrial morphological change was established on the observation that cells grown with galactose/glutamine instead of glucose resulted in more interconnected mitochondria with increased cristae (23). Here we demonstrate that mitochondria undergo rapid Mfn1- and Opa1-dependent elongation, mediated by two Drp1 posttranslational modifications after nutrient depletion (Fig. 5). The extent and rate of mitochondrial elongation increases upon additive nutrient starvation, with elongation protecting mitochondria from sequestration and subsequent degradation by autophagosomes.
Fig. 5.
Elongation of mitochondria is mediated by posttranslational modification of Drp1 and prevents mitophagy during nutrient starvation. Model shows the protective effect of tubulation on mitochondria upon nutrient deprivation. Tubulation of mitochondria is mediated through modifications of Drp1. Lack of Mfn1/2 or Opa1 inhibits the mitochondria's fusion capacity during starvation. Reference numbers are indicated in parentheses. (This figure was prepared by Medical Illustration, National Institutes of Health Medical Arts Design Section.)
Starvation-induced mitochondrial tubulation requires both Opa1 and Mfn1. Although Mfn2 appears capable of providing slight redundancy for Mfn1 fusion activity, the lack of a strong mitochondrial fusion phenotype in Mfn1KO cells indicates Mfn2 alone is insufficient to induce robust mitochondrial elongation after starvation. Our finding that Mfn2, but not Mfn1, is dispensable for starvation-induced mitochondrial fusion parallels the reported Mfn1 and Mfn2 requirements during stress-induced mitochondrial fusion (22). Mitochondrial elongation does not appear to be activated through increased expression levels of fusion proteins; however, it is unclear whether differences in the oligomerization states of either Mfn1 or Opa1 play a role. We also cannot rule out additional mechanisms like the involvement of SLP-2, which can stabilize the long isoforms of Opa-1 under low-stress conditions (22).
Our investigation of the mitochondrial fission machinery reveals two mechanisms by which Drp1 can be inhibited during nutrient depletion. First, Drp1 mitochondrial localization is decreased, likely by phosphorylation at S637. Second, Drp1 is dephosphorylated at S616. We demonstrate that expression of Drp1-S616E increases fission of starved cells by a small amount compared with the expression of Drp1-S637A. The partial effect of the Drp1-S616E may reflect the fact that Drp1 is regulated by a number of mechanisms. Indeed, a Drp1-S616E/S637A double mutant further reduces mitochondrial fusion capacity relative to each single mutant. It is likely that both Drp1-S616 dephosphorylation and Drp1-S637 phosphorylation are necessary for a robust and rapid mitochondrial elongation response after starvation. A recent publication by Scorrano and colleagues also demonstrated that Drp1-S637 modification can mediate starvation-induced mitochondrial elongation (24). The study did not address the role of Drp1-S616, and it is unclear whether they investigated this particular site. Additional mechanisms for Drp1 down-regulation, such as targeting of Drp1 on mitochondria by E3 ligases to decrease the mitochondrial pool of Drp1 (42, 43), may also be possible.
Our data suggest that mitochondrial elongation serves a protective function from mitophagy, in agreement with the findings of Gomes et al. (24) and consistent with the mitochondrial quality-control study published by Shirihai and colleagues showing dysfunctional mitochondria are unable to fuse into the mitochondrial network, making them susceptible to degradation (21). Another study shows that cytoplasmic components are degraded during the initial period of starvation (0–6 h), whereas mitochondria become a substrate much later (44). Mitochondrial fusion might be the basis for this specific exclusion of mitochondria from degradation during the initial period of starvation. Whether mitochondria cannot be targeted for autophagy because of their size or because the mitophagy machinery does not recognize them in a fused form is still unclear and requires further investigation.
The fact that starvation-induced mitochondrial fusion prevents mitophagy begs the question: Why are mitochondria elongated and spared from autophagy during starvation? Our group recently demonstrated that mitochondria are capable of providing membrane to autophagosomes during starvation (25). Perhaps one advantage of sparing mitochondria during starvation is to permit mitochondria to serve as an autophagosome membrane source. It is also possible that cells elongate and protect their mitochondria during starvation to use the efficient ATP-producing capacity of fused mitochondria. Mitochondrial fusion has been linked to increased ATP production during stress, starvation, and cell cycle (19, 22, 24). Therefore, elongated mitochondria during starvation might lead to increased mitochondrial ATP production important for meeting the cell's metabolic needs while allowing for recycling of other biological materials. Our data demonstrate that depletion of groups of nutrients, including glutamine and amino acids, can elicit the mitochondrial elongation response. It is possible that when deprived of nitrogen sources (i.e., amino acids and glutamine), cells maximize ATP production efficiency so that amino acids can be used exclusively for protein synthesis instead of for energy production. That is, protection of mitochondria from mitophagy combined with autophagic degradation of cytosolic material would provide for both maximal energy production and generation of macromolecular precursors to promote survival during starvation. The additive effect of depleting nutrients important for mitochondrial respiration further supports a model in which mitochondria are protected from autophagy specifically to ensure maximally efficient ATP production under nutrient-limiting conditions.
Finally, we show that Mfn2KO MEFs can be rescued from serum starvation-induced mitophagy by additional depletion of glucose and glutamine. These data demonstrate that starvation-induced autophagy is not as nonspecific as previously assumed, and substrate targeting depends on the type of nutrients that are depleted. Our data suggest that targeting of mitochondria by the autophagy machinery can be rapidly reversed depending on the stimulus and, presumably, the cell's needs. The fact that autophagy is capable of differentially targeting substrates during starvation is perhaps not surprising, given that peroxisomes (45), bacteria (46), and other substrates are also preferentially targeted under specific conditions. Further investigation of autophagic responses to specific stresses and determination of how substrate specificity is achieved will be crucial to gain a better understanding of the mechanisms promoting cell survival under diverse nutrient-limiting conditions.
Materials and Methods
Cell Culture and Transfection.
All cell lines were maintained in DMEM with 10% FBS. For starvation experiments, media was purchased from Invitrogen. Bafilomycin A1 (Sigma-Aldrich) was used at a concentration of 200 nM for the time points indicated. Cells were transfected with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions.
Cells were transfected with mito-YFP, mito-RFP, GFP-Drp1, and GFP-LC3 as described previously (19, 25). Drp1 mutants YFP-Drp1-S616E, YFP-Drp1-S637A, and YFP-Drp1-S616E/S616A (numbers correspond to human isoform 1; isoform 4 was used here) were generated by site-directed mutagenesis. Primers are listed in SI Materials and Methods.
Immunoblotting and Mitochondrial Isolation.
Antibody purchasing information can be found in SI Materials and Methods. Mitochondria were isolated with the Mitochondria Isolation Kit for Cultured Cells (Pierce) according to the manufacturer's instructions. Cells and isolated mitochondria were lysed with lysis buffer A [20 mM Tris (pH 7.4), 150 mM NaCl, 2 mM EDTA, and 1% Triton X100 with protease and phosphatase inhibitors] for 20 min at 4 °C. Protein concentrations were measured with bicinchoninic acid (Pierce) according the manufacturer's instructions, and 10 μg of protein was loaded on 4–12% NUPAGE Gels (Invitrogen). Primary antibodies were added for 16 h at 4 °C and secondary antibodies for 1 h at 24 °C. Antibody binding was detected with SuperSignal West Pico Chemiluminescent Substrate (Pierce).
Imaging and Statistics.
Cells were stained as described previously (47). Cells were seeded either in LabTek chambers (Thermo Scientific) or on coverslips for live-cell imaging and immunofluorescence, respectively. Live and fixed cells were imaged on an LSM 510 META Zeiss microscope using a 63× oil objective at 37 °C. Images were analyzed with Karl Zeiss MicroImaging Software. Brightness and contrast were adjusted in Adobe Photoshop CS. Data were expressed as means ± SD. Statistical analysis among groups was performed using Student's t test.
Supplementary Material
Acknowledgments
We thank R. Youle for contributing the Bax/BakDKO cells and the YFP-Drp1 construct and N. Mizushima and D. C. Chan for contributing the ATG5KO and Mfn-deficient cell lines, respectively. We thank K. Mitra for helpful discussions. A.R. was supported by a postdoctoral fellowship from the Deutsche Forschungsgemeinschaft.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1107402108/-/DCSupplemental.
References
- 1.Bereiter-Hahn J. Behavior of mitochondria in the living cell. Int Rev Cytol. 1990;122:1–63. doi: 10.1016/s0074-7696(08)61205-x. [DOI] [PubMed] [Google Scholar]
- 2.Nunnari J, et al. Mitochondrial transmission during mating in Saccharomyces cerevisiae is determined by mitochondrial fusion and fission and the intramitochondrial segregation of mitochondrial DNA. Mol Biol Cell. 1997;8:1233–1242. doi: 10.1091/mbc.8.7.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Santel A, et al. Mitofusin-1 protein is a generally expressed mediator of mitochondrial fusion in mammalian cells. J Cell Sci. 2003;116:2763–2774. doi: 10.1242/jcs.00479. [DOI] [PubMed] [Google Scholar]
- 4.Santel A, Fuller MT. Control of mitochondrial morphology by a human mitofusin. J Cell Sci. 2001;114:867–874. doi: 10.1242/jcs.114.5.867. [DOI] [PubMed] [Google Scholar]
- 5.Chen H, et al. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J Cell Biol. 2003;160:189–200. doi: 10.1083/jcb.200211046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meeusen S, et al. Mitochondrial inner-membrane fusion and crista maintenance requires the dynamin-related GTPase Mgm1. Cell. 2006;127:383–395. doi: 10.1016/j.cell.2006.09.021. [DOI] [PubMed] [Google Scholar]
- 7.Otsuga D, et al. The dynamin-related GTPase, Dnm1p, controls mitochondrial morphology in yeast. J Cell Biol. 1998;143:333–349. doi: 10.1083/jcb.143.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bleazard W, et al. The dynamin-related GTPase Dnm1 regulates mitochondrial fission in yeast. Nat Cell Biol. 1999;1:298–304. doi: 10.1038/13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoon Y, Krueger EW, Oswald BJ, McNiven MA. The mitochondrial protein hFis1 regulates mitochondrial fission in mammalian cells through an interaction with the dynamin-like protein DLP1. Mol Cell Biol. 2003;23:5409–5420. doi: 10.1128/MCB.23.15.5409-5420.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Otera H, et al. Mff is an essential factor for mitochondrial recruitment of Drp1 during mitochondrial fission in mammalian cells. J Cell Biol. 2010;191:1141–1158. doi: 10.1083/jcb.201007152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cribbs JT, Strack S. Reversible phosphorylation of Drp1 by cyclic AMP-dependent protein kinase and calcineurin regulates mitochondrial fission and cell death. EMBO Rep. 2007;8:939–944. doi: 10.1038/sj.embor.7401062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harder Z, Zunino R, McBride H. Sumo1 conjugates mitochondrial substrates and participates in mitochondrial fission. Curr Biol. 2004;14:340–345. doi: 10.1016/j.cub.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 13.Taguchi N, Ishihara N, Jofuku A, Oka T, Mihara K. Mitotic phosphorylation of dynamin-related GTPase Drp1 participates in mitochondrial fission. J Biol Chem. 2007;282:11521–11529. doi: 10.1074/jbc.M607279200. [DOI] [PubMed] [Google Scholar]
- 14.Yonashiro R, et al. A novel mitochondrial ubiquitin ligase plays a critical role in mitochondrial dynamics. EMBO J. 2006;25:3618–3626. doi: 10.1038/sj.emboj.7601249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cereghetti GM, et al. Dephosphorylation by calcineurin regulates translocation of Drp1 to mitochondria. Proc Natl Acad Sci USA. 2008;105:15803–15808. doi: 10.1073/pnas.0808249105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scorrano L, et al. A distinct pathway remodels mitochondrial cristae and mobilizes cytochrome c during apoptosis. Dev Cell. 2002;2:55–67. doi: 10.1016/s1534-5807(01)00116-2. [DOI] [PubMed] [Google Scholar]
- 17.Youle RJ, Karbowski M. Mitochondrial fission in apoptosis. Nat Rev Mol Cell Biol. 2005;6:657–663. doi: 10.1038/nrm1697. [DOI] [PubMed] [Google Scholar]
- 18.Park MK, Ashby MC, Erdemli G, Petersen OH, Tepikin AV. Perinuclear, perigranular and sub-plasmalemmal mitochondria have distinct functions in the regulation of cellular calcium transport. EMBO J. 2001;20:1863–1874. doi: 10.1093/emboj/20.8.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mitra K, Wunder C, Roysam B, Lin G, Lippincott-Schwartz J. A hyperfused mitochondrial state achieved at G1-S regulates cyclin E buildup and entry into S phase. Proc Natl Acad Sci USA. 2009;106:11960–11965. doi: 10.1073/pnas.0904875106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen H, Chomyn A, Chan DC. Disruption of fusion results in mitochondrial heterogeneity and dysfunction. J Biol Chem. 2005;280:26185–26192. doi: 10.1074/jbc.M503062200. [DOI] [PubMed] [Google Scholar]
- 21.Twig G, et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008;27:433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tondera D, et al. SLP-2 is required for stress-induced mitochondrial hyperfusion. EMBO J. 2009;28:1589–1600. doi: 10.1038/emboj.2009.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rossignol R, et al. Energy substrate modulates mitochondrial structure and oxidative capacity in cancer cells. Cancer Res. 2004;64:985–993. doi: 10.1158/0008-5472.can-03-1101. [DOI] [PubMed] [Google Scholar]
- 24.Gomes LC, Benedetto GD, Scorrano L. During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nat Cell Biol. 2011;13:589–598. doi: 10.1038/ncb2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hailey DW, et al. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell. 2010;141:656–667. doi: 10.1016/j.cell.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rabinowitz JD, White E. Autophagy and metabolism. Science. 2010;330:1344–1348. doi: 10.1126/science.1193497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schweers RL, et al. NIX is required for programmed mitochondrial clearance during reticulocyte maturation. Proc Natl Acad Sci USA. 2007;104:19500–19505. doi: 10.1073/pnas.0708818104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mizushima N, et al. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J Cell Biol. 2001;152:657–668. doi: 10.1083/jcb.152.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Song Z, Chen H, Fiket M, Alexander C, Chan DC. OPA1 processing controls mitochondrial fusion and is regulated by mRNA splicing, membrane potential, and Yme1L. J Cell Biol. 2007;178:749–755. doi: 10.1083/jcb.200704110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wei MC, et al. Proapoptotic BAX and BAK: A requisite gateway to mitochondrial dysfunction and death. Science. 2001;292:727–730. doi: 10.1126/science.1059108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hoppins S, et al. The soluble form of Bax regulates mitochondrial fusion via MFN2 homotypic complexes. Mol Cell. 2011;41:150–160. doi: 10.1016/j.molcel.2010.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karbowski M, Norris KL, Cleland MM, Jeong SY, Youle RJ. Role of Bax and Bak in mitochondrial morphogenesis. Nature. 2006;443:658–662. doi: 10.1038/nature05111. [DOI] [PubMed] [Google Scholar]
- 34.de Brito OM, Scorrano L. Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature. 2008;456:605–610. doi: 10.1038/nature07534. [DOI] [PubMed] [Google Scholar]
- 35.Ishihara N, Eura Y, Mihara K. Mitofusin 1 and 2 play distinct roles in mitochondrial fusion reactions via GTPase activity. J Cell Sci. 2004;117:6535–6546. doi: 10.1242/jcs.01565. [DOI] [PubMed] [Google Scholar]
- 36.Gomes LC, Scorrano L. High levels of Fis1, a pro-fission mitochondrial protein, trigger autophagy. Biochim Biophys Acta. 2008;1777:860–866. doi: 10.1016/j.bbabio.2008.05.442. [DOI] [PubMed] [Google Scholar]
- 37.Elmore SP, Qian T, Grissom SF, Lemasters JJ. The mitochondrial permeability transition initiates autophagy in rat hepatocytes. FASEB J. 2001;15:2286–2287. doi: 10.1096/fj.01-0206fje. [DOI] [PubMed] [Google Scholar]
- 38.Frank S, et al. The role of dynamin-related protein 1, a mediator of mitochondrial fission, in apoptosis. Dev Cell. 2001;1:515–525. doi: 10.1016/s1534-5807(01)00055-7. [DOI] [PubMed] [Google Scholar]
- 39.Lee YJ, Jeong SY, Karbowski M, Smith CL, Youle RJ. Roles of the mammalian mitochondrial fission and fusion mediators Fis1, Drp1, and Opa1 in apoptosis. Mol Biol Cell. 2004;15:5001–5011. doi: 10.1091/mbc.E04-04-0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yaffe MP. The machinery of mitochondrial inheritance and behavior. Science. 1999;283:1493–1497. doi: 10.1126/science.283.5407.1493. [DOI] [PubMed] [Google Scholar]
- 41.Chen H, et al. Mitochondrial fusion is required for mtDNA stability in skeletal muscle and tolerance of mtDNA mutations. Cell. 2010;141:280–289. doi: 10.1016/j.cell.2010.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Karbowski M, Neutzner A, Youle RJ. The mitochondrial E3 ubiquitin ligase MARCH5 is required for Drp1 dependent mitochondrial division. J Cell Biol. 2007;178:71–84. doi: 10.1083/jcb.200611064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nakamura N, Kimura Y, Tokuda M, Honda S, Hirose S. MARCH-V is a novel mitofusin 2- and Drp1-binding protein able to change mitochondrial morphology. EMBO Rep. 2006;7:1019–1022. doi: 10.1038/sj.embor.7400790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kristensen AR, et al. Ordered organelle degradation during starvation-induced autophagy. Mol Cell Proteomics. 2008;7:2419–2428. doi: 10.1074/mcp.M800184-MCP200. [DOI] [PubMed] [Google Scholar]
- 45.Iwata J, et al. Excess peroxisomes are degraded by autophagic machinery in mammals. J Biol Chem. 2006;281:4035–4041. doi: 10.1074/jbc.M512283200. [DOI] [PubMed] [Google Scholar]
- 46.Collins CA, et al. Atg5-independent sequestration of ubiquitinated mycobacteria. PLoS Pathog. 2009;5:e1000430. doi: 10.1371/journal.ppat.1000430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rambold AS, et al. Association of Bcl-2 with misfolded prion protein is linked to the toxic potential of cytosolic PrP. Mol Biol Cell. 2006;17:3356–3368. doi: 10.1091/mbc.E06-01-0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.