Abstract
Autosomal dominant polycystic kidney disease (ADPKD) is caused by mutations in TRPP2 and PKD1, which form an ion channel/receptor complex containing three TRPP2 and one PKD1. A TRPP2 C-terminal coiled-coil trimer, critical for the assembly of this complex, associates with a single PKD1 C-terminal coiled-coil. Many ADPKD pathogenic mutations result in the abolishment of the TRPP2/PKD1 coiled-coil complex. To gain molecular and functional insights into this heterotetrameric complex, we computationally constructed a structural model by using a two-step docking strategy, based on a known crystal structure of the TRPP2 coiled-coil trimer. The model shows that this tetrameric complex has a novel di-trimer configuration: An upstream trimer made of three TRPP2 helices and a downstream trimer made of two TRPP2 helices and one PKD1 helix. Mutagenesis and biochemical analysis identified critical TRPP2/PKD1 interface contacts essential for the heteromeric coiled-coil complex. Mutation of these interface positions in the full-length proteins showed that these interactions were critical for the assembly of the full-length complex in cells. Our results provide a means to specifically weaken the TRPP2 and PKD1 association, thus facilitating future in vitro and in vivo studies on the functional importance of this association.
Keywords: transient receptor potential channel, polycystin, protein docking, molecular dynamics simulation
The transient receptor potential (TRP) channel family constitutes a large group of cation-permeable channels expressed in diverse species and cell types (1, 2). TRPP2 (also known as PKD2 or polycystin-2), a member of the TRPP subfamily (3), coassembles with PKD1 (also known as polycystin-1), an integral membrane protein that presumably functions as a cell surface receptor (4), to form an ion channel/receptor complex (5–12). Both TRPP2 and PKD1 are widely distributed, with high expression levels in kidney epithelial cells (3, 4, 10, 11), where they appear to colocalize in the primary cilia and may function as mechanosensitive, Ca2+-permeable cation channels (5, 13). The functional importance of TRPP2 and PKD1 is demonstrated by the fact that mutations of these proteins are the prevalent causes of autosomal dominant polycystic kidney disease (ADPKD), one of the most common inherited human diseases (14–17). That mutations of TRPP2 and PKD1 produce similar pathological manifestations suggests that they may act through a disruption of a common complex of the two proteins.
TRPP2 and PKD1 also interact with numerous other proteins, with significant functional consequences (17). For example, PKD1 interacts with and activates heterotrimeric Gi/o proteins, potentially affecting diverse downstream signaling pathways (18–21). On the other hand, TRPP2 interacts and forms functional heteromeric cation channels with other TRP channel subunits, including TRPC1 and TRPV4 (9, 22–24). To fully understand the cellular and molecular mechanisms underlying ADPKD, it would be a major advantage to be able to selectively disrupt the TRPP2/PKD1 complex and thereby distinguish its function from those of other TRPP2- and PKD1-containing complexes. We therefore set out to elucidate structural details about the TRPP2/PKD1 interface.
TRPP2 and PKD1 associate directly through their C termini (25, 26), forming a complex containing three TRPP2 and one PKD1 (27). This association involves coiled-coil domains in the C termini of the two proteins (25, 27). The TRPP2 coiled-coil domain forms a homotrimer, both in solution (27–29) and in crystals (27). Disruption of this trimer abolishes the assembly of heteromeric TRPP2/PKD1 complexes (27). Although a high-resolution crystal structure of the TRPP2 coiled-coil trimer has been solved (27), how the PKD1 coiled-coil binds to this trimer is still unknown.
The potential importance of the TRPP2/PKD1 coiled-coil interaction is underscored by the fact that many ADPKD-causing mutations, including R742X, where X stands for a stop codon mutation, R807X, E837X, and R872X in TRPP2 and R4227X and Y4236X in PKD1 (15, 30, 31), result in the deletion of the coiled-coil domains. However, besides abolishing the TRPP2/PKD1 C-terminal interaction, these truncation mutations could produce global and detrimental effects on the folding, assembly, function, or regulation of the TRPP2/PKD1 complex. To probe the functional importance of the TRPP2/PKD1 coiled-coil interaction it would be helpful to have a high-resolution structure; however, no such structure is yet available despite extensive efforts (27).
In this study, we undertook a computational approach to construct a structural model for the TRPP2/PKD1 coiled-coil complex, starting with a 1.9-Å resolution crystal structure of the TRPP2 coiled-coil trimer and the knowledge that a single PKD1 coiled-coil binds to this trimer (27). Two sequential computational steps were taken. First, a rigid-body docking procedure termed iterative modular optimization (IMO), which was originally developed for refining secondary structure elements within homology models (32), was used to generate a model of the TRPP2/PKD1 coiled-coil complex. Second, molecular dynamics (MD) simulations in implicit solvent (33) were employed to optimize the TRPP2/PKD1 coiled-coil interface, using standard as well as replica-exchange MD (REMD) simulation protocols (34, 35). The resulting structural model was subsequently verified by extensive mutagenesis experiments at both the fragment and full-length protein levels.
Results
Construction of a TRPP2/PKD1 Coiled-Coil Docking Complex.
The region from L4214 to R4248 of PKD1 forms a coiled-coil domain and binds the TRPP2 C terminus (25), specifically to the G833–G895 region (denoted as TRPP2_G833–G895) (27). A crystal structure of TRPP2_G833–G895 shows that it forms an α helix and assembles into a homotrimer (27). This trimer contains two distinct regions: The upstream region, from F839 to A873, forms a classical coiled-coil domain and is tightly packed together, whereas the downstream region, from K874 to G895, splays open (27) (Fig. S1).
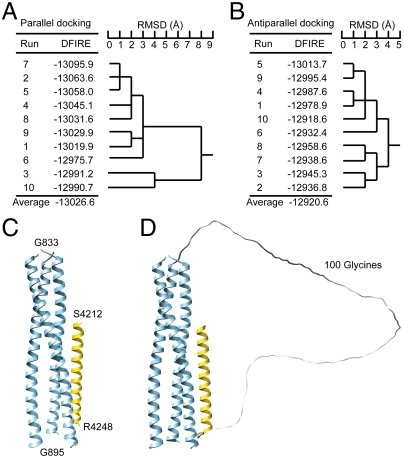
Based on the above information, we docked a PKD1 coiled-coil helix (PKD1_S4212–R4248) to the TRPP2 coiled-coil trimer to generate an initial model of the TRPP2/PKD1 coiled-coil complex by using an IMO-based docking procedure (32). Twenty docking calculations were carried out with a predetermined packing mode, parallel or antiparallel, each with a different sampling pathway (Fig. S2). The model complexes obtained from parallel docking were found to have a lower distance-scaled, finite, ideal-gas reference (DFIRE) statistical potential energy (36) than those from antiparallel docking, on average by 106 kcal•mol-1 (Fig. 1 A and B), suggesting that parallel packing is energetically more favorable and is likely the native form. A structural comparison revealed that eight of the ten parallel docking calculations with lower DFIRE energies converged to an almost identical conformation (Fig. S2A), with the backbone rmsd of the entire PKD1 helix within 3.0 Å. After minimization with a detailed physical energy function, one of these eight conformations that gave the lowest physical energy (Fig. 1A, complex 4 depicted in Fig. 1C) was selected as being representative and as the model for the following disulfide bond analysis. In this complex, the PKD1 helix binds to a groove formed by two of the three TRPP2 helices, located at the C-terminal open region of the TRPP2 coiled-coil trimer (Fig. 1C). In an alternative random docking procedure where both parallel and antiparallel packing were allowed, seven out of ten docking calculations converged to parallel packing conformations (Fig. S2C). The one of these that had the lowest DFIRE energy (Fig. 1D) was structurally very similar to the complex 4 model from the parallel docking procedure (Fig. 1C). The agreement of these two docking studies led us to test this model biochemically.
Fig. 1.
Conformations of the TRPP2/PKD1 coiled-coil domain complex obtained from docking. (A and B) DFIRE energy and structural similarity for TRPP2/PKD1 coiled-coil complex conformations generated from parallel docking (A) and antiparallel docking (B). (C) The complex conformation selected from parallel docking (run 4) and used in subsequent MD simulations. (D) The lowest-energy complex conformation obtained from random docking with an artificial connecting loop consisting of 100 glycines.
Testing the Docking Complex Model by Disulfide Bond Analysis.
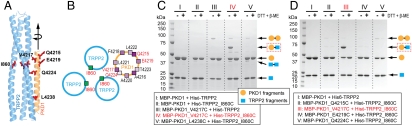
The docking complex is characterized by two coordinates of the PKD1 helix: the translation along the TRPP2 groove and the rotation around its helical axis (Fig. 2 A and B). To test the validity of this model, we examined the distance between a selected pair of amino acids, TRPP2_I860 and PKD1_V4217, which the model predicts to be in close proximity (Fig. 2A). These residues were mutated to cysteines in a TRPP2 fragment containing a six histidine (His6) tag (His6–TRPP2_K695–V968) and in a fusion protein of maltose-binding protein (MBP) and the PKD1 coiled-coil domain (MBP–PKD1_S4212–R4248). If these introduced cysteines were located in as close proximity as seen in the docking model then they should be able to form a disulfide bond under nonreducing conditions.
Fig. 2.
Disulfide bond analysis of a predicted close TRPP2/PKD1 contact in the docking complex. (A) The TRPP2/PKD1 coiled-coil docking complex, illustrating the side-chain positions of I860 of TRPP2 and five PKD1 residues that were individually mutated to cysteines and tested for disulfide bond formation. (B) Helical wheel representation of a portion of the TRPP2/PKD1 coiled-coil docking complex, showing the location and side-chain projection of I860 of TRPP2 and the tested PKD1 residues (red). (C and D) SDS-PAGE of various purified TRPP2/PKD1 C-terminal complexes (lanes I–V) in a reducing and nonreducing condition [with or without 100 mM DTT and 5% β-mercaptoethanol (β-ME) in a 3X SDS sample buffer]. WT protein fragments, His6–TRPP2_K695–V968 (simplified as His6–TRPP2) and MBP–PKD1_S4212–R4248 (simplified as MBP–PKD1), and their mutants, are annotated at the bottom. Putative protein composition of the major bands is indicated on the right, with the disulfide-bonded TRPP2/PKD1 complex framed with a red dashed line.
WT and/or mutant protein fragments were coexpressed in bacteria and copurified, and disulfide bond formation was examined by SDS-PAGE under reducing and nonreducing conditions (Fig. 2C). Another mutant of PKD1 in which cysteine was substituted for L4238 (PKD1_L4238C), far away from TRPP2_I860 in the structural model (Fig. 2A), was examined for comparison. WT TRPP2 and PKD1 fragments gave only monomer bands, with molecular masses of 20 and 43 kDa, respectively, in both nonreducing and reducing conditions (Fig. 2C, lane I). Under nonreducing conditions, the combination of TRPP2_I860C and PKD1_V4217C yielded major monomer bands for the two proteins, as well as a strong band at 65 kDa, corresponding in size to the disulfide-bonded heterodimer of the TRPP2 and PKD1 fragments (Fig. 2C, lane IV). This 65-kDa band disappeared under reducing conditions, as expected for a disulfide bonding dimer. It was not seen when the same cysteine version of TRPP2 (TRPP2_I860C) was combined with the other cysteine mutant of PKD1 (PKD1_L4238C) (Fig. 2C, lane V), supporting the model. Two other faint bands were seen in the heteromeric mixtures which disappeared under reducing conditions. One of these was a band at 40 kDa that was seen in all mixtures that included TRPP2_I860C and corresponded in mass to a dimer of His6–TRPP2_K695–V968 (Fig. 2C, lanes II, IV, and V). The other of these was a band at 90 kDa that was seen in every mixture that included either of the PKD1 cysteine mutants (V4217C or L4238C) and corresponded in mass to a dimer of MBP–PKD1_S4212–R4248 (Fig. 2C, lanes III, IV, and V).
To further test the specificity of disulfide bond formation and the validity of the docking complex in Fig. 2A, we examined whether TRPP2_I860C was able to form disulfide bonds with three other PKD1 cysteine mutants: PKD1_Q4215C, PKD1_E4219C, and PKD1_Q4224C. According to the docking complex, Q4215 and E4219 are located near I860 on TRPP2 (Fig. 2A), but their side chains point away from the TRPP2/PKD1 interface (Fig. 2 A and B); on the other hand, although the side chain of Q4224 faces the interface, it is located two α-helical turns away from I860 (Fig. 2 A and B). Thus, the docking complex predicts that PKD1_Q4215C, PKD1_E4219C, and PKD1_Q4224C would not dimerize with TRPP2_I860C through a disulfide bond. This prediction was fully borne out by experiment (Fig. 2D). These results indicate that our docking calculations are realistic and that the resulting docking complex (Fig. 1C and Fig. 2A) is a good starting point for MD refinement.
Structural Model of the TRPP2/PKD1 Coiled-Coil Complex.
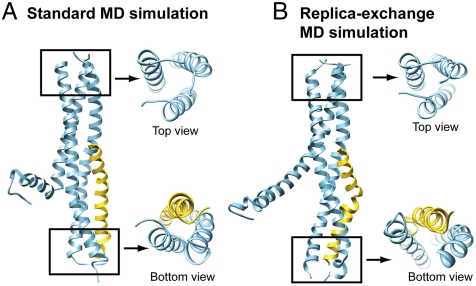
The docking complex was refined by two MD simulation protocols: standard MD and REMD. Ten 5-ns standard MD simulations and a 5-ns REMD simulation with 20 replicas were performed. A structure clustering algorithm was used to group the structures into clusters and select a small set of conformations to represent the conformational space sampled in the last 1 ns of the MD simulations. The top 10 clusters that had the lowest energy and contained at least 500 conformations were analyzed, with a representative conformation for each cluster displayed in Fig. S3. All the representative conformations shared the following structural features: (i) the upstream region of the TRPP2 trimer (up to G854) was stable during the MD simulation with virtually all the coiled-coil interactions preserved; (ii) the downstream region of one of the three TRPP2 helices that did not interact directly with the PKD1 helix became highly flexible and tended to peel away from the other two TRPP2 helices; (iii) in most cases the downstream region of the remaining two TRPP2 helices formed stable interactions with the one PKD1 helix.
Notably, conformer 2 from the standard MD protocol and conformer 1 from the REMD protocol were very similar and were characterized by low energy and a distinct di-trimer configuration: an upstream trimer formed by three TRPP2 helices and a downstream trimer formed by two TRPP2 helices and one PKD1 helix (Fig. 3). We consider the conformation in Fig. 3A from the standard MD to be a more suitable structural model for the TRPP2/PKD1 coiled-coil complex because it exhibits a more compact packing than the one obtained from REMD (Fig. 3B).
Fig. 3.
Structural models of the TRPP2/PKD1 coiled-coil domain complex. (A) Representative conformation of the second lowest-energy cluster obtained from the standard MD protocol (MD6_447 in Fig. S3A). Top and bottom views show a distinctive di-trimer configuration. (B) Representative conformation of the lowest-energy cluster obtained from the REMD protocol (RE0_999 in Fig. S3B), with top and bottom views showing a highly similar di-trimer configuration. Their coordinates are available as Datasets S1 and S2.
We also performed standard MD on the TRPP2 coiled-coil trimer itself. Clustering analysis shows that the upstream region of the TRPP2 trimer (up to G854) remains stable whereas the downstream region adopts various open conformations (Fig. S4).
Structural Model-Derived Mutations Disrupt the Assembly of the TRPP2/PKD1 Coiled-Coil Complex.
To analyze TRPP2/PKD1 coiled-coil interface contacts, four parameters were extracted from an ensemble of 1,000 conformations sampled in the last nanosecond of 10 standard MD runs or 10 lowest-temperature REMD replicas. These parameters included the Cα-Cα and Cβ-Cβ distances between pairs of interfacial contacting residues, the total number of simulation runs in which these contacts were observed, and the average frequency of occurrence (FOC) of these contacts in the runs where they were found (Table S1). These parameters show how stable a given residue-residue contact is in the simulations. However, they do not, on their own, provide information on the contribution of a contact to the binding energy, because an apparent stable contact with a high FOC can result from other interfacial interactions that contribute more significantly to binding. To confirm the structural model and determine which interfacial contacts were actually critical for binding, we designed a total of 26 mutations to disrupt a selected set of predicted stable interface contacts and examined the effect of these mutations on the assembly of the TRPP2/PKD1 coiled-coil complex by pull-down experiments, using coexpressed fusion protein fragments His6–TRPP2_K695–V968 and MBP–PKD1_S4212–R4248 (Table 1). Overall, the experimental results are in remarkable agreement with the predictions, with the majority of mutations attenuating or severely disrupting TRPP2/PKD1 binding (Table 1). As expected, the effect of mutating two or more contact residues was more severe than mutating a single contact residue (Table 1). In the following, we highlight three examples of critical interfacial contacts in a C terminus-to-N terminus order.
Table 1.
Mutations and their effects on the assembly of the TRPP2 and PKD1 coiled-coil domain complex
| PKD1 mutations | TRPP2 mutations | ||||
| No. | Mutation | Binding ability, % | No. | Mutation | Binding ability, % |
| WT | 100 | WT | 100 | ||
| 1 | R4213A/V4217A | 106 | 1 | D861A/I864A/V865A | 22 |
| 2 | V4217A/L4228A | 15 | 2 | V865A/I869A | 34 |
| 3 | F4225A/R4227A | 9 | 3 | K876A/R877A | 11 |
| 4 | F4225A/L4228A | 5 | 4 | K876A/R877A/R878A/E879A | 15 |
| 5 | L4228A/N4229A | 5 | |||
| 6 | D4234R | 25 | 5 | K876G/R877G/R878G/E879G | 5 |
| 7 | Y4236R | 39 | |||
| 8 | D4234A/Y4236A | 50 | 6 | R877A/R878A | 42 |
| 9 | L4238A | 22 | 7 | R877G/R878G | 1.5 |
| 10 | L4238A/E4239A | 1.3 | 8 | V880A/L881A/L885A | 2.9 |
| 11 | L4238A/L4242A | 3 | 9 | L881A/L885A | 3 |
| 12 | L4238A/L4242A/L4245A | 4 | 10 | L881D/L885D | 3.5 |
| 11 | R883A/L884A | 41 | |||
| 13 | Q4240A | 101 | |||
| 14 | L4245A | 72 | |||
| 15 | Q4246A | 83 | |||
| Negative control: | Negative control: | ||||
| 16 | Q4241A/H4243A | 111 | 12 | R883A/D886A | 115 |
| 17 | H4243A/S4244A | 128 | |||
Binding ability was tested by pull-down experiments in which His6–TRPP2_K695–V968 was used to pull down MBP–PKD1_S4212–R4248 after they were coexpressed in E. coli. Binding ability is defined as the normalized ratio of the intensity of MBP–PKD1_S4212–R4248 to His6–TRPP2_K695–V968 on SDS-PAGE gels. See SI Text for details.
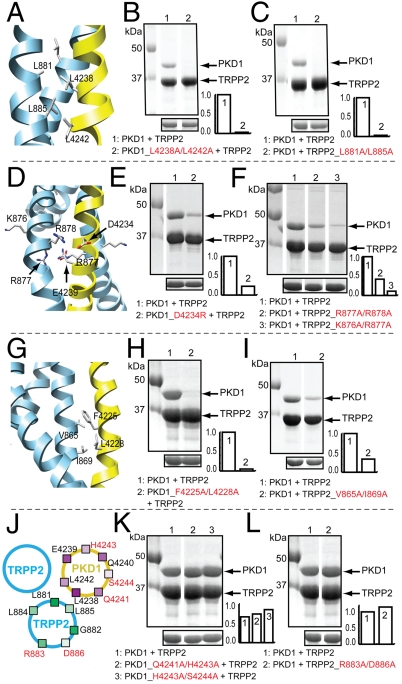
First, two hydrophobic interactions near the C-terminal end of the complex, involving PKD1_L4238, PKD1_L4242, TRPP2_L881, and TRPP2_L885, appeared to be most important for binding (Fig. 4 A–C). In the structural model, this cluster of leucines forms tightly packed hydrophobic contacts (Fig. 4A) with a Cβ-Cβ distance of 4.1–5.4 Å (Table S1), and their contacts have a high FOC ranking in the contact analysis (Table S1). A double alanine mutation of PKD1_L4238 and PKD1_L4242 or TRPP2_L881 and TRPP2_L885 abolished binding (Fig. 4 B and C and Table 1), and even a single mutation of PKD1_L4238 significantly weakened binding (Table 1).
Fig. 4.
Experimental test of selected TRPP2/PKD1 interface contacts. (A, D, G) Critical contacts in three different locations of the TRPP2/PKD1 coiled-coil domain complex: (A) near C terminus, (D) midway, and (G) near N terminus. (B, C, E, F, H, I, K, L) SDS-PAGE showing the effects of the indicated mutations (at the bottom of each panel) on the association between His6–TRPP2_K695–V968 (labeled as TRPP2) and MBP–PKD1_S4212–R4248 (labeled as PKD1). His6–TRPP2_K695–V968 fragments were used to pull-down the coexpressed MBP–PKD1_S4212–R4248 fragments (upper gel), which were expressed at a similar level in each case (lower gel). Bar graphs at the lower right corner of the gels show the normalized ratio of the intensity of PKD1 fragment∶TRPP2 fragment. (J) Cartoon of a portion of the TRPP2/PKD1 coiled-coil domain complex, showing the location of the indicated TRPP2 and PKD1 residues on a helical wheel representation. The red residues project away from the TRPP2/PKD1 interface and were tested by mutagenesis.
Second, the charge-charge interactions midway along the length of the coiled-coil interface were also important for binding. Four charged amino acids, K876/R877/R878/E879, form a KRRE charge cluster midway along the binding interface on TRPP2 (Fig. 4D). This charge cluster may facilitate the formation of the downstream trimer by increasing the flexibility of the C-terminal portion of the TRPP2 helix, allowing the displacement of one TRPP2 helix by the PKD1 helix. Mutating two consecutive positively charged residues in this cluster, R877/R878 or K876/R877, to alanine, or mutating one of their interacting partners on PKD1, D4234, to arginine, greatly weakened binding (Fig. 4 E and F and Table 1). A double glycine mutation in the TRPP2 charge cluster, R877G/R878G, completely disrupted binding (Table 1). The more potent effect of the glycine mutations could be due to a further increased flexibility and mobility of the C-terminal portion of the TRPP2 helix, which might reduce the stability of critical downstream contacts, including the hydrophobic contacts described above.
Third, like near C-terminal hydrophobic contacts, near N-terminal hydrophobic interactions also played a crucial role in binding (Fig. 4G and Table 1). A double alanine mutation of F4225 and L4228 on PKD1 nearly abolished binding (Fig. 4H and Table 1). Other double alanine mutations involving only one of these two residues, such as F4225A/R4227A, L4228A/N4229A and V4217A/L4228A, also significantly weakened or nearly abolished binding (Table 1). Similarly, the corresponding mutation of their binding partners on TRP2, V865A/I869A, also significantly weakened binding (Fig. 4I and Table 1).
Two mutations in PKD1 had no effect on TRPP2/PKD1 coiled-coil binding, one was R4213A/V4217A and the other Q4240A (Table 1). R4213 and V4217 are at the very N-terminal end of the PKD1 coiled-coil, and Q4240 is sandwiched between L4238 and L4242, two of the most critical PKD1 residues for binding. Thus, the high FOC of contacts involving R4213, V4217A, and Q4240 is likely a consequence of the strong interactions of other nearby interfacial contacts.
In the structural model, residues Q4241, H4243, and S4244 of PKD1 and R883 and D886 of TRPP2 project away from the TRPP2/PKD1 interface (Fig. 4J). If the model was correct then mutating these residues should not affect TRPP2/PKD1 binding. In agreement with this prediction, three sets of double alanine mutations, PKD1_Q4241A/H4243A, PKD1_H4243A/S4244A, and TRPP2_R833A/D886A, produced little effect on or a slight increase in binding (Fig. 4 K and L and Table 1). These results, together with those described above, strongly support the structural model.
Disruption of the TRPP2/PKD1 Coiled-Coil Complex Disrupts the Assembly of the Full-Length Complex in Cells.
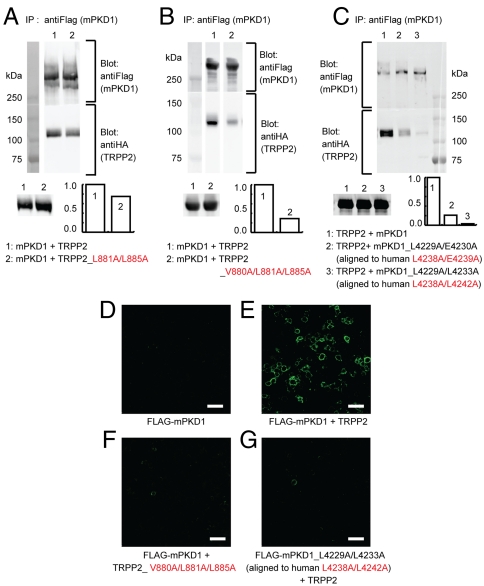
To examine the validity of our structural model in the context of full-length proteins, we investigated whether disruption of the TRPP2/PKD1 coiled-coil complex affects the assembly of the full-length TRPP2/PKD1 complex. To this end, we first examined the effect of selected mutations that disrupted the TRPP2/PKD1 coiled-coil binding on the coimmunoprecipitation (coIP) of mouse PKD1 with an N-terminal FLAG tag (FLAG–mPKD1) and human TRPP2 with an N-terminal HA tag (HA–TRPP2). When HA–TRPP2 was expressed in HEK 293T cells stably expressing FLAG–mPKD1 (27), it was abundantly coIPed with FLAG–mPKD1 by antiFLAG antibody (Fig. 5 A and B, lane 1), as shown previously (27). Expression of a TRPP2 mutant, L881A/L885A, reduced the coIP of HA_TRPP2 (Fig. 5A, lane 2), and expression of a more severe TRPP2 mutant, V880A/L881A/L885A, further reduced the ability of TRPP2 to bind PKD1 (Fig. 5B, lane 2). Similarly, when a PKD1 mutant, mPKD1_L4229A/E4230A (corresponding to human L4238A/E4239A), was expressed in HEK 293T cells stably expressing HA–TRPP2 (27), coIP of HA_TRPP2 was greatly reduced (Fig. 5C, lane 2). Expression of another mPKD1 mutant, mPKD1_L4229A/L4233A (corresponding to human L4238A/L4242A), nearly abolished the coIP of HA_TRPP2 (Fig. 5C, lane 3). These results indicate that the TRPP2/PKD1 coiled-coil interaction is critical for the assembly of the full-length TRPP2/PKD1 complex.
Fig. 5.
Effect of selected mutations on the assembly of the full-length TRPP2/PKD1 complex. (A, B, C) Western blotting showing the effect of the indicated mutations (bottom of each panel) on the coIP of HA–TRPP2 (labeled as TRPP2) and FLAG–mPKD1 (labeled as mPKD1). IP was preformed with an antiFLAG antibody, and TRPP2 and PKD1 were detected by an antiHA or antiFLAG antibody, respectively. HA–TRPP2 showed a similar expression level in each experiment (lower gel). Bar graphs at the lower right corner of the gels show the normalized ratio of the intensity of HA–TRPP2∶FLAG–mPKD1. (D–G) Confocal images of HEK 293T cells transfected with the indicated constructs. Surface PKD1 was detected by staining with an antiFLAG antibody without permeabilizing the cell membrane. None of the mutations affected the total expression of PKD1, as detected by antibody staining after membrane permeabilization (Fig. S5). Scale bar: 50 μm.
We next examined the effect of the above mutations on the surface expression of full-length TRPP2/PKD1 complexes (Fig. 5 D–G and Fig. S5). Previous work shows that in heterologous expression systems PKD1 does not reach the plasma membrane on its own but does so when coassembled with TRPP2 (27, 37, 38). In agreement with it, we found that FLAG–mPKD1 did not reach the cell surface when transfected alone into HEK 293T cells (Fig. 5D), but exhibited robust surface expression when cotransfected with TRPP2 (Fig. 5E). Cotransfection of FLAG–mPKD1 with TRPP2_V880A/L881A/L885A, which binds weakly to PKD1 (Fig. 5B), did not yield PKD1 surface expression (Fig. 5F). Likewise, cotransfection of WT TRPP2 with mPKD1_L4229A/L4233A, which binds weakly to TRPP2 (Fig. 5C), failed to stimulate PKD1 surface expression (Fig. 5G). These results further demonstrate the critical importance of the TRPP2/PKD1 coiled-coil interaction in the assembly of full-length TRPP2/PKD1 complexes and further support the validity of our structural model.
Discussion
In studying protein-protein interactions, docking can be employed to generate structural models of protein complexes of unknown structures. In such studies, prior experimental data are often required to guide the docking process (39–45). In this work, however, we constructed a structural model of the TRPP2/PKD1 coiled-coil complex solely based on a two-step docking strategy, and mutagenesis experiments were performed only to verify the computational predictions. The remarkable agreement between our computational predictions and the consequent experimental results at both the fragment and full-length protein levels suggests that the structural model in Fig. 3A represents a native-like structure in the TRPP2/PKD1 complex. Although the two-step computational strategy employed in this study was specially designed for the TRPP2/PKD1 coiled-coil complex, it may have general applications in generating structural models for other protein/protein or protein/peptide complexes.
The starting point of our modeling was the 3∶1 complex of the TRPP2/PKD1 coiled-coil domain. Using static light scattering, we determined that the PKD1 coiled-coil domain alone existed as a monomer (Fig. S6). In a previous study, we showed, by using static light scattering and X-ray crystallography, that the TRPP2 coiled-coil domain itself and four different TRPP2 C-terminal fragments containing the coiled-coil domain all form homomeric trimers, and that these trimers associate with a single PKD1 coiled-coil to form a 3∶1 complex (27). Two recent studies using a variety of biochemical and biophysical approaches also reported that the TRPP2 C terminus forms a trimer (28, 29). On the other hand, another recent study reported that the TRPP2 C terminus forms a dimer (46). This conclusion, however, is debatable (29) as the method (namely analytical ultracentrifugation) used for determining protein oligomeric states in this study depends critically on protein shape. Using single-molecule photobleaching, we further demonstrated that the full-length TRPP2/PKD1 complex heterologously expressed on the plasma membrane of Xenopus oocytes has a 3∶1 stoichiometry (27). No other reports exist concerning the stoichiometry of either the TRPP2/PKD1 coiled-coil domain complex or the full-length TRPP2/PKD1 complex. Thus, we consider the 3∶1 stoichiometry a reasonable starting point for our structural modeling. That the experimental results match well with the predictions of the final structural model validates this approach.
A unique feature of our structural model is the di-trimer configuration (Fig. 3A). The upstream trimer, formed by three TRPP2 helices, is stable during all MD simulation runs, either in the presence or absence of the PKD1 coiled-coil (Fig. S3 and Fig. S4). This result is expected because this region is packed together by extensive hydrophobic contacts (27). That the PKD1 coiled-coil binds to the C-terminal portion of the TRPP2 coiled-coil trimer is also understandable, as this region is not bundled together (27) and, due to the presence of the KRRE charge cluster, is likely more flexible and more adaptable. Further analysis suggests that the upstream TRPP2 coiled-coil trimer determines the formation and stability of the downstream TRPP2/PKD1 coiled-coil trimer (Figs. S7 and S8). This notion is consistent with a prior finding that the PKD1 coiled-coil interacts with the TRPP2 coiled-coil trimer but not the monomer (27). We speculate that during the biogenesis of the TRPP2/PKD1 complex, the formation of the upstream TRPP2 coiled-coil trimer precedes and determines the formation of the downstream TRPP2/PKD1 coiled-coil trimer.
The structural model provides a basis for understanding the pathogenic mechanism of ADPKD-causing mutations that disrupt the TRPP2/PKD1 coiled-coil association. A particularly illuminating example is TRPP2_R878del (31), which deletes a single amino acid in the TRPP2 KRRE charge cluster. This deletion should not affect the formation of the TRPP2 coiled-coil trimer, as deleting the entire KRRE charge cluster and downstream region has no such effect (27). However, according to our model, the R878del mutation would abolish the TRPP2/PKD1 coiled-coil association because it induces a rotation of the downstream region of the TRPP2 coiled-coil helix and, as a result, disrupts the downstream hydrophobic contacts (involving V880, L881, and L885) that are critical for the association of the PKD1 coiled-coil (Table 1). This example, in conjunction with previous studies showing that ADPKD pathogenic mutations that delete the TRPP2/PKD1 coiled-coils (such as TRPP2_742X and PKD1_4227X) abolish TRPP2/PKD1 assembly (6), indicate that the TRPP2/PKD1 coiled-coil association is crucial for the assembly and/or function of the TRPP2/PKD1 complex.
The only association between TRPP2 and PKD1 reported to date is through their C termini (25–27). A recent study reported that TRPP2 and PKD1 could still be expressed on the plasma membrane of HEK 293 cells even when the C terminus of one of the proteins was deleted (37). We also observed that in Xenopus oocytes, TRPP2 and PKD1 could still form complexes and reach the plasma membrane when their coiled-coil association was severely disrupted (Fig. S9). Although these studies were not quantitative and even significant reduction of the surface expression level might not have been detected, they suggest that other regions of TRPP2 and PKD1, probably the transmembrane segments, likely interact and contribute to the assembly of the TRPP2/PKD1 complex. The availability of a structural model of the TRPP2/PKD1 coiled-coil complex now makes it possible to weaken or abolish the TRPP2/PKD1 coiled-coil association by mutating as few as two amino acids (Table 1). This ability will help clarify the role of the TRPP2/PKD1 coiled-coil association in the assembly, membrane targeting, function, and regulation of the TRPP2/PKD1 complex. It also opens up the possibility to specifically weaken the TRPP2/PKD1 association without affecting the interaction of either protein with other partners, thereby facilitating future studies on the physiological functions of the TRPP2/PKD1 complex, both in vitro and in vivo.
Materials and Methods
A two-step docking strategy that combines IMO-based rigid-body docking (Fig. S10) and MD simulation was used for building and refining the structural model. Details for this computational strategy and all biochemistry experiments are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments.
We thank Dr. Stefan Somlo and Dr. Yiqiang Cai at Yale University for TRPP2 cDNA, Dr. Xing-Zhen Chen at University of Alberta for mPKD1 cDNA, and Dr. Hiroaki Matsunami at Duke University for 293T cells. We thank Kathryn Abele and Zafir Buraei at Columbia University for commenting on the manuscript. This work was supported by National Institutes of Health Grants NS045383 and GM085234 (to J.Y.), GM30518 (to B.H.), and NS035549 (to E.Y.I.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1017669108/-/DCSupplemental.
References
- 1.Ramsey IS, Delling M, Clapham DE. An introduction to TRP channels. Annu Rev Physiol. 2006;68:619–647. doi: 10.1146/annurev.physiol.68.040204.100431. [DOI] [PubMed] [Google Scholar]
- 2.Venkatachalam K, Montell C. TRP channels. Annu Rev Biochem. 2007;76:387–417. doi: 10.1146/annurev.biochem.75.103004.142819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mochizuki T, et al. PKD2, a gene for polycystic kidney disease that encodes an integral membrane protein. Science. 1996;272:1339–1342. doi: 10.1126/science.272.5266.1339. [DOI] [PubMed] [Google Scholar]
- 4.Hughes J, et al. The polycystic kidney-disease-1 (pkd1) gene encodes a novel protein with multiple cell recognition domains. Nat Genet. 1995;10:151–160. doi: 10.1038/ng0695-151. [DOI] [PubMed] [Google Scholar]
- 5.Nauli SM, et al. Polycystins 1 and 2 mediate mechanosensation in the primary cilium of kidney cells. Nat Genet. 2003;33:129–137. doi: 10.1038/ng1076. [DOI] [PubMed] [Google Scholar]
- 6.Hanaoka K, et al. Co-assembly of polycystin-1 and -2 produces unique cation-permeable currents. Nature. 2000;408:990–994. doi: 10.1038/35050128. [DOI] [PubMed] [Google Scholar]
- 7.Newby LJ, et al. Identification, characterization, and localization of a novel kidney polycystin-1-polycystin-2 complex. J Biol Chem. 2002;277:20763–20773. doi: 10.1074/jbc.M107788200. [DOI] [PubMed] [Google Scholar]
- 8.Delmas P, et al. Gating of the polycystin ion channel signaling complex in neurons and kidney cells. FASEB J. 2004;18:740–742. doi: 10.1096/fj.03-0319fje. [DOI] [PubMed] [Google Scholar]
- 9.Bai CX, et al. Formation of a new receptor-operated channel by heteromeric assembly of TRPP2 and TRPC1 subunits. EMBO Rep. 2008;9:472–479. doi: 10.1038/embor.2008.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Delmas P. Polycystins: Polymodal receptor/ion-channel cellular sensors. Pflugers Arch. 2005;451:264–276. doi: 10.1007/s00424-005-1431-5. [DOI] [PubMed] [Google Scholar]
- 11.Giamarchi A, et al. The versatile nature of the calcium-permeable cation channel TRPP2. EMBO Rep. 2006;7:787–793. doi: 10.1038/sj.embor.7400745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsiokas L. Function and regulation of TRPP2 at the plasma membrane. Am J Physiol Renal Physiol. 2009;297:F1–F9. doi: 10.1152/ajprenal.90277.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou J. Polycystins and primary cilia: Primers for cell cycle progression. Annu Rev Physiol. 2009;71:83–113. doi: 10.1146/annurev.physiol.70.113006.100621. [DOI] [PubMed] [Google Scholar]
- 14.Ong ACM, Harris PC. Molecular pathogenesis of ADPKD: The polycystin complex gets complex. Kidney Int. 2005;67:1234–1247. doi: 10.1111/j.1523-1755.2005.00201.x. [DOI] [PubMed] [Google Scholar]
- 15.Wu GQ, Somlo S. Molecular genetics and mechanism of autosomal dominant polycystic kidney disease. Mol Genet Metab. 2000;69:1–15. doi: 10.1006/mgme.1999.2943. [DOI] [PubMed] [Google Scholar]
- 16.Harris PC, Torres VE. Polycystic kidney disease. Annu Rev Med. 2009;60:321–337. doi: 10.1146/annurev.med.60.101707.125712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Torres VE, Harris PC. Autosomal dominant polycystic kidney disease: The last 3 years. Kidney Int. 2009;76:149–168. doi: 10.1038/ki.2009.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parnell SC, et al. The polycystic kidney disease-1 protein, polycystin-1, binds and activates heterotrimeric G-proteins in vitro. Biochem Biophys Res Commun. 1998;251:625–631. doi: 10.1006/bbrc.1998.9514. [DOI] [PubMed] [Google Scholar]
- 19.Delmas P, et al. Constitutive activation of G-proteins by polycystin-1 is antagonized by polycystin-2. J Biol Chem. 2002;277:11276–11283. doi: 10.1074/jbc.M110483200. [DOI] [PubMed] [Google Scholar]
- 20.Parnell SC, et al. Polycystin-1 activation of c-Jun N-terminal kinase and AP-1 is mediated by heterotrimeric G proteins. J Biol Chem. 2002;277:19566–19572. doi: 10.1074/jbc.M201875200. [DOI] [PubMed] [Google Scholar]
- 21.Yuasa T, Takakura A, Denker BM, Venugopal B, Zhou J. Polycystin-1L2 is a novel G-protein-binding protein. Genomics. 2004;84:126–138. doi: 10.1016/j.ygeno.2004.02.008. [DOI] [PubMed] [Google Scholar]
- 22.Kotttgen M, et al. TRPP2 and TRPV4 form a polymodal sensory channel complex. J Cell Biol. 2008;182:437–447. doi: 10.1083/jcb.200805124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsiokas L, et al. Specific association of the gene product of PKD2 with the TRPC1 channel. Proc Natl Acad Sci USA. 1999;96:3934–3939. doi: 10.1073/pnas.96.7.3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang P, et al. The multimeric structure of polycystin-2 (TRPP2): Structural-functional correlates of homo- and hetero-multimers with TRPC1. Hum Mol Genet. 2009;18:1238–1251. doi: 10.1093/hmg/ddp024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qian F, et al. PKD1 interacts with PKD2 through a probable coiled-coil domain. Nat Genet. 1997;16:179–183. doi: 10.1038/ng0697-179. [DOI] [PubMed] [Google Scholar]
- 26.Tsiokas L, Kim E, Arnould T, Sukhatme VP, Walz G. Homo- and heterodimeric interactions between the gene products of PKD1 and PKD2. Proc Natl Acad Sci USA. 1997;94:6965–6970. doi: 10.1073/pnas.94.13.6965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu Y, et al. Structural and molecular basis of the assembly of the TRPP2/PKD1 complex. Proc Natl Acad Sci USA. 2009;106:11558–11563. doi: 10.1073/pnas.0903684106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Behn D, et al. Quantifying the interaction of the C-terminal regions of polycystin-2 and polycystin-1 attached to a lipid bilayer by means of QCM. Biophys Chem. 2010;150:47–53. doi: 10.1016/j.bpc.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 29.Molland KL, Narayanan A, Burgner JW, Yernool DA. Identification of the structural motif responsible for trimeric assembly of the C-terminal regulatory domains of polycystin channels PKD2L1 and PKD2. Biochem J. 2010;429:171–183. doi: 10.1042/BJ20091843. [DOI] [PubMed] [Google Scholar]
- 30.Perrichot R, Mercier B, Carre A, Cledes J, Ferec C. Identification of 3 novel mutations (Y4236X, Q3820X, 11745+2 ins3) in autosomal dominant polycystic kidney disease 1 gene (PKD1) Hum Mutat. 2000;15:582. doi: 10.1002/1098-1004(200006)15:6<582::AID-HUMU20>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 31.Rossetti S, et al. Comprehensive molecular diagnostics in autosomal dominant polycystic kidney disease. J Am Soc Nephrol. 2007;18:2143–2160. doi: 10.1681/ASN.2006121387. [DOI] [PubMed] [Google Scholar]
- 32.Zhu J, Xie L, Honig B. Structural refinement of protein segments containing secondary structure elements: Local sampling, knowledge-based potentials, and clustering. Proteins Struct Funct Bioinf. 2006;65:463–479. doi: 10.1002/prot.21085. [DOI] [PubMed] [Google Scholar]
- 33.Zhu J, Alexov E, Honig B. Comparative study of generalized born models: Born radii and peptide folding. J Phys Chem B. 2005;109:3008–3022. doi: 10.1021/jp046307s. [DOI] [PubMed] [Google Scholar]
- 34.Mitsutake A, Sugita Y, Okamoto Y. Generalized-ensemble algorithms for molecular simulations of biopolymers. Biopolymers. 2001;60:96–123. doi: 10.1002/1097-0282(2001)60:2<96::AID-BIP1007>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 35.Zhu J, Fan H, Periole X, Honig B, Mark AE. Refining homology models by combining replica-exchange molecular dynamics and statistical potentials. Proteins Struct Funct Bioinf. 2008;72:1171–1188. doi: 10.1002/prot.22005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou HY, Zhou YQ. Distance-scaled, finite ideal-gas reference state improves structure-derived potentials of mean force for structure selection and stability prediction. Protein Sci. 2002;11:2714–2726. doi: 10.1110/ps.0217002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chapin HC, Rajendran V, Caplan MJ. Polycystin-1 surface localization is stimulated by polycystin-2 and cleavage at the G protein-coupled receptor proteolytic site. Mol Biol Cell. 2010;21:4338–4348. doi: 10.1091/mbc.E10-05-0407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grimm DH, et al. Polycystin-1 distribution is modulated by polycystin-2 expression in mammalian cells. J Biol Chem. 2003;278:36786–36793. doi: 10.1074/jbc.M306536200. [DOI] [PubMed] [Google Scholar]
- 39.Dominguez C, et al. Structural model of the UbcH5B/CNOT4 complex revealed by combining NMR, mutagenesis, and docking approaches. Structure. 2004;12:633–644. doi: 10.1016/j.str.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 40.Chen LJ, et al. Structural insight into the mechanisms of Wnt signaling antagonism by Dkk. J Biol Chem. 2008;283:23364–23370. doi: 10.1074/jbc.M802375200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sivasubramanian A, Chao G, Pressler HM, Wittrup KD, Gray JJ. Structural model of the mAb 806-EGFR complex using computational docking followed by computational and experimental mutagenesis. Structure. 2006;14:401–414. doi: 10.1016/j.str.2005.11.022. [DOI] [PubMed] [Google Scholar]
- 42.Lacy DB, et al. A model of Anthrax toxin lethal factor bound to protective antigen. Proc Natl Acad Sci USA. 2005;102:16409–16414. doi: 10.1073/pnas.0508259102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Volkov AN, Ferrari D, Worrall JAR, Bonvin A, Ubbink M. The orientations of cytochrome c in the highly dynamic complex with cytochrome b(5) visualized by NMR and docking using HADDOCK. Protein Sci. 2005;14:799–811. doi: 10.1110/ps.041150205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arnesano F, Banci L, Bertini I, Bonvin A. A docking approach to the study of copper trafficking proteins: Interaction between metallochaperones and soluble domains of copper ATPases. Structure. 2004;12:669–676. doi: 10.1016/j.str.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 45.van Dijk ADJ, Fushman D, Bonvin A. Various strategies of using residual dipolar couplings in NMR-driven protein docking: Application to Lys48-linked Di-ubiquitin and validation against N-15-relaxation data. Proteins Struct Funct Bioinf. 2005;60:367–381. doi: 10.1002/prot.20476. [DOI] [PubMed] [Google Scholar]
- 46.Giamarchi A, et al. A polycystin-2 (TRPP2) dimerization domain essential for the function of heteromeric polycystin complexes. EMBO J. 2010;29:1176–1191. doi: 10.1038/emboj.2010.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.