Abstract
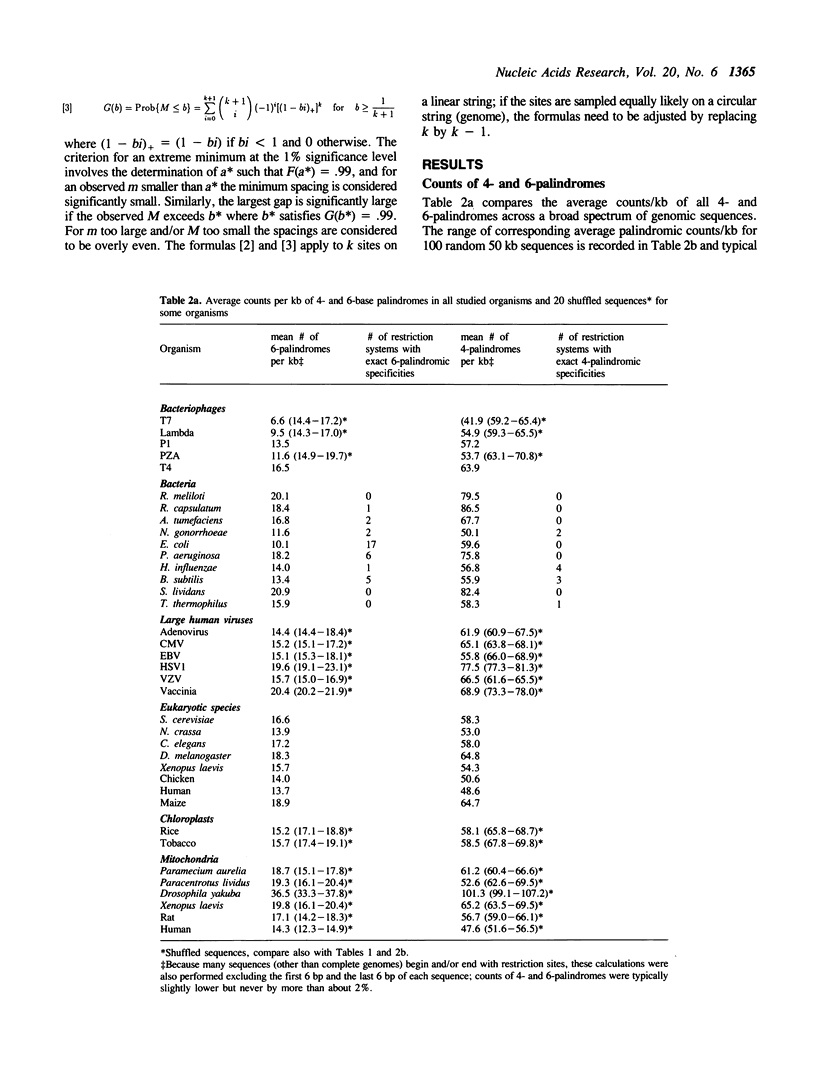
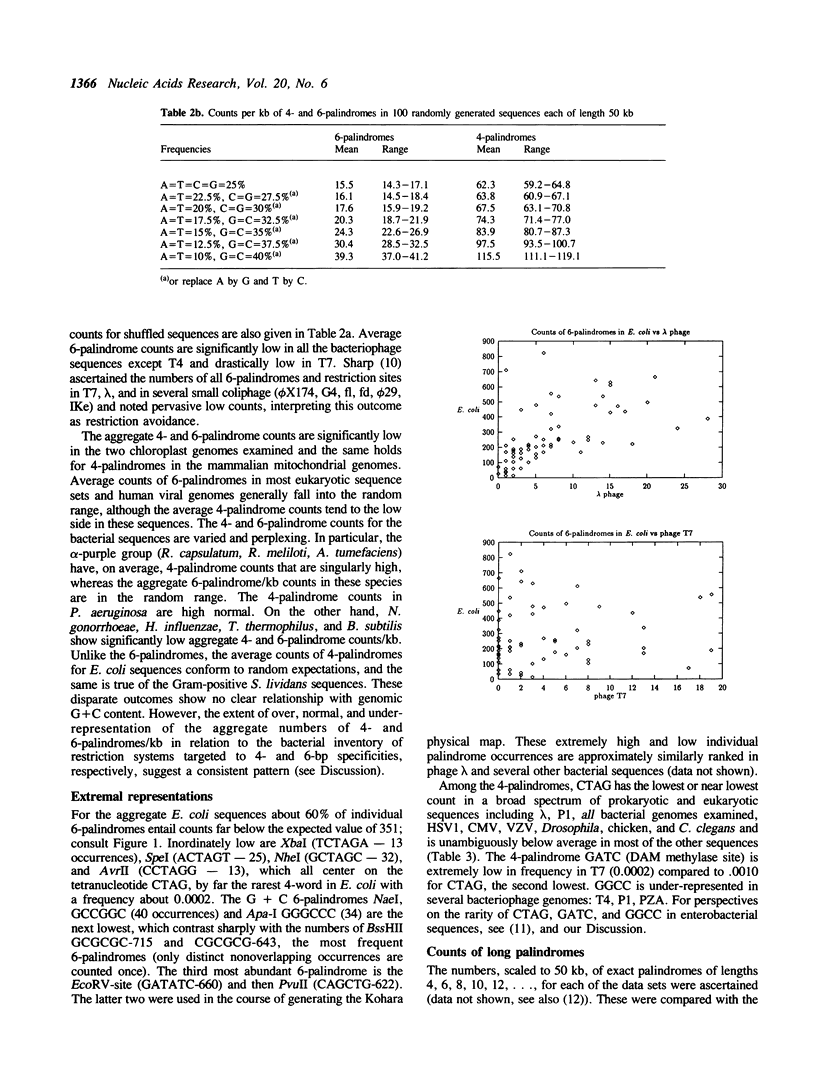
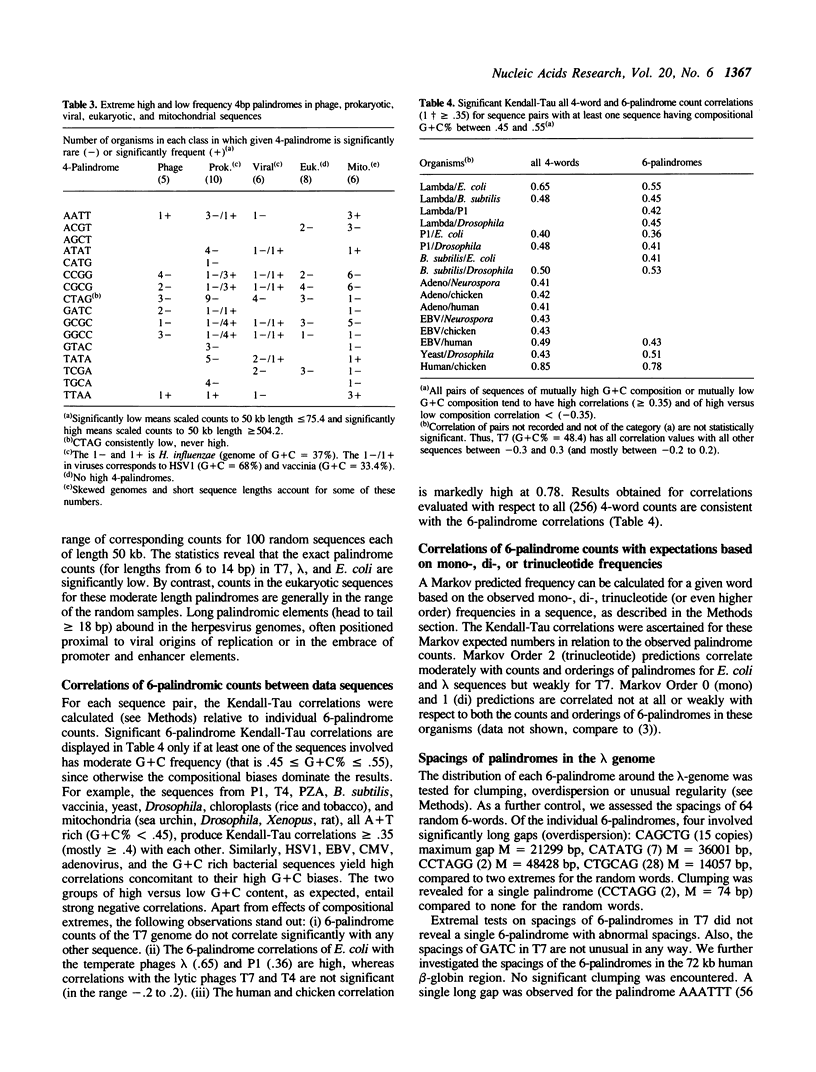
Counts and spacings of all 4- and 6-bp palindromes in DNA sequences from a broad range of organisms were investigated. Both 4- and 6-bp average palindrome counts were significantly low in all bacteriophages except one, probably as a means of avoiding restriction enzyme cleavage. The exception, T4 of normal 4- and 6-palindrome counts, putatively derives protection from modification of cytosine to hydroxymethylcytosine plus glycosylation. The counts and distributions of 4-bp and of 6-bp restriction sites in bacterial species are variable. Bacterial cells with multiple restriction systems for 4-bp or 6-bp target specificities are low in aggregate 4- or 6-bp palindrome counts/kb, respectively, but bacterial cells lacking exact 4-cutter enzymes generally show normal or high counts of 4-bp palindromes when compared with random control sequences of comparable nucleotide frequencies. For example, E. coli, apparently without an exact 4-bp target restriction endonuclease (see text), contains normal aggregate 4-palindrome counts/kb, while B. subtilis, which abounds with 4-bp restriction systems, shows a significant under-representation of 4-palindrome counts. Both E. coli and B. subtilis have many 6-bp restriction enzymes and concomitantly diminished aggregate 6-palindrome counts/kb. Eukaryote, viral, and organelle sequences generally have aggregate 4- and 6-palindromic counts/kb in the normal range. Interpretations of these results are given in terms of restriction/methylation regimes, recombination and transcription processes, and possible structural and regulatory roles of 4- and 6-bp palindromes.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andersson S. G., Kurland C. G. Codon preferences in free-living microorganisms. Microbiol Rev. 1990 Jun;54(2):198–210. doi: 10.1128/mr.54.2.198-210.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arndt K. M., Chamberlin M. J. RNA chain elongation by Escherichia coli RNA polymerase. Factors affecting the stability of elongating ternary complexes. J Mol Biol. 1990 May 5;213(1):79–108. doi: 10.1016/S0022-2836(05)80123-8. [DOI] [PubMed] [Google Scholar]
- Blaisdell B. E. Markov chain analysis finds a significant influence of neighboring bases on the occurrence of a base in eucaryotic nuclear DNA sequences both protein-coding and noncoding. J Mol Evol. 1984;21(3):278–288. doi: 10.1007/BF02102360. [DOI] [PubMed] [Google Scholar]
- Chalker A. F., Leach D. R., Lloyd R. G. Escherichia coli sbcC mutants permit stable propagation of DNA replicons containing a long palindrome. Gene. 1988 Nov 15;71(1):201–205. doi: 10.1016/0378-1119(88)90092-3. [DOI] [PubMed] [Google Scholar]
- Churchill G. A., Daniels D. L., Waterman M. S. The distribution of restriction enzyme sites in Escherichia coli. Nucleic Acids Res. 1990 Feb 11;18(3):589–597. doi: 10.1093/nar/18.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Günthert U., Trautner T. A. DNA methyltransferases of Bacillus subtilis and its bacteriophages. Curr Top Microbiol Immunol. 1984;108:11–22. doi: 10.1007/978-3-642-69370-0_2. [DOI] [PubMed] [Google Scholar]
- Hennecke F., Kolmar H., Bründl K., Fritz H. J. The vsr gene product of E. coli K-12 is a strand- and sequence-specific DNA mismatch endonuclease. Nature. 1991 Oct 24;353(6346):776–778. doi: 10.1038/353776a0. [DOI] [PubMed] [Google Scholar]
- Karlin S., Macken C. Assessment of inhomogeneities in an E. coli physical map. Nucleic Acids Res. 1991 Aug 11;19(15):4241–4246. doi: 10.1093/nar/19.15.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlin S., Morris M., Ghandour G., Leung M. Y. Algorithms for identifying local molecular sequence features. Comput Appl Biosci. 1988 Mar;4(1):41–51. doi: 10.1093/bioinformatics/4.1.41. [DOI] [PubMed] [Google Scholar]
- Kohara Y., Akiyama K., Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987 Jul 31;50(3):495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- Landick R., Yanofsky C. Stability of an RNA secondary structure affects in vitro transcription pausing in the trp operon leader region. J Biol Chem. 1984 Sep 25;259(18):11550–11555. [PubMed] [Google Scholar]
- Leach D. R., Stahl F. W. Viability of lambda phages carrying a perfect palindrome in the absence of recombination nucleases. 1983 Sep 29-Oct 5Nature. 305(5933):448–451. doi: 10.1038/305448a0. [DOI] [PubMed] [Google Scholar]
- Levin J. R., Chamberlin M. J. Mapping and characterization of transcriptional pause sites in the early genetic region of bacteriophage T7. J Mol Biol. 1987 Jul 5;196(1):61–84. doi: 10.1016/0022-2836(87)90511-0. [DOI] [PubMed] [Google Scholar]
- McClelland M., Jones R., Patel Y., Nelson M. Restriction endonucleases for pulsed field mapping of bacterial genomes. Nucleic Acids Res. 1987 Aug 11;15(15):5985–6005. doi: 10.1093/nar/15.15.5985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland M. Selection against dam methylation sites in the genomes of DNA of enterobacteriophages. J Mol Evol. 1984;21(4):317–322. doi: 10.1007/BF02115649. [DOI] [PubMed] [Google Scholar]
- Mizuuchi K., Kemper B., Hays J., Weisberg R. A. T4 endonuclease VII cleaves holliday structures. Cell. 1982 Jun;29(2):357–365. doi: 10.1016/0092-8674(82)90152-0. [DOI] [PubMed] [Google Scholar]
- Nelson M., McClelland M. Site-specific methylation: effect on DNA modification methyltransferases and restriction endonucleases. Nucleic Acids Res. 1991 Apr 25;19 (Suppl):2045–2071. doi: 10.1093/nar/19.suppl.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z., Schevitz R. W., Zhang R. G., Lawson C. L., Joachimiak A., Marmorstein R. Q., Luisi B. F., Sigler P. B. Crystal structure of trp repressor/operator complex at atomic resolution. Nature. 1988 Sep 22;335(6188):321–329. doi: 10.1038/335321a0. [DOI] [PubMed] [Google Scholar]
- Patel Y., Van Cott E., Wilson G. G., McClelland M. Cleavage at the twelve-base-pair sequence 5'-TCTAGATCTAGA-3' using M.Xbal (TCTAGm6A) methylation and DpnI (Gm6A/TC) cleavage. Nucleic Acids Res. 1990 Mar 25;18(6):1603–1607. doi: 10.1093/nar/18.6.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips G. J., Arnold J., Ivarie R. Mono- through hexanucleotide composition of the Escherichia coli genome: a Markov chain analysis. Nucleic Acids Res. 1987 Mar 25;15(6):2611–2626. doi: 10.1093/nar/15.6.2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall L. L., Josefsson L. G., Hardy S. J. Novel intermediates in the synthesis of maltose-binding protein in Escherichia coli. Eur J Biochem. 1980 Jun;107(2):375–379. doi: 10.1111/j.1432-1033.1980.tb06039.x. [DOI] [PubMed] [Google Scholar]
- Rudd K. E., Miller W., Werner C., Ostell J., Tolstoshev C., Satterfield S. G. Mapping sequenced E.coli genes by computer: software, strategies and examples. Nucleic Acids Res. 1991 Feb 11;19(3):637–647. doi: 10.1093/nar/19.3.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. M. Molecular evolution of bacteriophages: evidence of selection against the recognition sites of host restriction enzymes. Mol Biol Evol. 1986 Jan;3(1):75–83. doi: 10.1093/oxfordjournals.molbev.a040377. [DOI] [PubMed] [Google Scholar]
- Stückle E. E., Emmrich C., Grob U., Nielsen P. J. Statistical analysis of nucleotide sequences. Nucleic Acids Res. 1990 Nov 25;18(22):6641–6647. doi: 10.1093/nar/18.22.6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyman A. R., Wolfe L. B., Botstein D. Propagation of some human DNA sequences in bacteriophage lambda vectors requires mutant Escherichia coli hosts. Proc Natl Acad Sci U S A. 1985 May;82(9):2880–2884. doi: 10.1073/pnas.82.9.2880. [DOI] [PMC free article] [PubMed] [Google Scholar]


