Abstract
We present whole-genome assemblies of four divergent Arabidopsis thaliana strains that complement the 125-Mb reference genome sequence released a decade ago. Using a newly developed reference-guided approach, we assembled large contigs from 9 to 42 Gb of Illumina short-read data from the Landsberg erecta (Ler-1), C24, Bur-0, and Kro-0 strains, which have been sequenced as part of the 1,001 Genomes Project for this species. Using alignments against the reference sequence, we first reduced the complexity of the de novo assembly and later integrated reads without similarity to the reference sequence. As an example, half of the noncentromeric C24 genome was covered by scaffolds that are longer than 260 kb, with a maximum of 2.2 Mb. Moreover, over 96% of the reference genome was covered by the reference-guided assembly, compared with only 87% with a complete de novo assembly. Comparisons with 2 Mb of dideoxy sequence reveal that the per-base error rate of the reference-guided assemblies was below 1 in 10,000. Our assemblies provide a detailed, genomewide picture of large-scale differences between A. thaliana individuals, most of which are difficult to access with alignment-consensus methods only. We demonstrate their practical relevance in studying the expression differences of polymorphic genes and show how the analysis of sRNA sequencing data can lead to erroneous conclusions if aligned against the reference genome alone. Genome assemblies, raw reads, and further information are accessible through http://1001genomes.org/projects/assemblies.html.
In the past decade, there has been a growing appreciation that individuals of the same species are not only distinguished by small-scale differences such as single nucleotide polymorphisms (SNPs), but that copy number variants (CNVs) often account for an even greater difference in genetic material, both within and between closely related species. Since the advent of next generation sequencing (NGS) technologies, the main challenge in genome sequencing has shifted from data generation to reconstruction of genomes from short reads.
We have previously estimated that up to 7% of the Arabidopsis thaliana noncentromeric genome are highly diverged (1–3), and other plant species can be even more polymorphic (4, 5). The first NGS analysis of A. thaliana revealed a density of at least 1 SNP every 200 bp between random pairs of strains (3). This study followed the alignment-consensus approach and thus excluded most regions of high divergence or repetitiveness; the actual extent of genomic differences in A. thaliana is therefore expected to be substantially higher.
Prediction methods for structural variants (SVs) have been developed to annotate diverged regions on the basis of paired-end sequencing (6–16). Unfortunately, these predictions do not include the actual sequence of the variants, and they often miss larger rearrangements, complex changes, and small insertion/deletions (indels). In addition, regions of similar length but with dissimilar sequence will not reveal themselves in the form of paired alignments with unexpected distance or orientation to each other. To overcome these shortcomings, it has been suggested to locally assemble regions of high dissimilarity between sample and reference sequence (3, 17, 18). One way to reduce reference bias is to use multiple references as alignment target, each representing different strains or diverged regions in strains of the same species (19).
Perhaps the simplest way to bypass all problems specific to reference-based approaches is de novo assembly. This has been initially attempted with complex genomes analyzed with short NGS reads only, but the resulting contigs and scaffolds tended to be short and a substantial portion of the genome was not captured in these assemblies (20, 21). Different studies have tried to reduce the complexity by introducing reduced-representation libraries (22). Alternatively, jumping libraries with large insert sizes can greatly improve de novo assembly, but the production of such libraries is technically challenging (23).
Here we present a unique multitiered approach of de novo assembly guided by homology to a reference genome and compare it to complete de novo assembly. We demonstrate how information from the reference-guided assemblies can be used for more accurate annotation of the effects of sequence differences, as well as for improving estimates of strain-specific differences in expression of mRNAs and small RNAs.
Results
Reference-Guided Assembly.
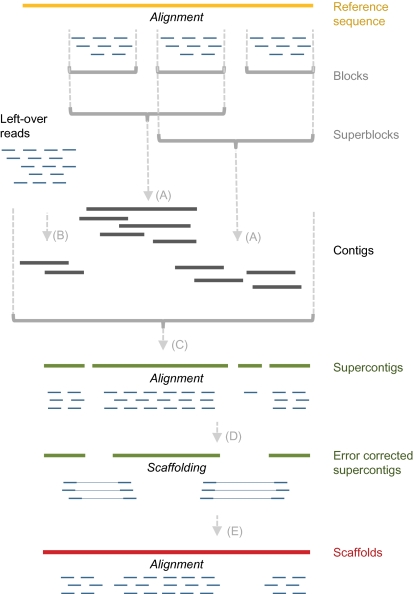
Our reference-guided assembly approach is outlined in Fig. 1. We used paired-end reads of 36–80 bp generated on the Illumina Genome Analyzer platform, with average library insert lengths from 177 to 4,700 bp (Table S1). Some of the reads had been produced previously (24, 25). Filtering and alignment of the short reads against the A. thaliana reference sequence were performed using GenomeMapper (19). We partitioned reads on the basis of their alignment locations and defined regions with constant coverage or adjacent regions connected by aligned mate pairs, i.e., two reads generated from the same fragment, as blocks. Adjacent blocks were combined into superblocks, with neighboring superblocks sharing at least one block. Each superblock contained all reads that aligned to the constituent blocks. We also included “dangling” reads where only one mate aligned to one of the constituent blocks (SI Materials and Methods). About 14 Mb of the reference sequence correspond to highly repetitive pericentromeric and centromeric sequences (1). Because they attract many erroneous mappings (3), we discarded superblocks overlapping with these regions.
Fig. 1.
Illustration of reference-guided assembly. Reads and their alignments are shown in blue. Regions of constant coverage were defined as blocks. Adjacent blocks were combined into superblocks until they reached a minimal length of 12 kb. Superblocks were defined in an overlapping fashion, such that blocks could belong to several superblocks. All reads of a superblock were assembled with reads that had not been aligned. Resulting contigs (dark blue) were merged into a nonredundant set of supercontigs (green). Short read alignments against the supercontigs allowed for error correction and scaffolding. Short read alignments against the scaffolds (red) enabled a final quality assessment and filtering.
Reads corresponding to each superblock were assembled separately using the de Bruijn graph-based tools ABYSS (26), VELVET (27), and EULER-SR (28) (Fig. 1A arrows). We executed each tool eight times per read set using eight different kmer sizes. The local assemblies were combined with an assembly of all leftover reads using SUPERLOCAS (SI Materials and Methods). Finally, we assembled all unmapped reads (including pairs with a dangling read) de novo with VELVET, to separately reconstruct long stretches of nonreference sequence (Fig. 1B arrow).
As single reads can contribute to different assemblies our workflow introduces redundancy into the contigs. As this redundancy generates overlaps between the contigs we used the homology guided Sanger assembler AMOScmp (29) to merge all contigs of each chromosome arm into a set of nonredundant supercontigs (Fig. 1C arrow).
To validate the supercontigs, we aligned all original reads against these. Consistent differences between supercontigs and reads were taken as indications of misassemblies. With this information we corrected or, in the most extreme cases, removed supercontigs (Fig. 1D arrow). Read pairs with ends that aligned to different supercontigs were used for scaffolding with BAMBUS (30) (Fig. 1E arrow).
BAMBUS scaffolds were used as alignment target for a third and final round of read mapping and consensus analysis. We developed a simple metric to assign per-base quality values according to the base qualities of the consensus analysis. Scaffolds shorter than 500 high-quality base pairs were discarded, and low-quality positions were masked. Additionally we ran a more stringent base masking to produce a high-quality, although less comprehensive assembly. Both types of assemblies can be downloaded at http://1001genomes.org/projects/assemblies.html.
Assembly Statistics.
We used a mix of single-end, paired-end, and mate-pair libraries for the genomes of the four A. thaliana strains Landsberg erecta (Ler-1), C24, Burren (Bur-0), and Krotzenburg (Kro-0) (Table S1). Total coverage was greater than 70× for all and greater than 320× for Ler-1. Common metrics to assess assembly quality are N50 and L50, which indicate the total number and minimum length, respectively, of all scaffolds that together account for 50% of the genome. After exclusion of centromeric regions, we had targeted sequences that correspond to around 105 Mb of the reference sequence. Based on this sequence, N50 and L50 for our assemblies ranged from 117 to 208, and 140 to 262 kb, respectively, with the longest scaffold in each assembly being between 1.1 and 2.2 Mb (Table 1). For comparison, contigs derived from concatenating consecutively called positions of an alignment consensus yielded N50 and L50 values of 6,147 and 4.1 kb, respectively, for Ler-1 (Table S2). The cumulative lengths of all scaffolds were about 5% shorter than the target of 105 Mb. This was mainly because of failure to assemble repetitive sequences, as indicated by the fact that the nonpericentromeric portions of the reference sequence not covered by assembly contigs had a 36–41% repeat content, compared with an average of 8% in noncentromeric regions, as assessed with 36-mers (3).
Table 1.
Assembly statistics
| Bur-0 | C24 | Kro-0 | Ler-1 | |
| Coverage | 83.2× | 75.0× | 72.7× | 322.4× |
| Libraries | 2 | 2 | 2 | 3 |
| N50 (intrinsic) | 193 | 109 | 161 | 113 |
| L50, kb | 147.3 | 273.2 | 163.5 | 272.5 |
| N50 (target) | 208 | 117 | 178 | 121 |
| L50, kb | 139.6 | 260.4 | 151.8 | 261.9 |
| Scaffolds | 2,526 | 2,052 | 2,670 | 1,528 |
| Total length, Mb | 101.0 | 101.3 | 99.9 | 100.8 |
| Longest scaffold, Mb | 1.12 | 2.18 | 1.48 | 1.09 |
| Ambiguous bases, % | 4.0 | 3.6 | 5.1 | 1.3 |
N50/L50 (intrinsic) using total length of the scaffolds as reference size. N50/L50 (target) using the expected genome size as reference (105.2 Mb).
Assembly Validation with 2 Mb of Dideoxy Data.
To assess the quality and error rate of the assemblies, we used 955 random Sanger reads of the Bur-0 genome (SI Materials and Methods; the “shotgun” set), and a published set of 3,388 fragments of the C24, Ler-1, and Bur-0 genomes produced by targeted Sanger resequencing of mostly unique, genic fragments (31) (the “MN2010 set”). Sanger reads were aligned against the respective assembly and the pericentromeric portions of the reference (32) (Table 2). About 4% of MN2010 and 28% of shotgun fragments aligned to organelles or pericentromeric regions. Our assemblies agreed very well with the remaining MN2010 data. At least 98.8% aligned uniquely and without any mismatch, and only 0.4–0.8% had mismatches. An additional 0.2–0.4% revealed short indel errors, all of which were associated with low sequence complexity including simple repeats. Only three MN2010 reads revealed long indel errors not associated with simple repeats, of up to 476 bp. The total per-base error measured across all MN2010 alignments (excluding the three with long indel errors) was less than 1 in 40,000 bp.
Table 2.
Assembly validation
| Ler-1 (MN2010) | C24 (MN2010) | Bur-0 (MN2010) | Bur-0 (shotgun) | |
| Sanger reads | 1,139 | 1,139 | 1,110 | 955 |
| Organelle/centromere hits | 48 | 48 | 49 | 267 |
| No significant hits | 12 | 4 | 6 | 52 (30)* |
| Euchromatic hits | 1,079 | 1,087 | 1,055 | 658 |
| Identical | 1,069 | 1,074 | 1,046 | 629 |
| With mismatching bases | 6 | 9 | 4 | 17 |
| With indels in simple repeats | 2 | 4 | 4 | 4 |
| With indels (up to 476 bp) | 2 | 0 | 1 | 8 |
| Nucleotides queried, kb | 580 | 584 | 563 | 285 |
| No. mismatching bases | 11 | 14 | 8 | 22 |
*Fifty-two reads were blasted against NCBI nonredundant database. Thirty reads did not feature alignments that were related to rDNA or human DNA.
The per-base error estimate with the shotgun set for Bur-0 was higher, but still less than 1 in 10,000 bp. Eight reads out of 658 revealed long indel errors. This was not unexpected, as the shotgun set was randomly sampled from the genome and included more intergenic and repetitive sequences, which are more difficult to assemble. In addition, the shotgun reads had not been subjected to similarly extensive manual curation as the MN2010 set and were thus likely to contain more errors themselves.
We compared all reads of the shotgun set without significant BLAST hit (E value < e−10) against National Center for Biotechnology Information (NCBI)’s nonredundant database (32). Twenty-one of 52 reads corresponded to rDNA, and one was the result of contamination with human DNA. The remaining reads, or 4.4% of all reads excluding organelles, centromeres, and contamination, present an upper boundary for the “unassembled space.” This is in agreement with the total scaffold length of 96.2% of the target (Tables 1 and 2), and less than what had been estimated to be inaccessible using alignment-consensus analysis (3).
Sequence Assemblies Capture Large-Scale Variations.
To determine the extent of large-scale sequence differences captured in the assemblies, we performed whole-genome alignments against the reference genome with MUMmer (33), using parameters that favored correctly placed alignments over sensitivity. The portion of the reference genome that could not be aligned against our assemblies was as low as 3.7%, whereas in the best case for the alignment-consensus approach, at least 10.3% of the reference could not be aligned (Table S3). In aligned regions, we annotated SNPs, indels, and highly diverged regions (HDRs) that are anchored within the whole-genome alignment by flanking sequences (SI Materials and Methods).
There was good concordance between SNPs and microindels (1–3 bp) predicted on the basis of either the whole-genome alignments or by the alignment-consensus approach (Table S3). The assemblies, however, revealed more small-scale changes: On average, 12% more SNPs, 29% more microdeletions, and 23% more microinsertions.
We also analyzed the length distributions of apparent deletions and insertions relative to the reference and HDRs (Table 3 for Ler-1, Table S4 for the other strains). Over 1.7 Mb of reference sequence was missing from the Ler-1 assembly, with the majority in deletions over 2 kb. As expected, deleted regions were significantly enriched for transposable elements (63.5%, compared with 13.7% of all noncentromeric positions). To assess the potential origin of novel, nonreference sequences, we selected 36 Ler-1 regions that were at least 500 bp long and at least 10 times longer than the reference allele. Of these, 14 sequences shared similarity with Arabidopsis lyrata (34) over at least half of their lengths, indicating that the reference genome lacks sequences present in the last common ancestor of A. thaliana and A. lyrata.
Table 3.
Variants of different lengths in Ler-1
| Deletions |
Insertions |
HDRs > ∼30 bp* |
||||
| Variant length (bp) | n | Length (bp)† | n | Length (bp)† | n | Length (bp)† |
| 1 | 35,370 | 35,370 | 34,261 | 34,261 | ||
| 2 | 9,861 | 19,722 | 10,060 | 20,120 | ||
| 3–4 | 8,305 | 28,221 | 7,963 | 27,148 | ||
| 5–8 | 5,816 | 36,809 | 5,677 | 35,766 | ||
| 9–16 | 3,757 | 43,673 | 3,505 | 40,435 | ||
| 17–32 | 1,824 | 41,552 | 1,238 | 27,800 | 66 | 1,752 |
| 33–64 | 663 | 30,310 | 579 | 26,413 | 165 | 8,133 |
| 65–128 | 296 | 26,190 | 340 | 29,810 | 379 | 35,178 |
| 129–256 | 219 | 40,825 | 127 | 21,676 | 406 | 76,128 |
| 257–512 | 204 | 74,045 | 63 | 22,600 | 359 | 129,491 |
| 513–1,024 | 240 | 176,491 | 20 | 12,823 | 217 | 155,935 |
| 1,025–2,048 | 160 | 223,702 | 2 | 3,376 | 138 | 192,553 |
| >2,048 | 208 | 996,542 | 4 | 16,129 | 99 | 538,179 |
*Length in reference genome.
†Cumulative length of all variants of the class in that row.
Even though they were too divergent to be aligned directly, the lengths of HDR alleles were strongly correlated (Fig. S1), with an overrepresentation of HDRs with a longer reference allele. This might again be due to the difficulty of assembling long insertions, also reflected in the smaller amount of sequence found only in one of the four new genomes, compared with reference-only sequences. Finally, regions with reverse complementary alignments revealed eighteen inversions (Table S5).
Comparison with Complete de Novo Assembly.
Some of the most impressive de novo assemblies of short-read data have been produced with ALLPATHS-LG in combination with sequencing libraries that had large insert sizes (23). Because ALLPATHS-LG requires overlapping paired-end reads, we generated an additional 64.5 million 101-bp paired-end reads from one of the Ler-1 libraries, which we knew to have an average clone length of 178 bp. We combined these new data with 8.7 million 40-bp mate-pair reads.
Different from our reference-guided assembly, the complete de novo assembly contained both noncentromeric and centromeric sequences, with a total length of 112.6 Mb in 1,705 scaffolds, compared with the 119 Mb of mostly noncentromeric reference sequence. Half of the assembly was contained in 102 scaffolds (N50) of minimum lenth 198 kb (L50). The N50 and L50 values were better than those of the reference-guided assembly, but a whole-genome alignment to the reference revealed that the scaffolds covered only 92 Mb of the noncentromeric regions. Thus, whereas only 3.7% of the noncentromeric reference genome sequence was absent from the reference-guided assembly, the complete de novo assembly lacked 12.6% of noncentromeric sequences.
Shared Polymorphisms and Their Effect on Genes.
When comparing only four individuals, a large fraction of polymorphisms is expected to be found in just a single strain, and many polymorphisms that segregate at low or intermediate frequency will be missed (1–3, 31) (Fig. S2). Of 27,929 genes (excluding transposable elements and pseudogenes), over 95% could be at least partially detected in our assemblies. Over half, 55%, had at least one nonsynonymous change (Table 4). In each accession, over 3% of the genes with completely aligned sequences featured large disruptions of their coding sequence (Table 4). Partial alignments indicated that between 4.3 and 5.5% of all genes were interrupted by an HDR.
Table 4.
Functional annotation of polymorphisms in 27,929 noncentromeric genes
| Bur-0 | C24 | Kro-0 | Ler-1 | |
| Accessible genes | 26,842 | 26,823 | 26,673 | 26,727 |
| Fully aligned | 23,220 | 23,262 | 23,448 | 23,770 |
| Conserved | 7,986 | 7,918 | 10,354 | 8,897 |
| Minor change | 14,320 | 14,438 | 12,306 | 14,007 |
| Nonsynonymous | 14,224 | 14,350 | 12,237 | 13,904 |
| Deletion* | 379 | 398 | 311 | 380 |
| Insertion* | 315 | 342 | 305 | 378 |
| Major change | 914 | 906 | 788 | 866 |
| Deletion* | 342 | 317 | 291 | 311 |
| Insertion* | 338 | 325 | 283 | 319 |
| Stop | 300 | 336 | 271 | 314 |
| Stop “reversion” | 99 | 92 | 73 | 83 |
| Partially aligned | 3,622 | 3,561 | 3,225 | 2,957 |
| HDR in genes | 1,369 | 1,540 | 1,212 | 1,461 |
| HDR in exons | 374 | 422 | 314 | 365 |
*Minor-effect indels have a length that is a multiple of 3 bp.
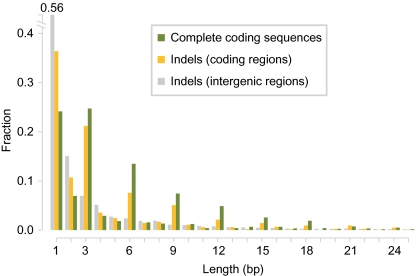
In humans, indels in coding regions occur preferentially in multiples of 3 bp, which avoids frameshifts (35). In our assemblies, 1-bp deletions were the most prevalent group, but there were distinct peaks at multiples of 3 bp, which were not seen in intergenic sequences. When considering all indels in a gene, the total variation in coding sequence length showed more pronounced peaks at multiples of 3 bp, indicating that additional indels could restore the ORF (Fig. 2 and Fig. S3). The pairwise alignments of all assembled genes can be accessed through our Web tool (http://1001genomes.org/projects/assemblies.html).
Fig. 2.
Frequency of indel lengths and variation of coding sequence lengths in Ler-1. Indels with lengths that are a multiple of three are enriched in coding regions (yellow), but not in noncoding regions (gray). This is even more apparent when comparing total length differences between orthologous coding sequences between Col-0 and Ler-1 (green). This trend can only be explained by complex changes in coding sequences that together restore the frame use. See Fig. S3 for other accessions.
Correcting Expression Estimates for Protein-Coding Genes.
Although RNA-seq is starting to eclipse microarray-based investigations of genomewide expression profile, both suffer from reliance on reference sequences, and lack of sequence conservation between individuals easily confounds expression estimates (36). We therefore investigated whether our genome assemblies would improve the interpretation of tiling array data (37) for Bur-0 and C24.
About 90% (27,607) of genes had poylmorphic probes. After probe removal, 8% (2,432) of genes could no longer be considered, because fewer than three probes had been retained. By excluding polymorphic probes, average estimates of expression levels increased slightly and were changed for many loci, especially for genes where half or more of the probes targeted polymorphic sequences (Fig. S4A). The variance in expression estimates for conserved genes, i.e., genes with less than 2.5% of exonic positions differing between Col-0, Bur-0, and C24, was also substantially lower than for polymorphic genes, even though the average estimates were the same (Fig. S4 B and C).
Correcting Expression Estimates for Small RNA Loci.
Loci that spawn populations of small RNAs (sRNAs) are more difficult to annotate than mRNA producing loci, because they are defined by a collection of molecules. Because sRNAs are short, even small-scale differences between the focal accession and the reference will greatly affect the number of correctly mapped sRNAs.
We sequenced sRNA libraries from C24 and Bur-0 inflorescences (38). We defined sRNA loci by consecutive and overlapping alignments of sequence reads from an sRNA library and used the normalized number of reads in such segments to estimate expression of the entire locus. For Bur-0 and C24, 6.5 + 5.6 and 5.8 + 6.8 million reads from two replicate sRNA libraries, respectively, were aligned against the reference genome with one mismatch. Unaligned reads were further aligned against the Bur-0 or C24 assemblies, again allowing for one mismatch. The second step increased the alignable reads by 7%.
On the basis of the reference alignments, we defined 30,787 segments with continuous coverage of at least 10 reads from each replicate for Bur-0 and 28,174 segments for C24. Taking not only the reads that aligned against the reference, but also those that aligned against the respective assembly into account, significantly changed the expression estimates of 348 segments (1.1%) for Bur-0 and 284 (1.0%) for C24 (Fig. S5). In addition, 1,283 (4.0%) of Bur-0 segments and 1,184 (4.0%) of C24 segments, could only be revealed by alignments against the nonreference genome. Finally, 579 (1.9%) of Bur-0 segments, and 556 (2.0%) of C24 segments defined by reference alignments alone were merged with neighboring segments by adding the alignments to the strain assemblies.
Discussion
Since the release of the A. thaliana reference genome sequence 10 y ago (39), no other whole-genome assemblies have been reported for this species. The reference genome was generated for some $70 million with a bacterial artificial chromosome (BAC)-by-BAC strategy using dideoxy sequencing (40). Differently from the first human genomes, a single individual, from the laboratory strain Col-0, was assembled.
The reference was essential for efforts to record sequence variation between natural strains of A. thaliana (41). However, simple alignment-based methods do not provide the complete picture to mine and exploit specieswide sequence diversity. Complete de novo assemblies would be the obvious alternative, but one of the best assembly methods available (23) resulted in significant underrepresentation of the targeted reference genome. Reasons for this might be the complexity of the A. thaliana genome, the length and quality of NGS reads as well as the limited insert sizes of our libraries compared with the human genome assemblies reported by Gnerre and colleagues (23). Previous work with plant genomes that achieved similar results to ours includes that of Huang and colleagues (42), who reported a whole-genome assembly of Illumina short read data for the 367-Mb cucumber genome. Whereas their L50 values were not too dissimilar from ours, they also found a larger fraction of the genome being completely missing from their assembly. Moreover, without independent validation of the cucumber results, a direct comparison with our work is difficult.
Notably, there was a limit to improving assembly statistics with additional short read data. We do not know whether this reflects an inability of the assembly tools to exploit more than about 70× coverage or whether this is an intrinsic property of read lengths and library insert sizes used and genomic repeat content. Assemblies also did not improve significantly with longer reads. For example, the C24 and Ler-1 assemblies had almost the same N50 and L50, despite C24 being sequenced mostly with 40-bp reads, and Ler-1 with 80-bp reads.
An obvious next application of reference-guided assemblies would be to improve draft genome sequences, through an iterative process, in which the initial draft genome is used as a reference for an assembly of the same strain. Two limitations are the current insert lengths of standard sequencing libraries, which do not span all classes of repetitive elements, as well as lack of automated software.
Our assemblies have also shed light on the functional consequences of sequence variants. For example, we have shown that a substantial fraction of 1- and 2-bp indels in coding regions are compensated by nearby indels that restore the coding frame (Fig. 2). Current genomewide association studies generally do not consider the nature of a variant, because not all variants are analyzed or even known. With better information, it will be possible to annotate the predicted effect of the combination of sequence variants in an allele, and subsequently base genomewide association studies on classes of alleles with reduced or increased activity, rather than ignoring such information.
In addition to comparing genome sequences, there is great interest in studying individual patterns of DNA methylation, chromatin modifications, and RNA expression. We have already demonstrated how our assemblies improve mRNA and sRNA expression studies, and we expect a similar impact on DNA methylation analyses.
In summary, our reference-guided assembly approach greatly reduces the bias introduced when next generation sequencing reads are only aligned against a reference genome. The availability of several whole-genome assemblies should thus improve the identification of variants in the 1,001 Genomes projects that is underway for A. thaliana, by exploiting all known variants as targets for mapping of short reads (19).
Materials and Methods
Combing the Contigs with AMOScmp.
The block assemblies computed by VELVET, ABYSS, and Euler-SR, the SUPERLOCAS run and the VELVET assembly of leftover reads introduced high levels of redundancy into the combined set of contigs. This was evident at five levels: (i) different runs of different tools assembling identical sets of input reads; (ii) reads used twice if their respective block was allocated to overlapping superblocks; (iii) unmapped reads contributing to multiple superblock assemblies; (iv) reads reused due to repetitive alignments to multiple blocks; and (v) assembly of leftover and dangling pairs also included in the block assemblies. To assemble the contigs and purge the redundancy, we used AMOScmp, which applies an alignment-layout-consensus approach using alignments against the reference to guide the overlap calculation of contigs (29). To reduce complexity and hardware requirements we ran AMOScmp on each chromosome arm separately (SI Materials and Methods).
Correction of Misassemblies.
We aligned all reads against the set of supercontigs using GenomeMapper and performed consensus analysis with SHORE. Differences between aligned reads and reference sequence revealed misassemblies in the supercontigs. Any supercontig shorter than 100 bp, featuring only low read coverage as well as supercontigs with mostly repetitive read alignments were removed. All remaining supercontigs were split at any region where variant predictions indicated misassemblies including uncovered regions, local clusters of differences, and regions with mate pairs that did not align in the expected order and orientation (SI Materials and Methods).
Scaffolding.
Read pairs with reads aligned to two different supercontigs defined a connection (bridge) between the respective supercontigs. Bridges suggested that two supercontigs were in local vicinity and had a defined order in the focal genome. We also used homology of supercontigs to the reference sequence to infer additional connections, as described in the BAMBUS (30) manual (SI Materials and Methods). As mate pair libraries suffer from high rates of potential clonal events we did not consider mate pair reads that aligned to the exact positions as others of the same library.
After running BAMBUS with the set of filtered bridges and connections based on homology as input, the final scaffolding graph was plotted, manually evaluated, and suspicious connections were removed (SI Materials and Methods). By default, BAMBUS connects contigs within scaffolds using a fixed number of 60 N's. Instead, we predicted the most likely distance between connected contigs on the basis of the alignment locations of read pairs mapped to two connected contigs, and introduced the corresponding number of N's into the scaffolds.
Base Quality Assessment and Masking.
To assign a final per-base quality value, we aligned reads against the respective assembly and used SHORE's resequencing pipeline for consensus analysis (SI Materials and Methods). On the basis of SHORE's positionwise quality values qref (reference) and qvar (variation) we assigned a per-base quality qass to each residue. If there was mere support for the reference allele, qass was set to qref. If only a nonreference allele was supported, qass was set to 0. If there was evidence for two alleles, qass was assigned the maximum of 0 and the difference of qref and qvar. Every base that was assigned a quality value of less than 10 was masked. Scaffolds with less than 500 unmasked bases were discarded. N's at the beginning of scaffolds were removed. For a more stringent, but less comprehensive assembly, we masked all bases with a quality of less than 15. Additionally we masked all unmasked regions that were shorter than 100 bp.
ALLPATHS-LG de Novo Assembly.
ALLPATHS-LG version allpathslg-35762 (23) was applied. Default parameters were used, except that USE_LONG_JUMPS was set to “false,” as long jumping libraries with size larger than 20 kb were not included in the analysis.
Supplementary Material
Acknowledgments
We thank all developers of the assembly tools used in this study. We thank Illumina for sequencing seven paired-end lanes of Bur-0. This work was supported by European Community Framework Programme 7 (FP7) Marie Curie Fellowship PIEF-GA-2008-221553 and a European Molecular Biology Organization (EMBO) long-term fellowship (to L.M.S.), Bundesministerium für Bildung und Forschung (BMBF) Genomanalyse im biologischen System Pflanze - Genome-wide Natural Diversity Exploitation in the Arabidopsis Population (GABI-GNADE) (D.H.H. and D.W.), European Community Framework Programme 6 (FP6) Integrated Programme (IP) Silencing RNAs: Organizers and Coordinators of Complexity in Eukaryotic Organisms (SIROCCO) (Contract LSHG-CT-2006-037900), Transnational Plant Alliance for Novel Technologies - Towards Implementing the Knowledge-based Bio-economy in Europe (PLANT-KBBE) project Transcriptional Networks and Their Evolution in the Brassicaceae (TRANSNET) (BMBF), a Gottfried Wilhelm Leibniz Award of the Deutsche Forschungsgemeinschaft (DFG), and the Max Planck Society (D.W.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1107739108/-/DCSupplemental.
References
- 1.Clark RM, et al. Common sequence polymorphisms shaping genetic diversity in Arabidopsis thaliana. Science. 2007;317:338–342. doi: 10.1126/science.1138632. [DOI] [PubMed] [Google Scholar]
- 2.Zeller G, et al. Detecting polymorphic regions in Arabidopsis thaliana with resequencing microarrays. Genome Res. 2008;18:918–929. doi: 10.1101/gr.070169.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ossowski S, et al. Sequencing of natural strains of Arabidopsis thaliana with short reads. Genome Res. 2008;18:2024–2033. doi: 10.1101/gr.080200.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Springer NM, et al. Maize inbreds exhibit high levels of copy number variation (CNV) and presence/absence variation (PAV) in genome content. PLoS Genet. 2009;5:e1000734. doi: 10.1371/journal.pgen.1000734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gore MA, et al. A first-generation haplotype map of maize. Science. 2009;326:1115–1117. doi: 10.1126/science.1177837. [DOI] [PubMed] [Google Scholar]
- 6.Quinlan AR, et al. Genome-wide mapping and assembly of structural variant breakpoints in the mouse genome. Genome Res. 2010;20:623–635. doi: 10.1101/gr.102970.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lam HY, et al. Nucleotide-resolution analysis of structural variants using BreakSeq and a breakpoint library. Nat Biotechnol. 2010;28:47–55. doi: 10.1038/nbt.1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hormozdiari F, et al. Next-generation VariationHunter: Combinatorial algorithms for transposon insertion discovery. Bioinformatics. 2010;26:i350–i357. doi: 10.1093/bioinformatics/btq216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee S, Hormozdiari F, Alkan C, Brudno M. MoDIL: Detecting small indels from clone-end sequencing with mixtures of distributions. Nat Methods. 2009;6:473–474. doi: 10.1038/nmeth.f.256. [DOI] [PubMed] [Google Scholar]
- 10.Korbel JO, et al. PEMer: A computational framework with simulation-based error models for inferring genomic structural variants from massive paired-end sequencing data. Genome Biol. 2009;10:R23. doi: 10.1186/gb-2009-10-2-r23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chiang DY, et al. High-resolution mapping of copy-number alterations with massively parallel sequencing. Nat Methods. 2009;6:99–103. doi: 10.1038/nmeth.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Campbell PJ, et al. Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nat Genet. 2008;40:722–729. doi: 10.1038/ng.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee S, Cheran E, Brudno M. A robust framework for detecting structural variations in a genome. Bioinformatics. 2008;24:i59–i67. doi: 10.1093/bioinformatics/btn176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang K, et al. PennCNV: An integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007;17:1665–1674. doi: 10.1101/gr.6861907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Korbel JO, et al. Paired-end mapping reveals extensive structural variation in the human genome. Science. 2007;318:420–426. doi: 10.1126/science.1149504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tuzun E, et al. Fine-scale structural variation of the human genome. Nat Genet. 2005;37:727–732. doi: 10.1038/ng1562. [DOI] [PubMed] [Google Scholar]
- 17.Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bentley DR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schneeberger K, et al. Simultaneous alignment of short reads against multiple genomes. Genome Biol. 2009;10:R98. doi: 10.1186/gb-2009-10-9-r98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dalloul RA, et al. Multi-platform next-generation sequencing of the domestic turkey (Meleagris gallopavo): Genome assembly and analysis. PLoS Biol. 2010;8:e1000475. doi: 10.1371/journal.pbio.1000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li R, et al. The sequence and de novo assembly of the giant panda genome. Nature. 2010;463:311–317. doi: 10.1038/nature08696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Young AL, et al. A new strategy for genome assembly using short sequence reads and reduced representation libraries. Genome Res. 2010;20:249–256. doi: 10.1101/gr.097956.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gnerre S, et al. High-quality draft assemblies of mammalian genomes from massively parallel sequence data. Proc Natl Acad Sci USA. 2011;108:1513–1518. doi: 10.1073/pnas.1017351108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mirouze M, et al. Selective epigenetic control of retrotransposition in Arabidopsis. Nature. 2009;461:427–430. doi: 10.1038/nature08328. [DOI] [PubMed] [Google Scholar]
- 25.Laitinen RA, Schneeberger K, Jelly NS, Ossowski S, Weigel D. Identification of a spontaneous frame shift mutation in a nonreference Arabidopsis accession using whole genome sequencing. Plant Physiol. 2010;153:652–654. doi: 10.1104/pp.110.156448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simpson JT, et al. ABySS: A parallel assembler for short read sequence data. Genome Res. 2009;19:1117–1123. doi: 10.1101/gr.089532.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zerbino DR, Birney E. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chaisson MJ, Pevzner PA. Short read fragment assembly of bacterial genomes. Genome Res. 2008;18:324–330. doi: 10.1101/gr.7088808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pop M, Phillippy A, Delcher AL, Salzberg SL. Comparative genome assembly. Brief Bioinform. 2004;5:237–248. doi: 10.1093/bib/5.3.237. [DOI] [PubMed] [Google Scholar]
- 30.Pop M, Kosack DS, Salzberg SL. Hierarchical scaffolding with Bambus. Genome Res. 2004;14:149–159. doi: 10.1101/gr.1536204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nordborg M, et al. The pattern of polymorphism in Arabidopsis thaliana. PLoS Biol. 2005;3:e196. doi: 10.1371/journal.pbio.0030196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 33.Kurtz S, et al. Versatile and open software for comparing large genomes. Genome Biol. 2004;5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu TT, et al. The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nat Genet. 2011;43:476–481. doi: 10.1038/ng.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pelak K, et al. The characterization of twenty sequenced human genomes. PLoS Genet. 2010;6:e1001111. doi: 10.1371/journal.pgen.1001111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Plantegenet S, et al. Comprehensive analysis of Arabidopsis expression level polymorphisms with simple inheritance. Mol Syst Biol. 2009;5:242. doi: 10.1038/msb.2008.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laubinger S, et al. At-TAX: A whole genome tiling array resource for developmental expression analysis and transcript identification in Arabidopsis thaliana. Genome Biol. 2008;9:R112. doi: 10.1186/gb-2008-9-7-r112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mosher RA, et al. Uniparental expression of PolIV-dependent siRNAs in developing endosperm of Arabidopsis. Nature. 2009;460:283–286. doi: 10.1038/nature08084. [DOI] [PubMed] [Google Scholar]
- 39.Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 40.Theologis A. Goodbye to 'one by one' genetics. Genome Biol. 2001;2:2004.1–2004.9. doi: 10.1186/gb-2001-2-4-comment2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rounsley SD, Last RL. Shotguns and SNPs: How fast and cheap sequencing is revolutionizing plant biology. Plant J. 2010;61:922–927. doi: 10.1111/j.1365-313X.2009.04030.x. [DOI] [PubMed] [Google Scholar]
- 42.Huang S, et al. The genome of the cucumber, Cucumis sativus L. Nat Genet. 2009;41:1275–1281. doi: 10.1038/ng.475. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.