Abstract
Most phagocytic protist viruses have large particles and genomes as well as many laterally acquired genes that may be associated with a sympatric intracellular life (a community-associated lifestyle with viruses, bacteria, and eukaryotes) and the presence of virophages. By subculturing Mimivirus 150 times in a germ-free amoebal host, we observed the emergence of a bald form of the virus that lacked surface fibers and replicated in a morphologically different type of viral factory. When studying a 0.40-μm filtered cloned particle, we found that its genome size shifted from 1.2 (M1) to 0.993 Mb (M4), mainly due to large deletions occurring at both ends of the genome. Some of the lost genes are encoding enzymes required for posttranslational modification of the structural viral proteins, such as glycosyltransferases and ankyrin repeat proteins. Proteomic analysis allowed identification of three proteins, probably required for the assembly of virus fibers. The genes for two of these were found to be deleted from the M4 virus genome. The proteins associated with fibers are highly antigenic and can be recognized by mouse and human antimimivirus antibodies. In addition, the bald strain (M4) was not able to propagate the sputnik virophage. Overall, the Mimivirus transition from a sympatric to an allopatric lifestyle was associated with a stepwise genome reduction and the production of a predominantly bald virophage resistant strain. The new axenic ecosystem allowed the allopatric Mimivirus to lose unnecessary genes that might be involved in the control of competitors.
Keywords: allopatry, experimental evolution, giant virus, virus evolution
Giant mimiviruses can replicate in amoebae (1). Mimivirus was the first virus whose genome was found to be larger than 1 Mbp (2). Its genome is composed of a complex repertoire of genes common to nucleocytoplasmic large DNA viruses (NCLDVs), as well as bacteria, eukaryotes, and archaea (3, 4). Moreover, its genome has many duplicated genes and exhibits many ORFans, i.e., ORFs that do not have a detectable homolog in current databases (5, 6). These characteristics contribute to the genome's high complexity. Viruses and intracellular bacteria, such as amoebae that are found in phagocytic protists, have a sympatric lifestyle because they replicate in communities with other species. These organisms have large chimeric genomes because they are able to exchange genes with each other inside amoebae, unlike those replicating in allopatric conditions (7, 8). There is evidence of lateral gene transfer between amoeba and its microbial parasites (9), between the phagocytosed protist viruses and bacteria (3, 4, 10) and between mimiviruses and their virophages (11).
To evaluate the role of the environment on the Mimivirus genome repertoire, we subcultured Mimivirus 150 times within the amoebal host that had been propagated in axenic conditions. We found that, as observed for allopatric bacteria growing in axenic media (12), Mimivirus subculturing leads to the emergence of morphologically and genetically different viruses (2, 13, 14), which in the case of Mimivirus M4 displays an ∼16% reduction in its genome size and lacks surface fibers.
Results
Monitoring Evolution of Mimivirus During 150 Passages.
We continuously cultured the first generation of wild-type Mimivirus (M1) 150 times in Acanthamoeba polyphaga and characterized its progeny virions after the 100th passage (M2 virus) and after the 150th passage (M3 virus). We observed three populations of virions: one with a structure identical to that of the wild-type M1 virus, a second that exhibited shorter fibers around its surface, and a third that lacked fibers (bald particles) (Fig. 1A). The proportion of long-haired mimiviruses decreased during the successive passages, and the bald form dominated the last passage (Table S1). To further characterize the bald subpopulation, we isolated a bald clone named M4 by filtering the 150th amoebal subculture supernatant through a 0.40-μm filter.
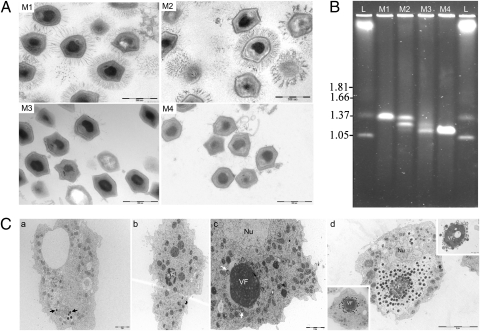
Fig. 1.
(A) Ultrastructure of the Mimivirus progeny virions stained by ruthenium red at the start of the experiment (M1), at the 100th passage (M2), at the 150th passage (M3), and after 0.40-μm filtering on the 150th amoebal subcultured supernatant (M4). (Scale bar, 500 nm.) (B) Comparative PFGE of intact genomic DNA from M1, M2, M3, and M4. Electrophoresis was performed in 1% agarose in 0.5× TBE buffer and the pulse time was ramped between 50 and 250 s for 22 h at a voltage of 5 V/cm at 14 °C. Hansenula wingei chromosomes (lane L) were used as size markers. Sizes are indicated on the Left in megabase pairs. (C) Ultrastructure of the Mimivirus M4 replication cycle in A. polyphaga at several time points postinfection. At 0.5 h p.i. (a), some viral particles (black arrows) were seen inside phagocytic vacuoles. At 4 h p.i. (b), the early virion factory (VF) appears as a dense structure with multilobed aspects. At 6 h p.i. (c), first steps of particle assembly (white arrows) were observed around the VF. Nu, cell nucleus. At 11 h p.i. (d), virions accumulated around the VF, sometimes imitating a wire (Lower Left Inset). According to our observations, this is the last step before particle release. After this point, free VF were often seen outside the cell (Upper Right Inset).
The genome size of the mimiviruses decreased over time, as evaluated by pulsed field gel electrophoresis (PFGE) (Fig. 1B) and by restriction enzyme digestions (Fig. S1). The M1 genome was a single species at 1.20 Mbp (as reported in ref. 2), and the M4 genome had only a 0.993-Mbp form. The M2 genome was a mixture of the M1 genome and an intermediate 1.10-Mbp form, whereas the M3 genome was a mixture of this 1.10-Mbp intermediate form and the 0.993-Mbp M4 form.
Early stages of the M4 replication cycle, between 30 min and 6 h postinfection (p.i.), were similar to that of the M1 strain (15) (Fig. 1 C, a–c) as observed by direct fluorescent staining (Fig. S2) or transmission electron microscopy (TEM). Between 4 and 6 h p.i., the electron-dense structure of the viral factory (VF) appeared and increased rapidly (Fig. 1 C, b and c), and a capsid assembly zone was observed at the periphery of the VF (Fig. 1 C, c), where viral particles were produced continuously until 11 h p.i. (Fig. 1 C, d). However, M4 virus assembly involved a repartitioning of the VF in which a fiber-like structure accumulated around the VF (Fig. 1 C, d, Lower Left Inset). Moreover, the peripheral electron-lucent zone, where newly formed M1 particles would acquire their fibers, was not observed. In contrast to M1 virus, lysis of M4-infected cells occurred earlier, before completely filling the amoebal cytoplasm with mature virions. Finally, some VFs could be observed outside the cells (Fig. 1 C, d, Upper Right Inset). In summary, allopatrically cultivated mimiviruses produced mixed subpopulations. This ecosystem selected viruses without fibers, with a reduced genome size and with a morphologically different VF.
Mimivirus M4 Structure.
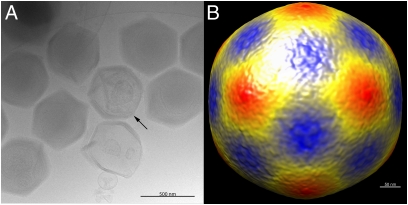
Samples of the passaged and 0.40-μm filtered M4 Mimivirus strain were frozen in vitreous ice on an electron microscope grid (Materials and Methods). The starfish-like structure associated with a special vertex seen on normal M1 Mimivirus particles (Fig. 2A) (13, 16) is visible on some of the M4 passaged particles. Many of the M4 passaged particles showed a densely packed genome compared with the M1 Mimivirus. The diameter of the M4 passaged particles was found to be ∼500 nm when measured from vertex to vertex. This is consistent with the diameter of fibered M1 particles. A reconstruction of ∼1,000 particles (Fig. 2B) assuming icosahedral symmetry, showed the flat faces that are also observed in M1 passaged mimiviruses. The starfish-like feature is not visible in this reconstruction due to icosahedral averaging.
Fig. 2.
Electron micrographs of frozen-hydrated Mimivirus M4 particles. (A) Individual particles, some of which show starfish-like structures (arrow) at a special vertex. The particles are lacking the fibrous layer of the M1 Mimivirus. (Scale bar, 500 nm.) (B) 3D cryo-EM reconstruction of the M4 passaged Mimivirus. (Scale bar, 50 nm.)
Mimivirus M4 Gene Loss.
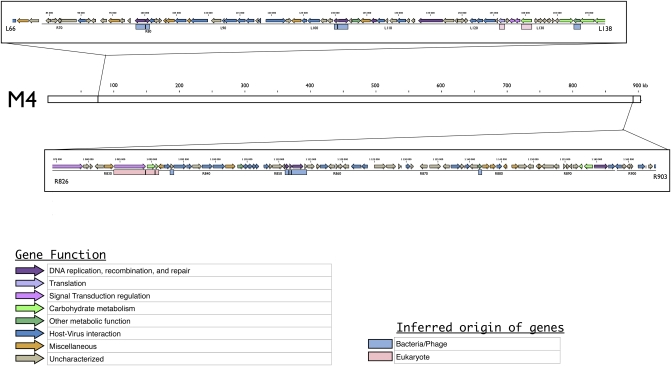
Enzymatic restriction digests using ApaI, EagI, and PmeI revealed different patterns by PFGE. There were two 80-kbp genomic deletions in the M4 strain compared with the M1 strain (Fig. S1). The deletions occurred at both ends of the M1 genome, as shown by M4 genome sequencing. The M4 genome has two large deletions between genes L67 and L137 (90.7 kb localized between positions 81,626 and 172,340 bp) and between genes L826 and R902 (95.6 kb localized between positions 1,074,695 and 1,170,290 bp) (Fig. 3). There were also putative deletions sporadically distributed along the genome affecting parts of genes as well as entire genes.
Fig. 3.
Genetic map of the two regions deleted at both ends of the M4 genome. The M4 genome is represented by a white box and its coordinates are indicated in kilobases. The two frames indicate the two regions deleted from the M1 virus. These are represented by genetic maps, including colored arrows representing CDS according to their putative function and colored boxes representing the phylogenetic origin of each gene inferred from previous phylogenetic analyses (9).
The two large deleted genomic regions are composed of 155 coding sequences (CDS) (Table S2) coding for proteins involved in diverse functions. We reanalyzed the deleted CDS, which allowed us to identify six proteins involved in carbohydrate metabolism, nine proteins involved in DNA recombination, a tyrosyl-tRNA synthetase, and three aminoacyl-tRNAs involved in translation. Moreover, 36 out of the 98 CDS in the Mimivirus genome encoding for proteins with ankyrin repeat domains, involved in intracellular host–virus interactions (17) were deleted. Two serine/threonine protein kinases and 69 other ORFans (6) were also deleted. Deletions occurred in regions with a high proportion of duplicated genes (5). Seven proteins detected in a previous proteomic study (18) and nine genes strongly expressed during the Mimivirus infectious cycle (19) were also lost. This suggested that the deleted genes were useful to M1 in its former sympatric lifestyle. The genes found in the two large deletions had varied phylogenetic origins. The R125 and R879 genes have no identified function, but they belong to the NCLDVs conserved core gene set (20). Additionally, there were six genes involved in DNA recombination and genes encoding a putative pyridoxal phosphate-dependent transferase, a putative lipocalin, and two other proteins of unknown function that were apparently acquired from bacteria (9). There were also two genes involved in carbohydrate metabolism, two genes involved in signaling, and one involved in translation, all of which were apparently acquired from eukaryotes. Overall, mimiviruses selected during allopatric growth showed a reduced genome, and the missing genes came from NCLDV, eukaryotic, and bacterial origins. Some of these genes were expressed and translated in the original M1 Mimivirus.
In addition to the two large deletions, the genome assembly showed gaps in 205 ORFs, suggesting that they were deleted or turned into pseudogenes. Twelve of these genes encoded proteins involved in DNA replication, recombination, and repair, 2 were involved in transcription and RNA processing, and 1 encoded an arginyl-tRNA synthetase involved in translation (Table S2). Moreover, we also found 3 proteins involved in carbohydrate metabolism, 10 proteins involved in diverse metabolic functions, 27 proteins with ankyrin domains, 5 putative serine/threonine-protein kinases, and 103 ORFans (Table S2). In addition, three genes encoding putative virus structure and morphogenesis proteins were also affected by potential deletions (Table S2). These modified genes recovered from the M4 sequencing suggested that rapid gene degradation was ongoing in this population.
Mimivirus M4 Fiber Loss.
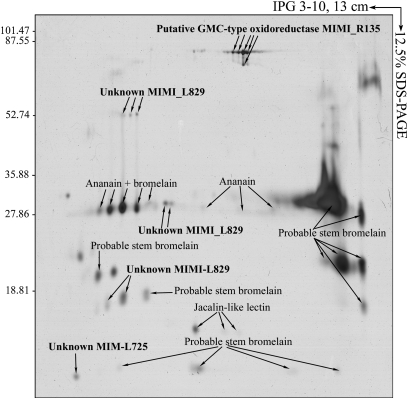
To determine whether M4 fiber loss was associated with modifications of the protein expression patterns between M1 and M4, we performed a comparative proteomic analysis. Using 2D difference-in-gel electrophoresis (DIGE), it was found that three proteins of unknown function (R641, L829, and L872), including two ORFans (R641 and L872), a putative GMC-type oxidoreductase (R135), and a putative tetratrico peptide repeat (TPR) containing protein (R856) were missing in M4 (Fig. 4 and Table S3). The R135, L829, R856, and L872 genes were deleted in the M4 strain, but the R641 gene apparently remained intact. Protein analysis from M1 purified fibers by 2D gel electrophoresis coupled with matrix-assisted laser desorption/ionization mass spectrometry (MALDI MS) (Fig. 5), allowed us to identify three fiber proteins: R135 and two proteins of unknown function (L725 encoded by an ORFan gene and L829). The absence of L829 and R135 in M4 particles can be linked to the absence of fibers surrounding the virions, and thus it appears that these two genes encode components of the viral fibers. We also identified glycosyltransferase-encoding genes that are deleted in the M4 genome. Therefore, we compared the protein glycosylation pattern in the M1 and M4 viruses (Fig. S3) and found that the L829 and R135 proteins were glycosylated in the M1 strain. Moreover, the fiber proteins are highly antigenic, because mouse monoclonal antibodies produced against Mimivirus (1) reacted with the M1 strain and not the M4 strain. This antibody was directed against L829, as detected by immunoblot analysis (Fig. S3). Serum from a patient with a laboratory infection caused by Mimivirus recognized the R135 and L829 proteins by Western blot analysis (21). Moreover, from previous work by immunogold staining, we found that the sera of patients with pneumonia that were also positive for Mimivirus by immunofluorescence analysis had antibodies directed against fibers (22). In conclusion, we identified two glycosylated proteins (L829 and R135) that are a part of the highly antigenic fibers. The absence of fibers in the subcultured Mimivirus was associated with a loss of antigenicity when tested against human sera (22, 23).
Fig. 4.
Representative 2D differential gel electrophoresis (DIGE) analysis of Mimivirus proteins. Each individual sample from the M1 and M4 strains and a pooled reference sample were labeled with Cy5, Cy3, and Cy2, respectively, and were then separated on the same gel using the 2D DIGE system. Three images were obtained from each gel and an overlay of dye scan images was also obtained. Selected protein spots (green) exhibiting an ANOVA score lower or close to 0.05 and a change of at least 1.9-fold intensity are indicated by circles and spot numbers as indicated in Table S3.
Fig. 5.
2D electrophoresis of the fiber fraction from the M1 Mimivirus strain. Purified fibers were resolved in pI values ranging from 3 to 10 in the first dimension followed by 12.5% SDS-PAGE in the second dimension, after which the proteins were detected by silver staining. Each polypeptide spot detected was identified by MALDI MS as previously described (18). Proteins identified as originating from M1 are indicated in bold.
Bald M4 Is Not Susceptible to Sputnik.
Sputnik is a small icosahedral virus that successfully parasitizes Mimiviridae (11). It was previously hypothesized that sputnik uses the Mimivirus fibers as a gateway to penetrate the amoeba (24). Another virophage (called sputnik 2) was recently found to be associated with a giant virus isolated from a liquid from a contact lens of a patient with keratitis (25). Coinfections of M1 and M4 with either sputnik 1 or sputnik 2 followed by real-time quantitative PCR showed that both sputnik 1 and 2 replicated well in M1 but not in M4 (Fig. S4). Immunofluorescence performed 6 h p.i. confirmed these results and showed that the M1 viral factories produce sputnik virions (24). However, in M4, only the VF was visible, and no production of sputnik virions could be observed (Fig. S5).
Discussion
Here, we demonstrate that serial passages of Mimivirus cultivated in axenic amoebae were associated with gene loss corresponding to ∼16% of its genome. Occurrence of large genomic deletions around the termini of DNA genomes was previously reported for other NCLDV members, including poxviruses, African swine fever virus (ASFV), and the chlorella viruses (26–30). A similar situation was reported for ASFV where large deletions occurred between 8 and 20 kb from the left ends of DNA (31). In chlorella virus PBCV-1, isolated mutants contained 27- to 37-kb deletions beginning at the left end of the 330-kb genome (26). These deletions occurred spontaneously, as is the case for the serial-passaged Mimivirus at the final step of the experimental procedure. Comparative genomic analyses performed with new NCLDV genomes have expanded our knowledge on this property of NCLDVs. Indeed, many poxvirus genomes showed that conserved genes are retained in a central location, whereas variable regions including putative deletions or putative recently acquired genes were located closer to the chromosome ends (27, 28). Furthermore, it should be emphasized that the terminal ends of the NCLDV genomes were particularly affected by lateral gene transfers probably involved in the terminal genomic variability (3, 32). The recent availability of the genome of CroV, a new giant virus related to the Mimiviridae that infects phagotrophic flagellate protists, gave us the opportunity to perform comparative genomic analysis with genomes of CroV and Mimivirus. We found that orthologs were centrally located, whereas genes unique to a given species were located more at the terminal regions of the genome (33).Taken together, our results along with previous comparative genomics studies showed that the ends of the NCLDV genomes are highly recombinogenic. The location of the deletions supports the hypothesis that this process is not random, but preferentially occurred in variable regions that are less subject to selective pressures than the central regions that contain conserved “core” genes. However, more investigations should be performed to decipher mechanisms involved in these particular genomic rearrangements.
Comparable results were obtained with some bacteria. For instance Salmonella enterica exhibited multiple and extensive genome reductions associated with loss of metabolic and regulatory capabilities following serial allotropic cultures (12). As suggested by comparative genomic analyses (3, 9), we hypothesized that the large genome size and particles of giant viruses (Mimivirus and Marseillevirus) resulted from a sympatric lifestyle conducted with other microorganisms inside phagocytic protists (8, 34, 35). This was further confirmed by genome analysis of CroV, which acquired a large genomic region that encoded proteins involved in LPS metabolism from bacteria (10). In both Mimiviridae and CroV, this lateral gene transfer might be increased by the presence of virophages (11, 36). In addition, comparative genome analysis of closely related NCLDVs infecting phagocytic protists showed that their genomes were highly dynamic and that they had probably undergone a high level of turnover of acquired/deleted genes during the course of evolution. Hence, we hypothesize that there is a balance between gene acquisition/deletion in sympatric conditions. In allopatric cultures, this balance is broken because the major sources of gene novelties (e.g., intracellular bacteria) are lacking. In light of our result, we therefore suggest that this balance and the selective pressures acted together, leading to large deletions in allopatric lifestyle. We cannot exclude that a serial passage of Mimivirus in simplified sympatric culture conditions would have generated similar results; however, experimental setting for such control is challenging and is not currently possible to set up. We performed a preliminary experiment that showed that the proportion of amoeba cells not infected by M4 when propagated with other intracellular bacteria was two times higher than when M4 was propagated alone (Table S4 and Fig. S6). As this result was not found for M1, we hypothesized that M4 shows a reduced infectivity in a sympatric lifestyle although alternative interpretations might be possible.
The lost genes in the M4 Mimivirus strain affect the expression of fiber proteins and their glycosylation. Presumably these proteins were unnecessary when competition decreased during protist infection. Furthermore, the putative fibrous proteins do not have any easily recognizable similarity to other known fibrous proteins. Fibers from the M1 form are highly immunogenic and are recognized by mouse and human antibodies following infection (21, 22). Even though we cannot rule out that some of the genes deleted in M4 are required for intracellular replication of the virophage, we assume that the loss of fibers could also affect the association between mimiviruses and sputnik virophages. It is likely that the sputnik particles are phagocytosed by amoebae because of their attachment to the Mimivirus fibers (24). So far, all characterized isolates of Mimiviridae exhibited fibers in early cultures (25). Additionally the Mimivirus fiber component R135 was always copurified and identified in the proteome of sputnik 1 (11), apparently the R135 protein has a high affinity for the virophage particles, showing that the fibers are important in allowing the virophage to coinfect amoeba.
A second group of genes that were lost in the M4 Mimivirus strain are ankyrin repeat encoding genes. These eukaryotic genes are found in many intracellular microorganisms, especially those living in amoebae (37). Bacteria living in protists, such as Legionella pneumophila, Coxiella burnetii, or Rickettsia bellii, as well as viruses like CroV (2, 10), have a very large number of these genes. This may be the result of convergent evolution, as they replicate in the same kind of ecological niche. Both C. burnetii and L. pneumophila manipulate their host by using ankyrin repeat proteins (17, 38). Therefore, we hypothesize that ankyrin repeats play a similar role in Mimivirus, and that the absence of competition with other intraamoebal microorganisms resulted in the loss of these genes without decreasing fitness. Finally, the genome reduction occurring in Mimivirus is reminiscent of the “use it or lose it” phenomenon often proposed for intracellular bacterial evolution (39).
We conclude that Mimivirus can undergo a transition from what can be described as a sympatric intracellular community-associated lifestyle to a simplified host–virus interaction after serial passages. This transition is associated with a dramatic genome reduction, which leads to the bald mutant virus. As for other microorganisms, our results suggested that ecosystem changes are associated with rapid loss of useless genes, especially those promoting amoeba internalization and those involved in the control of competitors.
Materials and Methods
Culture Conditions for Serial Passaging.
A. polyphaga used in the different experimental procedures to propagate Mimivirus were free of any intracellular microflora. Absence of bacterial endosymbionts was checked by PCR on the complete 16S rRNA gene using the external primers FD1 (5′ AGAGTTTGATCCTGGCTGAG 3′) and RP2 (5′ ACGGCTACCTTGTTACGACTT 3′). The thermal cycle consisted of an initial denaturation step at 94 °C for 2 min, followed by 35 cycles of denaturation at 94 °C for 30 s, annealing at 55 °C for 30 s, and extension at 72 °C for 2 min. The amplification was completed by extension for 7 min at 72 °C. Axenic A. polyphaga were seeded at a 5 × 104 cells/mL density in PYG medium (2% proteose peptone, 0.1% yeast extract, and 100 mM glucose), infected with titrated Mimivirus (Mimivirus M1) at a multiplicity of infection of 10 viruses/cell and centrifuged at 1,000 × g for 30 min. Mimiviruses were propagated until there was complete lysis of the amoebal culture. To achieve successive flask passaging, 5 mL of the viral supernatant was collected when lysis of amoebae was complete by low-speed (3,000 × g) centrifugation for 10 min. This was used to infect a fresh culture of A. polyphaga seeded at a 5 × 105 cells/mL density in 30 mL of PYG medium. This procedure was repeated up to 150 times. At the 100th and 150th passages (producing the M2 and M3 Mimivirus populations, respectively), viral supernatant was saved and analyzed by electron microscopy and pulsed-field gel electrophoresis. To isolate a clonal bald lineage of Mimivirus from the endpoint passage (M4 Mimivirus), viral supernatant from the 150th passage was filtered through a 0.40-μm filter, and the filtrate was used to perform endpoint dilutions. Viral supernatant of the lowest dilution displaying amoebal lysis was used as a clonal lineage for further characterization.
Electron Microscopy and Immunofluorescence.
Experiments were performed as previously described (15). For Mimivirus fiber observations by TEM, 0.15% ruthenium red was included in the fixing medium used for sample preparation as previously described (40).
Cryo-EM.
Samples for cryo-EM were prepared at Purdue University using the same procedure described above, but starting with a M4 inoculum. However, the filter used had an average pore size of 0.45 μm (Nalgene). Particles were frozen into liquid ethane using a Cryoplunge 3 (Gatan) instrument on C-flat CF-4/2–2C holey carbon grids (EMS). Images were obtained using 24,000 magnification with a CM300 FEG electron microscope recorded on a 4,000 by 4,000 CCD camera (Gatan). A total of 959 particles were boxed and normalized using EMAN (http://blake.bcm.tmc.edu/eman/) (41). Due to the low resolution of the final reconstruction, no attempt was made to apply a contrast transfer function correction. The original Mimivirus structure (13) was used to initiate the 3D reconstruction, assuming icosahedral symmetry. Essentially, all particles were used to calculate the final cryo-EM map. The resolution was estimated to be ∼30 nm based on the spatial frequency where the Fourier shell coefficient fell below 0.5 (42). The relatively poor resolution is undoubtedly due to the heterogeneous nature of the particles that were easily recognized by visual inspection. The particles varied in their vertex-to-vertex diameter by approximately ±20 nm. In addition, the large size of M4 Mimivirus would require a much larger number of particles to achieve better resolution.
Sequencing of the Mimivirus M4 Genome and Bioinformatic Analyses.
Phenol-chloroform extracted Mimivirus genomic DNA was pyrosequenced on a Roche 454 Life Sciences GS FLX titanium platform (43). The raw data (30.7 Mb) with an average length of 352 bp were assembled by Newbler software (the 454 Roche Assembler), generating a 901-kb molecule and by mapping against the Mimivirus reference genome (accession no. NC_014649). This allowed us to identify two large regions of M1 DNA missing in the M4 genome (90.7 kb localized between positions 81,626 and 172,340 bp along with 95.6 kb localized between positions 1,074,695 and 1,170,290 bp) that were overlapped by several reads. In addition, the M4 genome assembly produced 322 gaps compared with the M1 genome, which represented 94.2 kb of M1 sequence that were not covered by any M4 sequence. Annotation information of the CDS was retrieved from GenBank, except for R137, R138, and L142, which were assigned as members of the glycosyltransferase family GT2, and R654, which was assigned as a member of the glycosyltransferase family GT10, according to the CAZY database (www.cazy.org) (44).
Proteomic Analysis.
All proteomic analysis (sample preparation, 2D gel electrophoresis, silver staining, protein glycosylation analysis, protein identification, and Western blotting) were performed as previously described (18). Detailed experimental procedures on 2D DIGE analysis and protein identification are described in SI Materials and Methods.
Fiber Characterization.
The Mimivirus fibers were prepared by sequential application of bromelain (Sigma) and lysozyme from human milk (Sigma) as previously described (13). After treatment, the supernatant containing Mimivirus fibers and bromelain was precipitated using PlusOne 2D Clean-Up kit (GE Healthcare). The final pellet was resuspended in solubilizing buffer (7 M urea, 2 M thiourea, 4% CHAPS). These Mimivirus fibers were used for 2-dimensional electrophoresis (2-DE) analysis. We resolved proteins in the 3–10 pI range followed by a second dimension in 12.5% SDS-PAGE. The major polypeptide spots detected by silver staining were then subject to identification by MALDI MS as previously described (18).
SI Materials and Methods contains further detailed and experimental procedures on coinfection of Mimivirus with sputnik and PFGE analysis.
Supplementary Material
Acknowledgments
We are grateful to Marie Suzan-Monti for her contribution to the Mimivirus fiber characterization and for reading the manuscript, Thi Tien N. Guyen for technical assistance in genome sequencing, and Claude Nappez for anti-M4 polyclonal antibody production. This work was funded by the Centre National de la Recherche Scientifique (CNRS, credits récurrents).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The Mimivirus M4 genome has been deposited in the GenBank database (accession no. JN036606).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1101118108/-/DCSupplemental.
References
- 1.La Scola B, et al. A giant virus in amoebae. Science. 2003;299:2033. doi: 10.1126/science.1081867. [DOI] [PubMed] [Google Scholar]
- 2.Raoult D, et al. The 1.2-megabase genome sequence of Mimivirus. Science. 2004;306:1344–1350. doi: 10.1126/science.1101485. [DOI] [PubMed] [Google Scholar]
- 3.Filée J, Pouget N, Chandler M. Phylogenetic evidence for extensive lateral acquisition of cellular genes by nucleocytoplasmic large DNA viruses. BMC Evol Biol. 2008;8:320. doi: 10.1186/1471-2148-8-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Colson P, Raoult D. Gene repertoire of amoeba-associated giant viruses. Intervirology. 2010;53:330–343. doi: 10.1159/000312918. [DOI] [PubMed] [Google Scholar]
- 5.Suhre K. Gene and genome duplication in Acanthamoeba polyphaga Mimivirus. J Virol. 2005;79:14095–14101. doi: 10.1128/JVI.79.22.14095-14101.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyer M, Gimenez G, Suzan-Monti M, Raoult D. Classification and determination of possible origins of ORFans through analysis of nucleocytoplasmic large DNA viruses. Intervirology. 2010;53:310–320. doi: 10.1159/000312916. [DOI] [PubMed] [Google Scholar]
- 7.Thomas V, Greub G. Amoeba/amoebal symbiont genetic transfers: Lessons from giant virus neighbours. Intervirology. 2010;53:254–267. doi: 10.1159/000312910. [DOI] [PubMed] [Google Scholar]
- 8.Raoult D, Boyer M. Amoebae as genitors and reservoirs of giant viruses. Intervirology. 2010;53:321–329. doi: 10.1159/000312917. [DOI] [PubMed] [Google Scholar]
- 9.Moreira D, Brochier-Armanet C. Giant viruses, giant chimeras: The multiple evolutionary histories of Mimivirus genes. BMC Evol Biol. 2008;8:12. doi: 10.1186/1471-2148-8-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fischer MG, Allen MJ, Wilson WH, Suttle CA. Giant virus with a remarkable complement of genes infects marine zooplankton. Proc Natl Acad Sci USA. 2010;107:19508–19513. doi: 10.1073/pnas.1007615107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.La Scola B, et al. The virophage as a unique parasite of the giant mimivirus. Nature. 2008;455:100–104. doi: 10.1038/nature07218. [DOI] [PubMed] [Google Scholar]
- 12.Nilsson AI, et al. Bacterial genome size reduction by experimental evolution. Proc Natl Acad Sci USA. 2005;102:12112–12116. doi: 10.1073/pnas.0503654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiao C, et al. Structural studies of the giant mimivirus. PLoS Biol. 2009;7:e92. doi: 10.1371/journal.pbio.1000092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuznetsov YG, et al. Atomic force microscopy investigation of the giant mimivirus. Virology. 2010;404:127–137. doi: 10.1016/j.virol.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 15.Suzan-Monti M, La Scola B, Barrassi L, Espinosa L, Raoult D. Ultrastructural characterization of the giant volcano-like virus factory of Acanthamoeba polyphaga Mimivirus. PLoS ONE. 2007;2:e328. doi: 10.1371/journal.pone.0000328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zauberman N, et al. Distinct DNA exit and packaging portals in the virus Acanthamoeba polyphaga mimivirus. PLoS Biol. 2008;6:e114. doi: 10.1371/journal.pbio.0060114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pan X, Lührmann A, Satoh A, Laskowski-Arce MA, Roy CR. Ankyrin repeat proteins comprise a diverse family of bacterial type IV effectors. Science. 2008;320:1651–1654. doi: 10.1126/science.1158160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Renesto P, et al. Mimivirus giant particles incorporate a large fraction of anonymous and unique gene products. J Virol. 2006;80:11678–11685. doi: 10.1128/JVI.00940-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Legendre M, et al. mRNA deep sequencing reveals 75 new genes and a complex transcriptional landscape in Mimivirus. Genome Res. 2010;20:664–674. doi: 10.1101/gr.102582.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koonin EV, Yutin N. Origin and evolution of eukaryotic large nucleo-cytoplasmic DNA viruses. Intervirology. 2010;53:284–292. doi: 10.1159/000312913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raoult D, La Scola B, Birtles R. The discovery and characterization of Mimivirus, the largest known virus and putative pneumonia agent. Clin Infect Dis. 2007;45:95–102. doi: 10.1086/518608. [DOI] [PubMed] [Google Scholar]
- 22.La Scola B, Marrie TJ, Auffray JP, Raoult D. Mimivirus in pneumonia patients. Emerg Infect Dis. 2005;11:449–452. doi: 10.3201/eid1103.040538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berger P, et al. Ameba-associated microorganisms and diagnosis of nosocomial pneumonia. Emerg Infect Dis. 2006;12:248–255. doi: 10.3201/eid1202.050434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Desnues C, Raoult D. Inside the lifestyle of the virophage. Intervirology. 2010;53:293–303. doi: 10.1159/000312914. [DOI] [PubMed] [Google Scholar]
- 25.La Scola B, et al. A burden of new giant viruses from environment and their tentative characterization by MALDI-TOF mass-spectrometry. Intervirology. 2010;53:344–353. doi: 10.1159/000312919. [DOI] [PubMed] [Google Scholar]
- 26.Landstein D, Burbank DE, Nietfeldt JW, Van Etten JL. Large deletions in antigenic variants of the chlorella virus PBCV-1. Virology. 1995;214:413–420. doi: 10.1006/viro.1995.0051. [DOI] [PubMed] [Google Scholar]
- 27.Pickup DJ, Ink BS, Parsons BL, Hu W, Joklik WK. Spontaneous deletions and duplications of sequences in the genome of cowpox virus. Proc Natl Acad Sci USA. 1984;81:6817–6821. doi: 10.1073/pnas.81.21.6817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McLysaght A, Baldi PF, Gaut BS. Extensive gene gain associated with adaptive evolution of poxviruses. Proc Natl Acad Sci USA. 2003;100:15655–15660. doi: 10.1073/pnas.2136653100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blasco R, Agüero M, Almendral JM, Viñuela E. Variable and constant regions in African swine fever virus DNA. Virology. 1989;168:330–338. doi: 10.1016/0042-6822(89)90273-0. [DOI] [PubMed] [Google Scholar]
- 30.Songsri P, Hamazaki T, Ishikawa Y, Yamada T. Large deletions in the genome of Chlorella virus CVK1. Virology. 1995;214:405–412. doi: 10.1006/viro.1995.0050. [DOI] [PubMed] [Google Scholar]
- 31.Blasco R, de la Vega I, Almazán F, Agüero M, Viñuela E. Genetic variation of African swine fever virus: Variable regions near the ends of the viral DNA. Virology. 1989;173:251–257. doi: 10.1016/0042-6822(89)90241-9. [DOI] [PubMed] [Google Scholar]
- 32.Filée J, Chandler M. Gene exchange and the origin of giant viruses. Intervirology. 2010;53:354–361. doi: 10.1159/000312920. [DOI] [PubMed] [Google Scholar]
- 33.Colson P, Gimenez G, Boyer M, Fournous G, Raoult D. The giant Cafeteria roenbergensis virus that infects a widespread marine phagocytic protist is a new member of the fourth domain of Life. PLoS ONE. 2011;6:e18935. doi: 10.1371/journal.pone.0018935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boyer M, et al. Giant Marseillevirus highlights the role of amoebae as a melting pot in emergence of chimeric microorganisms. Proc Natl Acad Sci USA. 2009;106:21848–21853. doi: 10.1073/pnas.0911354106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moliner C, Raoult D, Fournier PE. Evidence of horizontal gene transfer between amoeba and bacteria. Clin Microbiol Infect. 2009;15(Suppl 2):178–180. doi: 10.1111/j.1469-0691.2008.02216.x. [DOI] [PubMed] [Google Scholar]
- 36.Fischer MG. A virophage at the origin of large DNA transposons in eukaryotes. First International Congress on Virus of Microbes. 2010 (Paris) June 21–25. [Google Scholar]
- 37.Moliner C, Raoult D, Fournier PE. Evidence that the intra-amoebal Legionella drancourtii acquired a sterol reductase gene from eukaryotes. BMC Res Notes. 2009;2:51. doi: 10.1186/1756-0500-2-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lührmann A, Nogueira CV, Carey KL, Roy CR. Inhibition of pathogen-induced apoptosis by a Coxiella burnetii type IV effector protein. Proc Natl Acad Sci USA. 2010;107:18997–19001. doi: 10.1073/pnas.1004380107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moran NA. Microbial minimalism: Genome reduction in bacterial pathogens. Cell. 2002;108:583–586. doi: 10.1016/s0092-8674(02)00665-7. [DOI] [PubMed] [Google Scholar]
- 40.Luft JH. Ruthenium red and violet. I. Chemistry, purification, methods of use for electron microscopy and mechanism of action. Anat Rec. 1971;171:347–368. doi: 10.1002/ar.1091710302. [DOI] [PubMed] [Google Scholar]
- 41.Ludtke SJ, Baldwin PR, Chiu W. EMAN: Semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 42.van Heel M, Schatz M. Fourier shell correlation threshold criteria. J Struct Biol. 2005;151:250–262. doi: 10.1016/j.jsb.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 43.Margulies M, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cantarel BL, et al. The Carbohydrate-Active EnZymes database (CAZy): An expert resource for Glycogenomics. Nucleic Acids Res. 2009;37:D233–D238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.