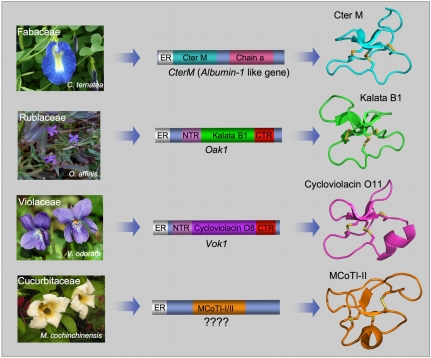
Cyclotides are fascinating circular proteins ranging from 28 to 37 aa residues that are naturally expressed in plants. They exhibit antimicrobial, insecticidal, antihelmintic, cytotoxic, and antiviral activities (1), and protease inhibitory activity (2), and can exert uterotonic effects (3). They all share a unique head-to-tail circular knotted topology of three disulfide bridges, with one disulfide bond penetrating through a macrocycle formed by the other two disulfides bonds and interconnecting peptide backbones, forming what is called a cystine knot topology (Fig. 1) (1). This cyclic cystine knot framework gives cyclotides a compact, highly rigid structure (4), which confers exceptional resistance to thermal/chemical denaturation and enzymatic degradation (5), thereby making cyclotides a promising molecular scaffold for drug discovery (6, 7). So far, cyclotides have been discovered in plants from the Rubiaceae (coffee), Violaceae (violet), and Cucurbitaceae (squash) families (8, 9), and more recently in the Fabaceae (legume) family (Fig. 1) (10). The discovery of cyclotides in the Fabaceae family of plants represents an important new development because this family of plants is the third largest on Earth, comprising approximately 18,000 different species. Some of these species are widely used as crops in human nutrition and food supply. This opens the intriguing possibility of using these plants for the large-scale production of cyclotides with pharmaceutical or agrochemical properties by using transgenic crops. The key to accomplishing that, however, is to have a better understanding of the mechanism that produces these interesting microproteins in this family of plants.
Fig. 1.
Genetic origin of cyclotides from different plant families. Rubiaceae (Oldenlandia affinis) and Violaceae (Viola odorata) plants have dedicated genes for the production of cyclotides (12). These cyclotide precursors comprise an ER signal peptide, an N-terminal Pro region, the N-terminal repeat (NTR), the mature cyclotide domain, and a C-terminal flanking region (CTR). In contrast, the CterM gene (C. ternatea, Fabaceae) shows an ER signal peptide immediately followed by the cyclotide domain, which is flanked at the C terminus by a peptide linker and the albumin a-chain. The Cter M cyclotide domain replaces albumin-1 b-chain. The genetic origin of the Cucurbitaceae cyclotides (found in the seeds of M. cochinchinensis) remains to be identified.
The report by Poth et al. in PNAS (11) brings us closer to that exciting possibility by describing the gene encoding the protein precursor of a unique cyclotide (Cter M) isolated from the leaf of butterfly pea (Clitoria ternatea), a representative member of the Fabaceae plant family. All the cyclotides reported so far from the Violaceae and Rubiaceae families are biosynthesized via processing from dedicated genes that, in some cases, encode multiple copies of the same cyclotide, and in others, mixtures of different cyclotide sequences (Fig. 1) (12). Poth et al. (11) reveal that the sequence encoding the cyclotide Cter M, however, is embedded within the albumin-1 gene of C. ternatea (Fig. 1). Plant albumins are part of the nutrient reservoir, but they also play a role in host defense. Generic albumin-1 genes are comprised of an ER signal sequence followed by an albumin chain-b, a linker, and an albumin chain-a. In the precursor of cyclotide Cter M, the cyclotide domain replaces the albumin chain-b domain. This interesting finding raises the question of how this replacement took place in evolution. There are two possibilities: (i) gradual evolution of the chain-b domain into the cyclotide domain or (ii) rapid lateral transfer of the cyclotide gene into the albumin gene. Poth et al. (11) present evidence supporting a gradual evolutionary path, whereby the albumin-1 chain-b slowly evolved into a more stable cyclotide domain. For example, the pea albumin-1 subunit-b (PA1b), one of the best-studied Fabaceae albumin components, is a 37-aa peptide from pea seeds (Pisum sativum), which also contains a cystine-knot structure (13). Remarkably, the cystine-knot core of PA1b overlays extremely well with that of the cyclotide Cter M. The composition and size of the PA1b loops are, however, totally different from those of the cyclotide Cter M. Recent mutagenesis studies on PA1b have also recently shown that this albumin domain is highly tolerant to mutations outside the cystine knot core (14). These observations support the possibility of divergent evolution of cyclotides from ancestral albumin domains, wherein evolution and natural selection provided an alternative loop decoration of the original cystine knot albumin core.
A final question remains. Linear cystine knot proteins such as PA1b are not backbone cyclized like cyclotides. What made the cyclization process possible, allowing the final transformation of an evolved cystine-knot albumin domain into a cyclotide? As indicated by Poth et al. (11), the structural analysis of PA1b may reveal some clues about how this could have happened. The NMR structure of PA1b (13) reveals that the N and C termini are very close to each other, and it is possible that mutations in the albumin genes predisposed them to cyclization during the evolution process.
However, what type of mutations could allow the backbone cyclization of a linear cystine knot albumin domain? Although the complete mechanism of how cyclotide precursors are processed and cyclized has not been fully characterized yet, recent studies indicate that an asparaginyl endopeptidase (AEP; also known as vacuolar processing enzyme or legumain) is a key element in the cyclization of cyclotides (15, 16). It has been proposed that the cyclization step mediated by AEP takes place at the same time as the cleavage of the C-terminal propeptide from the cyclotide precursor protein through a transpeptidation reaction (15). The transpeptidation reaction involves an acyl-transfer step from the acyl-AEP intermediate to the N-terminal residue of the cyclotide domain (16). A similar process has been used for the chemical (17), chemoenzymatic (18), and recombinant (19) production of cyclotides. AEPs are Cys proteases that are very common in plants and are able to specifically cleave the peptide bond at the C terminus of Asn and, less efficiently, Asp residues. All the cyclotide precursors identified so far, including those from C. ternatea, contain a well conserved Asn/Asp residue at the C terminus of the cyclotide domain, which is consistent with the idea that cyclotides are cyclized by a transpeptidation reaction mediated by AEP (15).
Despite these similarities, C. ternatea cyclotides also show some differences regarding the residue immediately following the mechanistically conserved Asn. In the cyclotide precursors from the Violaceae and Rubiaceae families, the C-terminal Asn/Asp residue is always followed by a small amino acid, either Gly or Ser. However, the Cter M precursor reported by Poth et al. (11) indicates that a small amino acid is not always required in that position. Moreover, some C. ternatea cyclotides also have a His residue at the N terminus of the cyclotide precursor rather than the most common Gly residue found in most cyclotide domains (10). These observations seem to indicate that, at least in the Fabaceae family, the AEP-mediated transpeptidation step may be more tolerant than previously recognized.
The finding that albumin genes can evolve into protein precursors that can be subsequently processed to become cyclic was described in a recent report on the biosynthesis of the sunflower trypsin inhibitor peptide, SFTI-1 (20). SFTI-1 is a 14-residue peptide isolated from sunflower seeds with a head-to-tail cyclic backbone structure having only a single disulfide bond. In this case, the SFTI-1 linear precursor is embedded within a “napin-type” 2S albumin.
The report by Poth et al. (11) indicates that the biosynthetic origin of some cyclotides are very different from others, which could suggest that cyclic peptides might be more widely distributed than is currently realized. The exceptional stability of backbone-cyclized peptides may give them an evolutionary advantage, which may provide the driving force for the evolution of multiple biosynthetic pathways including the use of dedicated or recycled genes, with albumins now being implicated in the biosynthesis of two different classes of cyclic peptides.
In this context, it is worth noting that the protein precursors of the only two cyclotides isolated so far from the Cucurbitaceae plant family, Momordica cochinchinensis trypsin inhibitor I and II (MCoTI-I/II; Fig. 1), remain yet to be identified. These cyclotides are found in the seeds of M. cochinchinensis (a tropical squash plant) and are potent trypsin inhibitors. MCoTI cyclotides do not share significant sequence homology with the other cyclotides beyond the presence of the three-cystine bridges that adopt a similar backbone-cyclic cystine-knot topology (Fig. 1) and are more related to linear cystine-knot squash trypsin inhibitors. In fact, an acyclic version of MCoTI-cyclotides (known as MCoTI-III) can also be found in the seeds of M. cochinchinensis. This situation, in which the cyclic and linear versions of the cys-knot protein coexist in the same organism, provides a unique opportunity to study the genetic origin and evolution of these interesting molecules. Identification of the protein precursors for the cyclic and linear versions of these cystine-knot trypsin inhibitors should provide a unique snapshot in the evolutionary process of plant cyclic cystine-knot proteins.
In summary, the work by Poth et al. (11) provides critical information on the origin, evolution, and processing of cyclotides from a plant of the Fabaceae family. The discovery of unique cyclotides as well as other cyclic peptides from a wide range of plants is key to define and fully understand the different cyclization mechanisms used by plants. So far, the expression of cyclotides in transgenic plants has been attempted only in Arabidopsis and tobacco (15, 16), in which cyclotide expression is highly inefficient, giving rise to mostly acyclic or truncated proteins. The proven ability of C. ternatea to produce fully folded cyclotides seems to suggest that other species of the Fabaceae family could also be used for the production of cyclotides. Several members of this large family of plants are agricultural crops, which opens the intriguing possibility of generating genetically engineered crops for the large-scale production of cyclotides with useful pharmacological or agrochemical properties in the near future.
Acknowledgments
This work was supported by National Institutes of Health Research Grant R01-GM090323 and Department of Defense Congressionally Directed Medical Research Program Grant PC09305.
Footnotes
The author declares no conflict of interest.
See companion article on page 10127.
References
- 1.Daly NL, Rosengren KJ, Craik DJ. Discovery, structure and biological activities of cyclotides. Adv Drug Deliv Rev. 2009;61:918–930. doi: 10.1016/j.addr.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 2.Avrutina O, et al. Trypsin inhibition by macrocyclic and open-chain variants of the squash inhibitor MCoTI-II. Biol Chem. 2005;386:1301–1306. doi: 10.1515/BC.2005.148. [DOI] [PubMed] [Google Scholar]
- 3.Gran L, Sandberg F, Sletten K. Oldenlandia affinis (R&S) DC. A plant containing uteroactive peptides used in African traditional medicine. J Ethnopharmacol. 2000;70:197–203. doi: 10.1016/s0378-8741(99)00175-0. [DOI] [PubMed] [Google Scholar]
- 4.Puttamadappa SS, Jagadish K, Shekhtman A, Camarero JA. Backbone dynamics of cyclotide MCoTI-I free and complexed with trypsin. Angew Chem Int Ed Engl. 2010;49:7030–7034. doi: 10.1002/anie.201002906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Colgrave ML, Craik DJ. Thermal, chemical, and enzymatic stability of the cyclotide kalata B1: The importance of the cyclic cystine knot. Biochemistry. 2004;43:5965–5975. doi: 10.1021/bi049711q. [DOI] [PubMed] [Google Scholar]
- 6.Jagadish K, Camarero JA. Cyclotides, a promising molecular scaffold for peptide-based therapeutics. Biopolymers. 2010;94:611–616. doi: 10.1002/bip.21433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garcia AE, Camarero JA. Biological activities of natural and engineered cyclotides, a novel molecular scaffold for peptide-based therapeutics. Curr Mol Pharmacol. 2010;3:153–163. doi: 10.2174/1874467211003030153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gruber CW, et al. Distribution and evolution of circular miniproteins in flowering plants. Plant Cell. 2008;20:2471–2483. doi: 10.1105/tpc.108.062331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiche L, et al. Squash inhibitors: From structural motifs to macrocyclic knottins. Curr Protein Pept Sci. 2004;5:341–349. doi: 10.2174/1389203043379477. [DOI] [PubMed] [Google Scholar]
- 10.Poth AG, et al. Discovery of cyclotides in the Fabaceae plant family provides new insights into the cyclization, evolution, and distribution of circular proteins. ACS Chem Biol. 2011;6:345–355. doi: 10.1021/cb100388j. [DOI] [PubMed] [Google Scholar]
- 11.Poth AG, Colgrave ML, Lyons RE, Daly NL, Craik DJ. Discovery of an unusual biosynthetic origin for circular proteins in legumes. Proc Natl Acad Sci USA. 2011;108:10127–10132. doi: 10.1073/pnas.1103660108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dutton JL, et al. Conserved structural and sequence elements implicated in the processing of gene-encoded circular proteins. J Biol Chem. 2004;279:46858–46867. doi: 10.1074/jbc.M407421200. [DOI] [PubMed] [Google Scholar]
- 13.Jouvensal L, et al. PA1b, an insecticidal protein extracted from pea seeds (Pisum sativum): 1H-2-D NMR study and molecular modeling. Biochemistry. 2003;42:11915–11923. doi: 10.1021/bi034803l. [DOI] [PubMed] [Google Scholar]
- 14.Da Silva P, et al. Molecular requirements for the insecticidal activity of the plant peptide pea albumin 1 subunit b (PA1b) J Biol Chem. 2010;285:32689–32694. doi: 10.1074/jbc.M110.147199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saska I, et al. An asparaginyl endopeptidase mediates in vivo protein backbone cyclization. J Biol Chem. 2007;282:29721–29728. doi: 10.1074/jbc.M705185200. [DOI] [PubMed] [Google Scholar]
- 16.Gillon AD, et al. Biosynthesis of circular proteins in plants. Plant J. 2008;53:505–515. doi: 10.1111/j.1365-313X.2007.03357.x. [DOI] [PubMed] [Google Scholar]
- 17.Craik DJ, Conibear AC. The chemistry of cyclotides. J Org Chem. 2011 doi: 10.1021/jo200520v. in press. [DOI] [PubMed] [Google Scholar]
- 18.Thongyoo P, Roqué-Rosell N, Leatherbarrow RJ, Tate EW. Chemical and biomimetic total syntheses of natural and engineered MCoTI cyclotides. Org Biomol Chem. 2008;6:1462–1470. doi: 10.1039/b801667d. [DOI] [PubMed] [Google Scholar]
- 19.Austin J, Wang W, Puttamadappa S, Shekhtman A, Camarero JA. Biosynthesis and biological screening of a genetically encoded library based on the cyclotide MCoTI-I. ChemBioChem. 2009;10:2663–2670. doi: 10.1002/cbic.200900534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mylne JS, et al. Albumins and their processing machinery are hijacked for cyclic peptides in sunflower. Nat Chem Biol. 2011;7:257–259. doi: 10.1038/nchembio.542. [DOI] [PubMed] [Google Scholar]