Abstract
Cyclotides are plant-derived proteins that have a unique cyclic cystine knot topology and are remarkably stable. Their natural function is host defense, but they have a diverse range of pharmaceutically important activities, including uterotonic activity and anti-HIV activity, and have also attracted recent interest as templates in drug design. Here we report an unusual biosynthetic origin of a precursor protein of a cyclotide from the butterfly pea, Clitoria ternatea, a representative member of the Fabaceae plant family. Unlike all previously reported cyclotides, the domain corresponding to the mature cyclotide from this Fabaceae plant is embedded within an albumin precursor protein. We confirmed the expression and correct processing of the cyclotide encoded by the Cter M precursor gene transcript following extraction from C. ternatea leaf and sequencing by tandem mass spectrometry. The sequence was verified by direct chemical synthesis and the peptide was found to adopt a classic knotted cyclotide fold as determined by NMR spectroscopy. Seven additional cyclotide sequences were also identified from C. ternatea leaf and flower, five of which were unique. Cter M displayed insecticidal activity against the cotton budworm Helicoverpa armigera and bound to phospholipid membranes, suggesting its activity is modulated by membrane disruption. The Fabaceae is the third largest family of flowering plants and many Fabaceous plants are of huge significance for human nutrition. Knowledge of Fabaceae cyclotide gene transcripts should enable the production of modified cyclotides in crop plants for a variety of agricultural or pharmaceutical applications, including plant-produced designer peptide drugs.
Keywords: cyclic peptides, structure, cystine knot, kalata B1
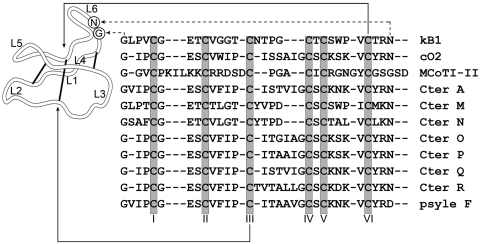
Cyclotides (1) are topologically unique plant proteins that are exceptionally stable. They comprise approximately 30 amino acids arranged in a head-to-tail cyclized peptide backbone that additionally is restrained by a cystine knot motif. The cystine knot is built from two disulfide bonds, and their connecting backbone segments form an internal ring in the structure that is threaded by a third disulfide bond to form an interlocked and cross-braced structure (Fig. 1). Superimposed on this cystine knot core are a β-sheet and a series of turns displaying surface-exposed loops.
Fig. 1.
Structures and sequences of cyclotides. The structure of the prototypical cyclotide kB1 from O. affinis is illustrated. The conserved Cys residues are labeled with Roman numerals and various loops in the backbone between them are labeled loops 1–6. The sequences of kB1 (PDB code 1NB1) (38), cycloviolacin O2 (1), MCoTI-II (39), and Cter A (21) represent examples of cyclotides isolated from the Rubiaceae, Violaceae, Cucurbitaceae, and Fabaceae plant families. The conserved cysteines are boxed and their location on the structure is indicated by the solid arrows. The putative processing points by which mature cyclotides are excised from precursor proteins are indicated and correspond to an N-terminal Gly and a C-terminal Asn (N) or Asp (D) residue (indicated by dashed arrows). The novel cyclotides identified in this study (Cter M-R) are aligned with representative cyclotides. Psyle F (24) was also identified in C. ternatea flower.
Cyclotides express a diversity of peptide sequences within their backbone loops and have a broad range of biological activities, including uterotonic (2), anti-HIV (3), antimicrobial (4), and anticancer activities (5). Accordingly, they are of great interest for pharmaceutical applications. Some plants from which they are derived are used in indigenous medicines, including kalata-kalata, a tea from the plant Oldenlandia affinis, which is used for accelerating childbirth in Africa and contains the prototypic cyclotide kalata B1, kB1 (2). This ethnobotanical use and recent biophysical studies (6) illustrate the remarkable stability of cyclotides; i.e., they survive boiling and ingestion, observations unprecedented for conventional peptides. Their exceptional stability has led to their use as templates in peptide-based drug design applications (7), where the grafting of bioactive peptide sequences into a cyclotide framework offers a new approach to stabilize peptide-based therapeutics, thereby overcoming one of the major limitations of peptides as drugs. Chemical (8, 9), chemo-enzymatic (10), and recombinant (11) approaches to the synthesis of cyclotides have been developed to facilitate these pharmaceutical applications.
The natural function of cyclotides appears to be in plant defense, based on their pesticidal activities, including insecticidal (12, 13), nematocidal (14), and molluscicidal (15) activities. These activities appear to be mediated by selective membrane binding and disruption (12, 16) that occurs as a result of cyclotides having a surface-exposed patch of hydrophobic residues. Individual plants typically contain dozens of cyclotides, expressed in multiple tissues, including flowers, leaf, and seeds, leading to their description as a natural combinatorial template (17). Plants presumably use this combinatorial strategy to target multiple pests or to reduce the possibility of an individual pest species developing resistance to the protective cyclotide armory. More than 170 cyclotides have been sequenced (18), although it is estimated that the family probably comprises around 50,000 members, making it a particularly large family of plant proteins (19).
Until recently cyclotides had been found only in the Rubiaceae (coffee) and Violaceae (violet) plant families, as well as in two atypical members in the Cucurbitaceae family (20). We recently discovered cyclotides in the seeds of a plant from the Fabaceae (legume) family, the third largest family of plants on Earth, comprising 18,000 species, many of which (e.g., peas and beans) are centrally involved in human food supply (21). Their discovery in the Fabaceae broadens interest in cyclotides because it facilitates the possibility of expressing genetically modified cyclotide sequences in crop plants from the Fabaceae for large-scale production. Legumes, including soybeans, are more widely used as transgenic vectors than members of the Rubiaceae or Violaceae. A range of traits could be incorporated in engineered cyclotides, including for host plant protection against pests or pharmaceutical attributes. In this paper we describe the nature of a gene transcript encoding a cyclotide in the Fabaceae to facilitate these applications.
Cyclotides from the Rubiaceae and Violaceae are biosynthesized via processing from dedicated precursor proteins encoded by multidomain genes, which contain one, two, or three cyclotide domains (22). The genetic origin of the cyclotides from Fabaceae was expected to be similar, but we here reveal an unexpected biosynthetic mechanism in which a cyclotide domain is embedded within an albumin precursor. This finding suggests that ribosomally synthesized cyclic peptides (23) might be much more common than has previously been realized and probably have evolved multiple alternative mechanisms for their production. The exceptional stability of cyclic peptides in harsh biological milieu appears to be the driving force for their multiple biosynthetic pathways.
Methods
(Detailed methods are given in SI Text S1.)
Plant Extraction and MS.
C. ternatea leaf material was ground prior to extraction with 50% acetonitrile and 2% formic acid in water. The extract was centrifuged and the supernatant filtered before lyophilization. Before MS analyses, cyclotides were reduced, alkylated, and linearized by digestion with endoproteinase Glu-C, trypsin, chymotrypsin, or a combination of these. Digestion was quenched with formic acid. MALDI-TOF analyses were done using an Applied Biosystems 4700 TOF-TOF and UltrafleXtreme TOF-TOF instrument. Linearized cyclotide-containing crude leaf extract was analyzed on a QStar® Elite hybrid LC-MS/MS system. MS/MS spectra were searched against a custom-built database of cyclotides with ProteinPilot.
RNA Extraction and cDNA Generation.
Total RNA was extracted from C. ternatea leaf using TRIzol® LS reagent (Invitrogen). RNA was DNAse-treated (Ambion), and complementary DNA was generated using random hexamers and Superscript III reverse transcriptase (Invitrogen). A degenerate primer (Ct-For1A, 5′-CCiACNTGYGGNGARACNTG- 3′) and an oligo-dT primer (5′- GCCCGGG T20-3′) were initially used to amplify products from cDNA. PCR products were cloned into pGEM-T Easy Vector System (Promega) and independently amplified clones were sequenced. Rapid amplification of cDNA ends (RACE) was performed using the FirstChoice® RLM-RACE kit (Applied Biosystems) according to manufacturer’s instructions. First strand cDNA synthesis was performed on leaf-derived RNA. Sequence-specific primers (Cter M-RACE-Rev1, 5′-GGAAACACCAACCAAAATGGATGT-3′; Cter M-RACE-Rev2, 5′-TCACTGTTTTTGCATTAGCTGCAA-3′) were used for first and second round PCR amplifications respectively. PCR products were cloned and sequenced. Primers (Cter M-SpecFor, 5′-TCCTTATTTTCATCAACTATGGCTTA-3′; Cter M-SpecRev, 5′-TCATACATGATCACTTTTAGTTGG-3′) were designed to the 5′ and 3′-untranslated regions and including the first and last bases of the coding region and were used to amplify full-length transcript from leaf-derived cDNA.
Synthesis.
Cter M was synthesized using solid phase peptide chemistry. RP-HPLC was used to purify the crude peptide, which was oxidized in 50% isopropanol, 1 mM reduced glutathione in 0.1 M ammonium bicarbonate, and further purified. Mass and purity were determined by ESI-MS and RP-HPLC, respectively.
NMR.
TOCSY, COSY, and NOESY spectra were recorded at 600 or 900 MHz on 1 mM Cter M in 10% D2O/20% CD3CN/70% H2O. Distance restraints were obtained from a NOESY spectrum with a 200 ms mixing time. A family of structures consistent with the experimental restraints was calculated using CYANA and CNS; 50 structures were calculated and the 20 lowest energy structures selected for analysis.
Proteolytic Stability.
Native Cter M and Cter M after reduction and alkylation were incubated at 37 °C with trypsin and chymotrypsin (20∶1 peptide∶enzyme). Aliquots were taken over 8 h and analyzed by comparing the intensity of the cyclic form of each peptide to that at the beginning of the incubation.
Surface Plasmon Resonance (SPR) Studies.
Peptide and lipid (POPC) samples were prepared in buffer 10 mM HEPES, pH 7.4. Lipids were deposited on a L1 chip in a BIAcore 3000 system. Association of Cter M and kB1 to the lipid bilayer was evaluated by injection of peptide over the lipid surface for 180 s (5 μL min-1) at 25 °C and the dissociation was followed for 600 s after injection. The chip surface was regenerated after each injection cycle.
Hemolytic Assay.
Serially diluted peptide solutions were incubated with human red blood cells and UV absorbance was measured. Hemolysis was calculated as the percentage of maximum lysis (1% Triton X-100 control) after adjusting for minimum lysis (PBS control). Melittin was used as a positive control. The dose necessary to lyse 50% of the RBCs (HD50) was calculated from the linear portion of the hemolytic titration curve.
Insecticidal Assay.
A feeding trial was conducted on H. armigera larvae for 48 h, with larvae maintained at 25 °C. Larvae were given diets consisting of wheat germ, yeast, and soy flour. Test diets contained Cter M or kB1 (used as a positive control) and the control diet did not have any added peptide. Larvae were weighed at 0, 24, and 48 h and photographed. Statistical differences were analyzed using a paired t-test or ANOVA test.
Results
Leaf tissue from C. ternatea was extracted with acetonitrile and the extract subsequently treated by reduction to break disulfide bonds, alkylation to block reactive cysteine residues, and digestion with endoproteinase Glu-C to linearize any cyclic peptides present in the extract. MALDI-TOF MS analysis of the crude leaf extract revealed a major peptide peak at m/z 3058.57 and following chemical treatment, an increase in mass of 366 Da corresponding to alkylation of six cysteines and linearization of the peptide backbone was observed, yielding an ion at m/z 3424.33 (Fig. S1). A combination of MALDI-TOF/TOF and LC-MS/MS analyses enabled the sequence of the peptide to be determined as TCTLGTCYVPDCSCSWPICMKNGLPTCGE. Two forms of this peptide were observed, one in which the Met was oxidized. We recently discovered 12 cyclotides in seed extracts from C. ternatea (21), all of which belong to the bracelet cyclotide subfamily. The current study reports the presence of a cyclotide belonging to the Möbius subfamily from Fabaceous plants. Using similar methods an additional five previously undescribed peptide sequences were deduced (Fig. 1 and Table S1 and Dataset S1). Known cyclotides Cter A (21) and Psyle F (24) were also identified in leaf and flower extracts.
One of the difficulties in using MS/MS spectra for de novo peptide sequencing is an inability to distinguish isobaric residues Ile and Leu. Amino acid analysis can yield the amino acid composition, but when both residues are present in a sequence it is not possible to distinguish their location. With this constraint in mind and with the aim of exploring biosynthesis of cyclotides within the Fabaceae we proceeded with gene transcript sequence determination. A degenerate primer was designed based upon the PTCGETC motif frequently observed in Möbius cyclotides, and used in combination with oligo-dT to isolate partial transcripts from cDNA derived from leaf total RNA. Analysis of PCR products revealed a single 402-bp band. Following cloning, the DNA sequences of independently amplified clones predicted a partial cyclotide sequence was attached to a second novel domain.
Cyclotide gene transcripts elucidated to date predict mature cyclotide domains followed by a C-terminal region (CTR) of 3–11 amino acids comprising a small amino acid (Gly or Ala) in the first position and a conserved Leu in the second position. This Leu has been postulated to play a critical role in docking to a binding pocket of asparaginyl endoprotease (AEP) during peptide excision and ligation (25). In the case of the C. ternatea-derived sequence, the mature peptide is flanked on the C terminus by a 74 amino acid tail, in which the Gly and the “critical” Leu are notably absent. BLAST searching of this C-terminal tail region revealed that it had high sequence homology to the C-terminal portion of albumin-1 proteins from a variety of Fabaceous species.
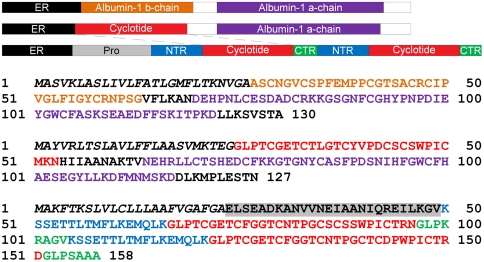
To ensure that this unusual sequence was not an artifact of cDNA synthesis or PCR, we undertook 5′ RACE studies. Following 5′ RACE amplification and alignment to previous sequences, a 514-bp consensus sequence was obtained. To confirm that this sequence represented a single mRNA expressed in C. ternatea leaf, primers were designed within the 5′ and 3′ untranslated regions, and a single 418 bp PCR product was amplified. Sequence analysis revealed this product, as anticipated, encoded a predicted protein of 127 amino acids (Fig. 2 and Fig. S2). The full protein sequence of the novel Fabaceae cyclotide precursor was aligned to the homologous albumin proteins identified in the initial BLAST search (Fig. S3). In the precursor protein from Oldenlandia affinis that encompasses the prototypic cyclotide, kB1, the mature peptide sequence is flanked by an endoplasmic reticulum (ER) signal and prodomains of 65aa at the N terminus and 7aa at the C terminus, with all of the six cysteines in the precursor located within the mature kB1 sequence (13). In contrast, the Cter M precursor protein has a typical ER signal sequence that immediately precedes the N terminus of the mature cyclotide. The cyclotide peptide sequence is then linked via 10aa to an albumin a-domain (Fig. 2).
Fig. 2.
Schematic of Cter M precursor protein (middle) alongside a typical Fabaceae albumin precursor (top), and a typical two-domain cyclotide precursor (bottom). Violaceae and Rubiaceae cyclotide mRNAs encode an ER signal peptide, an N-terminal Pro region, the N-terminal repeat (NTR), the mature cyclotide domain, and a C-terminal flanking region (CTR). There may be up to three repeats of the NTR, cyclotide domain and CTR within a typical cyclotide gene. In contrast, the CterM transcript shows an ER signal peptide immediately followed by the cyclotide domain and is flanked at the C terminus by a linking peptide and the albumin a-chain. The Cter M cyclotide domain replaces the PA1b subunit-b present in typical albumin-1 genes. The sequences of the precursor proteins are illustrated (bottom) using the color scheme from the schematic representation to indicate the location of the domains.
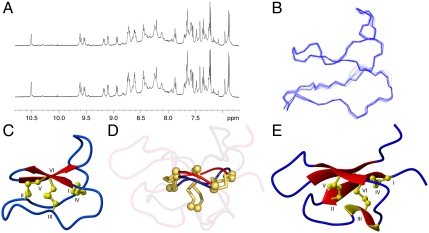
Cter M was chemically synthesized and was identical to the native form by MS and HPLC (Fig. S4) and, like the native peptide, had low solubility in water. The addition of 20% acetonitrile improved solubility for NMR analysis. NMR spectra of native and synthetic Cter M were identical. Cter M is extremely stable, as indicated by its resistance to heat denaturation; spectra were recorded before and after heating the peptide to 95 °C and no changes were observed upon return to ambient temperature (Fig. 3A). As for kB1, Cter M in its native oxidized form was impervious to trypsin and chymotrypsin, with no hydrolysis detected after 8 h incubation at 37 °C. By contrast the disulfide-reduced form of Cter M was susceptible to proteolysis (Fig. S5).
Fig. 3.
NMR spectra and 3D structures. (A) 1H spectra of Cter M recorded before (top) and after (bottom) heating to 95 °C for 5 minutes. (B) Superposition of the 20 lowest energy structures of Cter M. (C) Overview structure of Cter M. Strands are shown as arrows, helical turns as thickened ribbons, and disulfide bonds in ball-and-stick format. (D) Superposition of Cter M and PA1b highlighting the cystine knot motif; disulfide bonds are yellow and αC atoms are represented by spheres. (E) Structure of PA1b. The PDB ID code for Cter M is 2LAM and for PA1b is1P8B.
The 3D structure of Cter M was determined based on 398 NMR-derived distance restraints and 14 angle restraints. The derived family of structures had excellent geometric and energetic statistics (Table S2). An ensemble and ribbon representation is shown in Fig. 3B,C along with a comparison with PA1b (Fig. 3 D and E), a pea albumin whose precursor shares high homology with the Cter M precursor protein. Although variation in the loop regions of the two peptides is apparent, the eight-membered ring formed between loops 1 and 4 and their interconnecting disulfide bonds at the core of the cystine knot shows striking similarities.
Analysis of the structure of Cter M with PROMOTIF identified a type I β-turn at residues 9–12, a type II β-turn at residues 16–19, and a type VIa1 β-turn at residues 22–25. A β-hairpin occurs over residues 20–27, as typically seen in cystine knot proteins (26). Further examination of Cter M reveals the surface exposure of hydrophobic residues to be 51% of the solvent accessible surface (Fig. S6), which might explain why addition of acetonitrile is required for its solubilization in water, whereas at similar concentrations kB1 does not require extra solubilization (38% surface exposure).
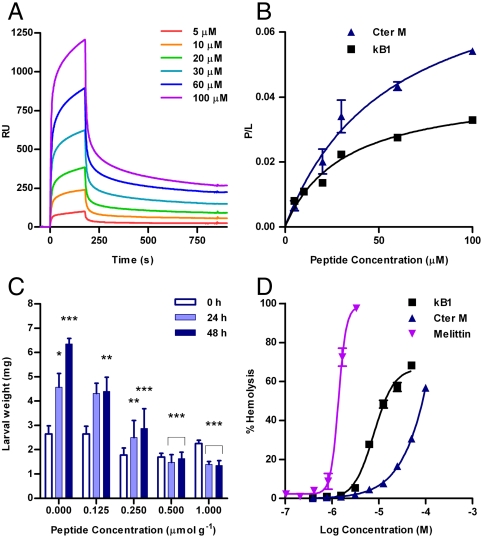
The properties and functions of Cter M were assessed using biophysical and biological assays. First, we undertook SPR studies to determine if the surface-exposed hydrophobic patch of residues identified in the structural study predisposed Cter M to membrane binding. Indeed, Cter M interacted with phospholipid vesicles in a concentration-dependent manner, similar to kB1 (Fig. 4 A and B). The calculated peptide-to-lipid ratio at 100 μM peptide concentration was 0.055 for Cter M and 0.032 for kB1. Second, the effect of Cter M on larval growth was assessed in a feeding trial where the peptide was incorporated into the diet of L3 larvae of H. armigera, a highly polyphagous species that is a pest of cotton, tomato, maize, chickpea, peanuts, and tobacco (27). Control larvae increased in weight in a monotonic manner with time whereas retardation of growth occurred in a dose-dependent manner for insects fed peptide-containing diets (Fig. 4C). At the highest concentration tested (1 μmol peptide g-1 diet), larval mortality occurred. This concentration is in a physiologically relevant range, as Cter M peptide was detected in C. ternatea at levels up to 5 μmol g-1 of leaf tissue. The insecticidal activity of Cter M was similar to kB1, which was tested in parallel (Fig. S6) and had a potency consistent with previous results (28). Finally, the hemolytic activity of Cter M was compared with kB1 and the pore-forming agent from bee venom, melittin. Cter M was only mildly hemolytic to human erythrocytes; the HD50 was 1.4 μM for melittin, 7.8 μM for kB1 and > 100 μM for Cter M (Fig. 4D). Thus, despite the increase in hydrophobicity of Cter M compared to kB1, a lower hemolytic activity was observed, indicating that an increase in surface-exposed hydrophobic residues in cyclotides does not necessarily correlate with increased hemolytic activity.
Fig. 4.
Biological and biophysical data for cyclotides. (A) Sensorgram for Cter M binding to immobilized POPC vesicles. Peptide samples were injected from 0 to 180 s; otherwise buffer was flowing. (B) Equilibrium binding curves for Cter M and kB1 binding to immobilized lipid vesicles. Fit to the single site binding model is shown as a solid line. (C) The weight of larvae at 0, 24, and 48 h is plotted versus Cter M concentration. Statistical differences were analyzed using a paired t-test for control growth and two way ANOVA for cyclotide treated growth values: *p < 0.05, **p < 0.01, ***p < 0.001. For control larvae, the weight at 24 and 48 h was compared to that at the start of the assay. For Cter M-treated larvae, the weight was compared to control larvae at the same time in the assay period. (D) Hemolytic activity of Cter M, kB1, and melittin from bee venom; HD50 was 1.4 μM for melittin, 7.8 μM for kB1 and > 100 μM for Cter M.
Discussion
Here we report a precursor transcript sequence encoding a previously undescribed cyclotide from the Fabaceae plant family and show that the peptide comprises a CCK motif, is ultrastable and insecticidal like other cyclotides, but has an unexpected biosynthetic origin in that it is embedded within an albumin precursor. It represents one of a suite of cyclotides in this plant that appear to have evolved differently from previously known cyclotides. Fabaceae plants are economically important in agriculture as nitrogen sources and in human nutrition. The discovery of an alternative biosynthetic origin for cyclotides in legumes potentially enables a wide range of new applications of cyclotides in agriculture and medicine.
Cter M and the other cyclotides reported here, despite occurring in a different plant family to previously characterized cyclotides, have sequences not dissimilar to known cyclotides. They have six conserved Cys residues arranged in a CCK motif and accordingly are exceptionally temperature stable and resistant to proteolysis. The most abundant of these cyclotides, Cter M, has the biological hallmarks of classic cyclotides in that it is insecticidal and binds to lipid membranes. Thus, we conclude that the natural function of cyclotides is host defense in Fabaceae plants. Furthermore, as Cter M has the characteristic high stability of other cyclotides, it has potential applications as a template in drug design. Significantly, Cter M is only mildly hemolytic to human erythrocytes compared to kB1 and thus, if utilized as a peptide template for pharmaceutical applications, would represent a safer alternative.
The tissue-specific expression of C. ternatea cyclotides, demonstrated by the detection of Cter M in leaf but not seed material (21) mirrors similar observations in Viola species (29), and might reflect plants defending against a spectrum of pathogens in a compartmentalized fashion. Interestingly, seed-borne cyclotides from C. ternatea, by contrast with the leaf Cter peptides reported here, are quite different in their sequences and contain atypical placements of otherwise rare amino acids in cyclotides, such as His (21). This observation led to the suggestion that cyclotide sequences are more diverse than has earlier been realized and that there might be a wider range of processing mechanisms available for the production of these circular proteins. The current study clearly shows that this is the case, based on different amino acids that flank putative processing points in Fabaceae precursors.
Despite variation in the residues flanking the processing sites for the cyclotides isolated from leaf, flower, and seed of C. ternatea, all contain a conserved Asx residue (i.e., Asn or Asp) at the C terminus of the mature cyclotide domain, consistent with the hypothesis that they are processed from precursor proteins by transpeptidation activity of AEP enzymes, like other cyclotides (25, 30). AEPs have been shown to cleave after both Asn and Asp, albeit with lower relative activity for Asp residues (31). However there are two significant surprises associated with the C. ternatea precursor protein. The first is the nature of the precursor protein itself. All cyclotides reported until now are processed from precursor proteins encoded by dedicated genes. By contrast, the Cter M precursor involves the recycling of an unrelated gene, notably via the substitution of part of an albumin-1 gene with a cyclotide domain (Fig. 2). Albumins form part of the nutrient reservoir in plants and are well known to be susceptible to processing by AEPs (31). Generic albumin-1 genes comprise an ER signal followed by an albumin chain-b domain, a linker region and then the albumin chain-a. In the Cter M precursor, the albumin chain-b domain is replaced with a cyclotide domain. How did this replacement occur? One scenario is successive evolution from one host-defense protein domain to another, and an alternative is sudden lateral transfer of a foreign element into the albumin gene. We undertook structural comparisons of cyclotide and albumin components to address this question.
The best studied Fabaceae albumin component is pea albumin 1 subunit-b (PA1b), a 37-amino acid peptide from pea seeds, Pisum sativum (32). PA1b is a potent insecticidal agent and contains a cystine knot but is larger than cyclotides and does not have a cyclic backbone. We determined the structure of Cter M and showed that its cystine knot core overlays remarkably well with that of PA1b (Fig. 3), with the respective α-carbons having an rmsd of only 1.07 Å. By contrast, the backbone loops of the two peptides are very different. This difference might reflect a case of convergent evolution to a particularly stable structural core motif (the cystine knot), or divergent evolution via alternative decoration of a cystine knot albumin core—i.e., are cyclotides ultrastabilized versions of an albumin subunit? In this regard it is interesting to note that the termini of PA1b are close to one another and it is possible that mutations in ancestral albumin genes predisposed them to cyclization. A phylogenetic analysis of the Fabaceae shows C. ternatea occupying a recent branch, consistent with recent acquisition of a cyclic motif if this mechanism applies.
Chemical mutagenesis studies provide additional insights into the potential evolution of albumin and cyclotide domains. Like kB1 (33), PA1b recently has been demonstrated to be highly tolerant to modification outside the cystine knot core (32). This observation supports the possibility of divergent evolution of cyclotides from ancestral albumin domains. Additional support comes from the similar biological activities of the two classes of molecule. The insecticidal activity of PA1b is dependent on a cluster of hydrophobic residues located on one face of the molecule (32), strikingly paralleling the importance of a hydrophobic patch of kB1 in modulating pesticidal activities (16, 33). Despite their common insecticidal activities, cyclotides and albumins appear to act via different mechanisms. SPR studies done here show that Cter M interacts with membrane lipids, a mechanism common to other cyclotides, which appear to not have stereospecific receptors and are instead pore-forming membrane disruptors (34). In contrast, PA1b interacts specifically with a membrane-based receptor in insect cells (32).
Although the function of the sulfur-rich 53-residue PA1a polypeptide of pea albumin-1 gene is unknown (35), recent studies of PA1a in transgenic grains revealed that even those that only expressed peptide PA1a (and not PA1b) were toxic to weevils. Thus, PA1a could be on its own a new type of entomotoxic peptide, or may act in synergy with PA1b (36). A similar synergistic arrangement between PA1a and Cter M is possible. PA1 is a multigene family, with at least four different genes encoding for PA1 in peas (35). Several PA1b-like peptides have been identified in other members of the Fabaceae family.
A second major difference between the cyclotide precursor reported here and previously known precursors concerns the residue immediately following the mechanistically conserved Asx. His has not been seen at this position in any other cyclotide precursors, which exclusively contain a small amino acid (usually Gly). We previously speculated that the tripeptide repeat GLP (or SLP), which frequently flanks both ends of cyclotide domains in their precursors might be important in AEP recognition. However, the Cter M precursor suggests that a small amino acid is not necessary at the start of the C-terminal flanking triplet and that the transpeptidation reaction mediated by AEP is much more tolerant than previously recognized. Furthermore, in two peptides from C. ternatea seed, Cter K and L, His also occurs as the N-terminal residue of the cyclotide domain and thus Fabaceae plants appear to be able to make an Asn-His link as well as the more conventional Asn-Gly link as part of the transpeptidation process.
The finding that an albumin gene has apparently evolved to embed a peptide domain that is subsequently processed to become cyclic has precedent in a recent report on the biosynthesis of the sunflower trypsin inhibitor peptide, SFTI-1. This 14 amino acid cyclic peptide, and the related peptide SFT-L1, are embedded within a “napin-type” 2S albumin (37). The PawS1 gene encodes a 151 amino acid protein comprising an ER signal peptide, the mature SFTI-1 domain, and a 2S albumin in which the small subunit is linked to the large subunit by intermolecular disulfide bonds. The high cysteine content of 2S albumins makes them a rich source of sulfur for plant development. A second gene (PawS2) differs from PawS1 in that there is no proregion linking the ER sequence to the mature cyclic peptide domain. In the case of PawS2 and SFT-L1, excision of the signal peptide frees the N termini of the cyclic peptide readying it for C-terminal transpeptidation and cyclization. In an analogous manner, the predicted signal peptide of the Cter M precursor identified in this study immediately precedes the cyclotide domain. Despite these similarities, there are clear differences: (i) The cyclic peptide domain is embedded upstream of the albumin region, rather than replacing the small subunit; (ii) the PawS1 and PawS2 genes encode an albumin that forms a heterodimer; and (iii) the albumins belong to distinct families with Cter M derived from an albumin-1 compared to the sunflower peptides arising from a 2S-albumin.
Cyclotides and SFTI-I are just two examples of ribosomally synthesized cyclic peptides that have been discovered in bacteria, fungi, plants, and animals over the last decade (23). The finding here that the biosynthetic origin of some cyclotides are very different from others supports a hypothesis that cyclic peptides might be more widely distributed than is currently realized. Their common feature of exceptional stability, which presumably gives them functional advantages, might be a driving force for evolution of diverse biosynthetic pathways, including the use of either dedicated or recycled precursors, with albumins now being implicated in two different classes of cyclic peptides.
This study provides impetus for the exploration of cyclotides and precursors from a range of Fabaceae species to define the mechanistic capabilities of processing enzymes. The discovery of cyclotides and other cyclic peptides from a wide range of plants is important for defining cyclization strategies employed by plants, and this knowledge may potentiate in planta synthesis of novel bioactive molecules. So far, attempts at the transgenic expression of cyclotides in plants have been confined to Arabidopsis and tobacco, where cyclotide expression gives rise to mostly acyclic or truncated proteins (25, 30). The demonstrated capacity of C. ternatea to produce fully formed cyclotides suggests that cyclotides with pharmaceutically or agriculturally relevant traits might be readily expressed in a functional form within Fabaceae species known for their suitability for large scale cropping.
Supplementary Material
Acknowledgments.
We thank Maurice Conway, Department of Employment, Economic Development and Innovation, for plant material; Bill James of Commonwealth Scientific and Industrial Research Organisation Division of Ecosystems Sciences for Helicoverpa larvae; and Andrew Kotze and Angela Ruffell (Commonwealth Scientific and Industrial Research Organisation Division of Livestock Industries) for helpful discussions. Yen-Hua Huang for SPR experiments, Quentin Kaas for the cystine knot superimposition, and Phillip Walsh and Philip Sunderland (University of Queensland) for peptide synthesis. This work was supported by the Australian Research Council. We acknowledge financial support by the Queensland State Government to the Queensland NMR Network facilities. N.L.D. is a Queensland Smart State Fellow. D.J.C. is a National Health and Medical Research Council Professorial Research Fellow.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: NMR, atomic coordinates, chemical shifts, and restraints. The Protein Data Bank (www.pdb.org) ID code for Cter M is 2LAM. The GenBank accession no. for Cter M is JF501210. The UniProtKB accession codes for C. ternatea cyclotides are: P86899 for Cyclotide Cter M, P86900 for Cyclotide Cter N, P86901 for Cyclotide Cter O, P86902 for Cyclotide Cter P, P86904 for Cyclotide Cter Q, and P86903 for Cyclotide Cter R.
See Commentary on page 10025.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1103660108/-/DCSupplemental.
References
- 1.Craik DJ, Daly NL, Bond T, Waine C. Plant cyclotides: A unique family of cyclic and knotted proteins that defines the cyclic cystine knot structural motif. J Mol Biol. 1999;294:1327–1336. doi: 10.1006/jmbi.1999.3383. [DOI] [PubMed] [Google Scholar]
- 2.Gran L. An oxytocic principle found in Oldenlandia affinis DC. An indigenous, Congolese drug “Kalata-Kalata” used to accelerate the delivery. Medd Norsk Farm Selskap. 1970;32:173–180. [Google Scholar]
- 3.Gustafson KR, et al. Circulins A and B: Novel HIV-inhibitory macrocyclic peptides from the tropical tree Chassalia parvifolia. J Am Chem Soc. 1994;116:9337–9338. [Google Scholar]
- 4.Tam JP, Lu Y-A, Yang J-L, Chiu K-W. An unusual structural motif of antimicrobial peptides containing end-to-end macrocycle and cystine-knot disulfides. Proc Natl Acad Sci USA. 1999;96:8913–8918. doi: 10.1073/pnas.96.16.8913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Svangård E, et al. Mechanism of action of cytotoxic cyclotides: Cycloviolacin O2 disrupts lipid membranes. J Nat Prod. 2007;70:643–647. doi: 10.1021/np070007v. [DOI] [PubMed] [Google Scholar]
- 6.Colgrave ML, Craik DJ. Thermal, chemical, and enzymatic stability of the cyclotide kalata B1: The importance of the cyclic cystine knot. Biochemistry. 2004;43:5965–5975. doi: 10.1021/bi049711q. [DOI] [PubMed] [Google Scholar]
- 7.Gunasekera S, et al. Engineering stabilized vascular endothelial growth factor-A antagonists: Synthesis, structural characterization, and bioactivity of grafted analogues of cyclotides. J Med Chem. 2008;51:7697–7704. doi: 10.1021/jm800704e. [DOI] [PubMed] [Google Scholar]
- 8.Daly NL, Love S, Alewood PF, Craik DJ. Chemical synthesis and folding pathways of large cyclic polypeptides: Studies of the cystine knot polypeptide kalata B1. Biochemistry. 1999;38:10606–10614. doi: 10.1021/bi990605b. [DOI] [PubMed] [Google Scholar]
- 9.Tam JP, Lu YA. A biomimetic strategy in the synthesis and fragmentation of cyclic protein. Protein Sci. 1998;7:1583–1592. doi: 10.1002/pro.5560070712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thongyoo P, Roque-Rosell N, Leatherbarrow RJ, Tate EW. Chemical and biomimetic total syntheses of natural and engineered MCoTI cyclotides. Org Biomol Chem. 2008;6:1462–1470. doi: 10.1039/b801667d. [DOI] [PubMed] [Google Scholar]
- 11.Camarero JA, Kimura RH, Woo YH, Shekhtman A, Cantor J. Biosynthesis of a fully functional cyclotide inside living bacterial cells. ChemBioChem. 2007;8:1363–1366. doi: 10.1002/cbic.200700183. [DOI] [PubMed] [Google Scholar]
- 12.Barbeta BL, Marshall AT, Gillon AD, Craik DJ, Anderson MA. Plant cyclotides disrupt epithelial cells in the midgut of lepidopteran larvae. Proc Natl Acad Sci USA. 2008;105:1221–1225. doi: 10.1073/pnas.0710338104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jennings C, West J, Waine C, Craik D, Anderson M. Biosynthesis and insecticidal properties of plant cyclotides: The cyclic knotted proteins from Oldenlandia affinis. Proc Natl Acad Sci USA. 2001;98:10614–10619. doi: 10.1073/pnas.191366898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Colgrave ML, et al. Cyclotides: Natural, circular plant peptides that possess significant activity against gastrointestinal nematode parasites of sheep. Biochemistry. 2008;47:5581–5589. doi: 10.1021/bi800223y. [DOI] [PubMed] [Google Scholar]
- 15.Plan MR, Saska I, Cagauan AG, Craik DJ. Backbone cyclised peptides from plants show molluscicidal activity against the rice pest Pomacea canaliculata (golden apple snail) J Agric Food Chem. 2008;56:5237–5241. doi: 10.1021/jf800302f. [DOI] [PubMed] [Google Scholar]
- 16.Huang YH, et al. The biological activity of the prototypic cyclotide kalata B1 is modulated by the formation of multimeric pores. J Biol Chem. 2009;284:20699–20707. doi: 10.1074/jbc.M109.003384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Craik DJ, Cemazar M, Wang CK, Daly NL. The cyclotide family of circular miniproteins: Nature’s combinatorial peptide template. Biopolymers. 2006;84:250–266. doi: 10.1002/bip.20451. [DOI] [PubMed] [Google Scholar]
- 18.Wang CKL, Kaas Q, Chiche L, Craik DJ. CyBase: A database of cyclic protein sequences and structures, with applications in protein discovery and engineering. Nucleic Acids Res. 2008;36:D206–D210. doi: 10.1093/nar/gkm953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gruber CW, et al. Distribution and evolution of circular miniproteins in flowering plants. Plant Cell. 2008;20:2471–2483. doi: 10.1105/tpc.108.062331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chiche L, et al. Squash inhibitors: From structural motifs to macrocyclic knottins. Curr Protein Pept Sci. 2004;5:341–349. doi: 10.2174/1389203043379477. [DOI] [PubMed] [Google Scholar]
- 21.Poth AG, et al. Discovery of cyclotides in the Fabaceae plant family provides new insights into the cyclization, evolution and distribution of circular proteins. ACS Chem Biol. 2011 doi: 10.1021/cb100388j. 10.1021/cb100388j. [DOI] [PubMed] [Google Scholar]
- 22.Dutton JL, et al. Conserved structural and sequence elements implicated in the processing of gene-encoded circular proteins. J Biol Chem. 2004;279:46858–46867. doi: 10.1074/jbc.M407421200. [DOI] [PubMed] [Google Scholar]
- 23.Craik DJ. Seamless proteins tie up their loose ends. Science. 2006;311:1563–1564. doi: 10.1126/science.1125248. [DOI] [PubMed] [Google Scholar]
- 24.Gerlach SL, Burman R, Bohlin L, Mondal D, Göransson U. Isolation, characterization, and bioactivity of cyclotides from the Micronesian plant Psychotria leptothyrsa. J Nat Prod. 2010;73:1207–1213. doi: 10.1021/np9007365. [DOI] [PubMed] [Google Scholar]
- 25.Gillon AD, et al. Biosynthesis of circular proteins in plants. Plant J. 2008;53:505–515. doi: 10.1111/j.1365-313X.2007.03357.x. [DOI] [PubMed] [Google Scholar]
- 26.Craik DJ, Daly NL, Waine C. The cystine knot motif in toxins and implications for drug design. Toxicon. 2001;39:43–60. doi: 10.1016/s0041-0101(00)00160-4. [DOI] [PubMed] [Google Scholar]
- 27.Wu KM, Lu YH, Feng HQ, Jiang YY, Zhao JZ. Suppression of cotton bollworm in multiple crops in China in areas with Bt toxin-containing cotton. Science. 2008;321:1676–1678. doi: 10.1126/science.1160550. [DOI] [PubMed] [Google Scholar]
- 28.Jennings CV, et al. Isolation, solution structure, and insecticidal activity of Kalata B2, a circular protein with a twist: Do Möbius strips exist in nature? Biochemistry. 2005;44:851–860. doi: 10.1021/bi047837h. [DOI] [PubMed] [Google Scholar]
- 29.Trabi M, Craik DJ. Tissue-specific expression of head-to-tail cyclized miniproteins in Violaceae and structure determination of the root cyclotide Viola hederacea root cyclotide 1. Plant Cell. 2004;16:2204–2216. doi: 10.1105/tpc.104.021790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saska I, et al. An asparaginyl endopeptidase mediates in vivo protein backbone cyclization. J Biol Chem. 2007;282:29721–29728. doi: 10.1074/jbc.M705185200. [DOI] [PubMed] [Google Scholar]
- 31.Hiraiwa N, Nishimura M, Hara-Nishimura I. Vacuolar processing enzyme is self-catalytically activated by sequential removal of the C-terminal and N-terminal propeptides. FEBS Lett. 1999;447:213–216. doi: 10.1016/s0014-5793(99)00286-0. [DOI] [PubMed] [Google Scholar]
- 32.Da Silva P, et al. Molecular requirements for the insecticidal activity of the plant peptide Pea Albumin 1 subunit b (PA1b) J Biol Chem. 2010;285:32689–32694. doi: 10.1074/jbc.M110.147199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simonsen SM, et al. Alanine scanning mutagenesis of the prototypic cyclotide reveals a cluster of residues essential for bioactivity. J Biol Chem. 2008;283:9805–9813. doi: 10.1074/jbc.M709303200. [DOI] [PubMed] [Google Scholar]
- 34.Huang Y-H, Colgrave ML, Clark RJ, Kotze AC, Craik DJ. Lysine-scanning mutagenesis reveals an amendable face of the cyclotide kalata B1 for the optimisation of nematocidal activity. J Biol Chem. 2010;285:10797–10805. doi: 10.1074/jbc.M109.089854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Higgins TJ, et al. Gene structure, protein structure, and regulation of the synthesis of a sulfur-rich protein in pea seeds. J Biol Chem. 1986;261:11124–11130. [PubMed] [Google Scholar]
- 36.Petit J, et al. WO/2009/056689 (07/05/2009) Bureau IP. 2009
- 37.Mylne JS, et al. Albumins and their processing machinery are hijacked for cyclic peptides in sunflower. Nat Chem Biol. 2011 doi: 10.1038/nchembio.542. (in press) 10.1038/nchembio.542. [DOI] [PubMed] [Google Scholar]
- 38.Saether O, et al. Elucidation of the primary and three-dimensional structure of the uterotonic polypeptide kalata B1. Biochemistry. 1995;34:4147–4158. doi: 10.1021/bi00013a002. [DOI] [PubMed] [Google Scholar]
- 39.Hernandez J-F, et al. Squash trypsin inhibitors from Momordica cochinchinensis exhibit an atypical macrocyclic structure. Biochemistry. 2000;39:5722–5730. doi: 10.1021/bi9929756. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.