Abstract
The understanding of protein–ligand binding is of critical importance for biomedical research, yet the process itself has been very difficult to study because of its intrinsically dynamic character. Here, we have been able to quantitatively reconstruct the complete binding process of the enzyme-inhibitor complex trypsin-benzamidine by performing 495 molecular dynamics simulations of free ligand binding of 100 ns each, 187 of which produced binding events with an rmsd less than 2 Å compared to the crystal structure. The binding paths obtained are able to capture the kinetic pathway of the inhibitor diffusing from solvent (S0) to the bound (S4) state passing through two metastable intermediate states S2 and S3. Rather than directly entering the binding pocket the inhibitor appears to roll on the surface of the protein in its transition between S3 and the final binding pocket, whereas the transition between S2 and the bound pose requires rediffusion to S3. An estimation of the standard free energy of binding gives ΔG° = -5.2 ± 0.4 kcal/mol (cf. the experimental value -6.2 kcal/mol), and a two-states kinetic model kon = (1.5 ± 0.2) × 108 M-1 s-1 and koff = (9.5 ± 3.3) × 104 s-1 for unbound to bound transitions. The ability to reconstruct by simple diffusion the binding pathway of an enzyme-inhibitor binding process demonstrates the predictive power of unconventional high-throughput molecular simulations. Moreover, the methodology is directly applicable to other molecular systems and thus of general interest in biomedical and pharmaceutical research.
Keywords: binding affinity, distributed computing, Markov state model, transition-state kinetics, association rates
Understanding protein–ligand binding processes is undoubtedly of critical importance in structure-based drug design, and much effort is being invested in experimental and computational methods to resolve binding. The focus has generally resided on predicting the lowest energy binding pose of a ligand (1, 2), but resolving the kinetic mechanisms and structure activity relationships of the ligand has increasingly been recognized to provide additional mechanisms for elucidating therapeutically safe and differentiated responses (3, 4). The kinetics of binding depend on the characteristic transition states of a system. Hence, characterizing the binding pathway is crucial to understanding how to control and reengineer the process of binding.
Commonly used methods to experimentally determine kinetic data for biomolecular interactions are available (5), but fast time-scale resolution of a binding mechanism with atomic resolution remains difficult due to the intrinsic dynamic and volatile nature of the process of binding. From a computational perspective, the difficulty lies in accurately measuring binding affinities and kinetic parameters, but it has become easier to try to predict binding free energies on a limited number of targets and to qualitatively interpret binding mechanisms using molecular dynamics. Although it still requires substantial computational resources, the use of special molecular dynamics (MD) engines running on graphical processing units (GPUs) have greatly reduced its cost (6, 7). In this work we introduce complete reconstruction of the binding process for an enzyme-inhibitor complex by free diffusion molecular dynamics simulations. Not only do we reproduce with atomic resolution the crystallographic mode of binding, but we also provide the kinetically and energetically meaningful transition states of the process.
Free ligand binding has been used in the past to describe computational experiments in which, typically, a ligand is placed at a certain distance from the target protein and first by diffusion and then by specific interactions binds to one or more sites in the protein. These works can be classified into two groups: those mainly trying to predict binding sites and modes for one or more ligands, and those adding some degree of mechanistic information about the process of binding. Nonetheless, previous attempts to perform free ligand binding to proteins could not recover more than a few binding events due to the computational cost, therefore providing some qualitative information on the process but lacking a quantitative validation of the results with experimental data. Proper validation is necessary to make sure that the results provide the correct strategy for understanding biological function.
Wu et al. (8) reported possible binding modes of thioflavin-T (ThT) to β-rich peptide self-assemblies complementing previous experimental work in which ThT binding sites could not be determined (9). Other predictions of single or multiple binding sites were carried out in nicotinic acetylcholine receptors where Brannigan et al. (10) proposed multiple binding sites for anesthetic isoflurane, and the more recent description of different binding modes of agonists to β-adrenergic receptors also with all-atom molecular dynamics simulations (11). In ref. 12, we could recover the experimental binding site for sodium ions obtaining several hundred binding events on D2-dopaminergic receptors. On the more mechanistic description, there has been the work on the fast recognition of proline-rich peptides by SH3 domains (13) as well as the pH-dependent mechanism of NO transport by nitrophorins (14). Finally, results on initial conformational changes upon binding are reported for glycerol 3-phosphate transporter (GlpT), which mediates the import of glycerol 3-phosphate using a phosphate gradient (15). Despite such progress having been made, none of the aforementioned studies have provided a complete reconstruction of a ligand binding process in terms of pathway and quantitative information of the energetics and kinetics.
An ideal method for resolving the binding process would provide not only the binding affinity and kinetics of the reaction, but also atomic resolution information on its pathway. Binding sites, transition states, and metastable states are potentially useful to broaden the probability of success in the context of drug design. Here, we present a kinetic model for the binding process of serine protease β-trypsin inhibitor benzamidine obtained from extensive high-throughput all-atom MD simulations of free ligand binding using the ACEMD (accelerating biomolecular dynamics) (6) software on the GPUGRID.net distributed computing network (7). An aggregate of 50 μs of trajectory data have been used to construct a Markov state model (MSM) (16) of the binding process of benzamidine to trypsin. Previous computational studies on trypsin date back to the 1980s with work by Warshel et al. (17) on binding free energies computed from potentials of mean force for trypsin and subtilisin. More recent efforts on the trypsin–benzamidine system, in particular, include studies of the conformational variability of the bound complex (18, 19) and benzamidinium-derivatives binding affinity calculations via end-point methods (20, 21) and via potential of mean force free-energy biased methods such as umbrella sampling (22) and metadynamics (23). In this work, we report 187 events of binding from bulk to bound based on free diffusion simulations, a quantitative analysis of the process of binding, and the identification of the rate-limiting step in the pathway of binding, which consists of a set of transient hydrogen-bond interactions on the surface of trypsin close to the binding pocket.
Results and Discussion
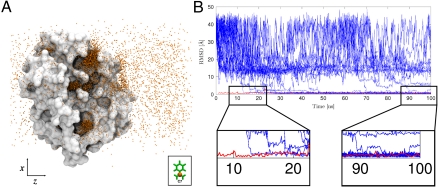
The results of this work are based on the analysis of 495 trajectories of free diffusion of benzamidine around trypsin each of 100 ns of length (see Fig. 1 for a schematic representation of the system). Among these, 187 trajectories (37%) successfully recovered the bound pose in the binding pocket with an rmsd compared to the crystal structure of less than 2 Å (Movie S1 shows a typical binding event). As shown in Fig. 2A, benzamidine placed at 35 Å from its binding pocket explores the entire simulation box (as monitored by its C7 carbon). Interestingly, several clusters of benzamidine on the surface of trypsin can be observed, which indicates that a rather complex mechanism of binding takes place instead of a simple pathway directly from the bulk. The rmsd of each trajectory in Fig. 2B shows the time evolution for a sample (one every ten) of the trajectories. Some trajectories reach the bound crystallographic pose just after 10–15 ns of simulation, whereas some reach the binding pocket only after 90 ns. Finally, the majority of the trajectories do not enter the binding site within 100 ns, as should be expected in such a short time frame. Nevertheless, there is more than enough data to carry out a detailed quantitative analysis of the binding pathway. In fact, being able to obtain 187 bound trajectories at less than 2 Å rmsd by high-throughput molecular dynamics is a compelling result.
Fig. 1.
(A) Crystallographic bound structure of benzamidine and trypsin (PDB ID code 3PTB). Parameters for benzamidine were derived from the generalized Amber force field (GAFF), and restrained electrostatic potential (RESP) partial charges fitted with Gaussian. The C7 atom was used as a reference to define the reaction coordinate. (B) System setup. Trypsin (T) and benzamidine (B) are separated by a distance of 35 Å along z first, and then solvated with a TIP3P water box extending 12 Å from any atom in all directions.
Fig. 2.
Raw data and binding events. (A) Visualization showing snapshots of the benzamidine C7 atom position during multiple trajectories. Some clusterings of benzamidine can already be seen on trypsin’s surface. The kinetic relationship between these clusters is analyzed with the construction of a Markov state model. (B) Time evolution of rmsd for benzamidine C3, C6, and C7 atoms. The plot shows only a fraction (5%) of the total runs for simplicity. Binding trajectories are considered as those 2 Å rmsd from the crystallographic pose, also calculated for reference (red line). Although some trajectories are seen to bind as soon as 10 ns after the start of the simulation, others do it beyond the 90 ns or did not bind.
In order to formalize the observable conformations of the system and its kinetic relationships, we built several Markov state models of the process (16): (i) a two-dimensional projection in the xz reaction plane, (ii) a simple five metastable-state model, and finally (iii) a full three-dimensional projection to explicitly compute kinetic and energetic information.
Identification of Metastable States.
Starting from the two-dimensional model of the process, we chose the projection of the coordinates of benzamidine C7 atom on the xz plane, which is the plane that better captures the binding process (see Fig. S1 for other projections). A two-dimensional 2,500-state MSM was constructed with a lag time of 50 ns (see Fig. S2). The clustering of the coordinates was performed on a matrix of 50 × 50 bins of 1.44 Å2 each. From the first eigenvector of the transition probability matrix we obtained the potential of mean force (PMF) of the binding process (Fig. 3A). Several metastable binding sites are identifiable by this energetic map. These are respectively named as follows: S0, to represent the bulk, i.e., all those conformations with z greater than 30 Å; S1, where a 1.0 kcal/mol minima indicates the first interactions of benzamidine with trypsin; S2, the isolated 2.5 kcal/mol minima in the lower-left corner of the PMF plane; S3, the second deepest minima of 3.0 kcal/mol at the top-left corner of the PMF plane; and finally S4, the bound conformation with relative free energy of 6.0 kcal/mol, centered at the origin. The metastable states are structurally characterized by visual inspection to highlight the underlying molecular interactions, as shown in Fig. 3B. In state S1, several mild hydrophobic interactions between benzamidine and Y59, Y94, and V90 are identified as representing the first-recognition contacts of the complex in our experiments. In S2, benzamidine is stabilized by a stronger π-π stacking interaction with Y151 and Y39 side chains. In S3, several hydrogen bonds are present between the amidine moiety of benzamidine and Q175, T98, L99, and T177 main chains in a rather electronegative region of trypsin. Finally, S4 shows the bound conformation of benzamidine and its strong interaction with D189 (24). Additionally, the crystal waters reported with the original structure in the binding pocket were reproduced by the binding simulations (a video of water molecules in the binding pocket is available in Movie S2).
Fig. 3.
Identification of metastable states. (A) PMF in the xz plane. Five different metastable states can be identified from the different free-energy minima (S0 to S4). The relative free energy between the unbound state S0 and the bound state S4 is -6 kcal/mol. The most probable transition to the bound state S4 may be from S3 from the fact that the barrier between the two states is just 1.5 kcal/mol. (B) Structural characterization of metastable states. In states S1 and S2, benzamidine is stabilized by π-π stacking interactions with Y151 and Y39 side chains. In S3, a hydrogen bond may be formed between NH2 groups of benzamidine (only heavy atoms shown for clarity) and Q175 side chain, or by a cation-π interaction between the Q175 side chain again, and benzamidine’s benzene ring.
Characterization of Binding Pathway.
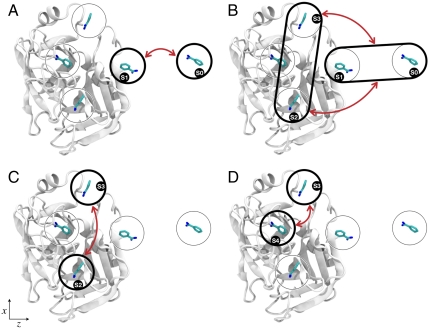
The two-dimensional PMF is further coarse-grained into five states using a Voronoi tessellation, with the centroids selected to represent the different metastable states of the system. This simple model of the process provides a direct interpretation of the metastable binding sites through analysis of the structure of the eigenvectors of the five-state MSM (available in Fig. S3), which describe the characteristic modes between the states as well as their implied time scales. Considering the slowest modes, we see transitions from site S0 to S1 (Fig. 4A) and collectively from sites S0/S1 to sites S3/S4 (Fig. 4B) on an implied time scale of between 6 and 10 ns, corresponding to the diffusion of the ligand from bulk to the first structural contact with the protein. With an implied time scale of ≈20 ns, the model shows the transitions between sites S2 and S3 (Fig. 4C). Therefore, the ligand is more likely to diffuse back and bind to the S3 site rather than directly transit from S2 to the binding pocket. Site S2 is a secondary binding pocket but not directly involved in the binding pathway. Finally, the rate-limiting step of the process is the transition to the bound site S4 (Fig. 4D), which is seen to happen at an implied time scale of around 60 ns in the approximate five-state model, and preferentially coming from S3 (the complete probability transition matrix is available in Table S1). This is consistent with the energetics in Fig. 3A, because the energetic barrier between the two states is only 1.5 kcal/mol high, lower than that to any other metastable state.
Fig. 4.
Characteristic transition modes for the metastable-states model. Each mode corresponds to a left eigenvector of the transition matrix in the five-state coarse-grained model. Considering the slowest modes, on an “implied time scale” (τ∗) between 6 and 10 ns we have transitions from state S0 to S1 (A) and collectively from states S0/S1 to states S3/S4 (B). With τ∗ ≈ 20 ns, the model shows the transitions between states S2 and S3 (C). Transitions from lower energy states take longer times on average. The rate-limiting step of the process is the transition to the bound state S4, which is seen to happen at τ∗ ≈ 60 ns and preferentially from state S3 (D).
To further understand the rate-limiting transition S3–S4, we inspected the trajectories that underwent binding to define a consensus pathway. As shown in Fig. 5A, benzamidine appears to “roll” over the surface of trypsin in its rate-limiting transition where three transition states are identified. TS1, TS2, and TS3 are found to be three path-defining interactions along which most of the binding trajectories transit. In higher detail, Fig. 5B displays TS1 as a single hydrogen bond of benzamidine with N89 main chain. TS2 is a stronger hydrogen-bond interaction with S210 and H57, the latter belonging to trypsin’s catalytic triad with S195 and N102. The last transition state identified prior to binding is TS3 corresponding to a hydrogen bond formed with the S214 side chain. Not all binding trajectories follow sequentially the transition through TS1, TS2, and TS3. Nonetheless, the vast majority bind passing through TS2 and TS3 or directly to TS3 before burying into the pocket.
Fig. 5.
Rate-limiting transition. (A) Transition between states S3–S4 often occurs as pictured. Benzamidine rolls over trypsin’s surface by a set of successive transient interactions (transition states) TS1, TS2, and TS3 that canalizes the penetration of the inhibitor to its binding pocket. (B) Transition states are defined by the hydrogen-bond interactions formed between benzamidine and N89 main chain (TS1), with H57 main chain and S210 side chain (TS2), and finally with S214 main chain (TS3). Far from all binding trajectories following exactly this path; some trajectories bind passing only from TS2 and TS3, or directly to TS3, prior to burying into the binding pocket at the known bound conformation. As a side note, H57 belongs to the trypsin catalytic triad, responsible for the peptide bond cleavage activity of serine proteases.
Estimation of Energetics and Kinetics.
In order to provide a more precise quantitative interpretation of binding, we compute both the binding affinity and the kinetics of the overall process of binding. For this, a higher resolution three-dimensional MSM model was built with 9,700 states after clustering the coordinates into 18 × 18 × 30 bins with a volume of 36.33 Å3 each. Using the new model, it was possible to isolate with higher precision the binding site by making a single binary distinction of the conformations into bound and unbound. After a sensitivity analysis in the three-dimensional PMF (Fig. S4), we defined the bound conformation as those states with GPMF ≤ 3 kcal/mol and the unbound as GPMF > 3 kcal/mol.
The standard free energy of binding (ΔG°) is calculated using the following expression (derivation available in SI Text S1):
 |
where ΔW3D is the depth of the PMF, kB is the Boltzmann constant, T is the temperature, Vb is the bound volume calculated as the integral of the PMF, and V° is the standard-state volume. The three-dimensional PMF (shown in Fig. S5) yields a standard free energy of binding of ΔG° = -5.2 ± 0.4 kcal/mol, where the error is calculated by block averaging over five sets of data of 10 μs each. The value of Vb (defined in SI Text S1) is constant for large enough integration domains (Fig. S6). Our calculated free energy differs by 1 kcal/mol from the experimentally determined value of -6.2 kcal/mol (25). Previous computational attempts to determine trypsin-benzamidine standard free-energy of binding reported the ample range of -5.5 to -9.0 kcal/mol via pathway-based free-energy methods (22), whereas ref. 23 recovered a PMF depth similar to ours, although obtained in a different setting and without reporting the standard free energy of binding.
In order to compare the reconstructed binding pathway with experimental kinetics data, we mapped the binding process onto a simple two-state model for the reaction of an inhibitor (I) with an enzyme (E), i.e.,
 |
where, assuming first-order kinetics, kon and koff directly relate to the mean times of binding and unbinding, respectively (26, 27). In a binary view of the states of the system as having bound and unbound conformations, computing the mean first passage time (MFPT) for the on and off reactions would directly give us the constant rates for the reactions as
where kon, measured in units of M-1 s-1, is inversely dependent on the ligand concentration, Ccomp = 0.015 M. The calculated mean times for the process are MFPTon = 444 ± 59 ns and MFPToff = (1.17 ± 0.43) × 104 ns. The derived kinetic parameters for the binding process therefore are kon = (1.5 ± 0.2) × 108 M-1 s-1 and koff = (9.5 ± 3.3) × 104 s-1. Both results deviate from the experimental reference values (28) by at least an order of magnitude, with a higher error on koff ( and
and  ). A larger uncertainty is coupled to the precision of koff because a larger extrapolation is done by inferring long-timed events from various short-time transitions. The complete unbinding rate is recovered by the MSM from the probability of transitions moving up the binding well in the PMF. So, although it is not necessary to produce a full unbinding event, the error in the estimation of the transitions may be higher in the off direction rather than on (29). For higher accuracy in the computation of the kinetic parameters, a better sampling of the slowest mode of the MSM might improve the estimations. Methodologies that increase the unbiased sampling on the states involved in the slowest modes could be used to improve the efficiency of sampling and possibly yield more accurate estimation of quantitative parameters (30, 31). Finally, it could also be that these results are simply within the limitations of the force field employed, as our results are in agreement with previous estimations (32).
). A larger uncertainty is coupled to the precision of koff because a larger extrapolation is done by inferring long-timed events from various short-time transitions. The complete unbinding rate is recovered by the MSM from the probability of transitions moving up the binding well in the PMF. So, although it is not necessary to produce a full unbinding event, the error in the estimation of the transitions may be higher in the off direction rather than on (29). For higher accuracy in the computation of the kinetic parameters, a better sampling of the slowest mode of the MSM might improve the estimations. Methodologies that increase the unbiased sampling on the states involved in the slowest modes could be used to improve the efficiency of sampling and possibly yield more accurate estimation of quantitative parameters (30, 31). Finally, it could also be that these results are simply within the limitations of the force field employed, as our results are in agreement with previous estimations (32).
Conclusions
This study proposes an approach to reconstruct a complete enzyme-inhibitor binding process from free ligand-binding MD simulations. The quantitative reconstruction is achieved through the construction of Markov state models of the process. In 187 of the trajectories (37% of total), benzamidine was found to bind with rmsd less than 2 Å from the crystallographic pose. Investigation of the energetics for the interaction revealed two additional metastable states of the complex, S2 and S3, in addition to the bound state S4 (Fig. 3A). Structural characterization of these states revealed persistent interactions with key residues. Interestingly, transitions to the bound state S4 tend to come from the metastable binding site S3 and not from the bulk. Further investigation of the transition revealed how benzamidine “rolls” over the surface of trypsin. Mechanistically, this is explained by the existence of three representative transition states (TS1, TS2, and TS3) along this transition corresponding to hydrogen-bond interactions between amidine moiety of benzamidine and trypsin residues. In particular, TS2 and TS3 characterized as interactions with catalytic triad residue H57 and S210 for TS2 and S214 for TS3. We propose that putative mutations to the transition-state residues may alter the binding kinetics of benzamidine. Further examples and validation of this approach need to be carried out comparing the binding of several ligands to several proteins (33). We plan to validate and improve this methodology in the future to extend its applicability, also taking advantage of improved computational capabilities. In a broader sense, the present approach should be useful in studying the binding pathways of complexes of similar size. According to our results, 20% of the data generated in the current study could be sufficient to reconstruct similar protein–ligand binding processes. Surely, not all cases would be amenable to this methodology—larger, flexible ligands and proteins would present more complex binding processes requiring higher computational effort. We do not think that this approach is that relevant for high-throughput screening for several reasons. First, the amount of calculations required for a single compound is not scalable to thousands or millions of ligands, and second, because the degree of detail provided by the methodology is probably not needed during the screening phase. Instead, we envisage a successful application in the lead optimization phase on possible tens of lead compounds to completely reconstruct and understand their binding pathways before moving forward in the pipeline. The method, although computationally expensive, is certainly feasible if performed on just a few ligands as indicated. Here, we have made use of our volunteer GPU project GPUGRID.net (7) based on ACEMD (6). Extrapolating from the current study, it would be possible to reconstruct the binding pathways of 5 to 10 ligands per week in-house on a moderately sized GPU cluster of 32–64 GPUs. This level of complexity should be already within the reach of many interested in using the methodology, whereas future hardware, software, and improved protocols could further reduce the computational costs.
Materials and Methods
System Preparation.
The input model was based on the crystallographic structure of the inhibitor benzamidine binding to the enzyme bovine trypsin (PDB ID code 3PTB, Fig. 1A) (24). The protein was modeled using the AMBER 99SB force field (34) and the ligand with the general amber force field (GAFF) force field (35) as described by Doudou et al. (22). The complex was rotated to align the vector defined by the C7 atom of benzamidine and the Cγ atom of ASP 189 to the z axis of the box. The ligand was placed in the bulk at z = 35 Å from the binding pocket, on the z axis (see Fig. 1B). The complex was then solvated with 10604 TIP3P water molecules (36) and neutralized with the addition of 9 Cl- ions. The setup resulted in a system of 35,060 atoms in a simulation box of initial dimensions 71 × 64 × 82 Å3.
The system was minimized and equilibrated under constant pressure and temperature (NPT) conditions at 1 atm and 298 K using a time step of 2 fs, nonbonded cutoff of 9 Å, rigid bonds and particle-mesh Ewald long-range electrostatics with a grid of 72 × 64 × 88. During minimization and equilibration, the heavy protein atoms were restrained by a 10 kcal mol-1 Å-2 spring constant. Two rounds of velocity reinitialization and 2-ps simulation were performed under constant volume and temperature (NVT) conditions. The magnitude of the restraining spring constant was then reduced to 1 kcal mol-1 Å-2 during 10 ps of NVT before the barostat was switched on at 1 atm for a further 10 ps of NPT simulation. A final 40 ps of NPT simulation was conducted with a restraint constant of 0.05 kcal mol-1 Å-2. Finally, the volume was allowed to relax for 10 ns under NPT conditions reaching a final box size of 69.7 × 63.2 × 80.2 Å3. During this run, Cα atoms of the complex were restrained with a 1 kcal mol-1 Å-2 harmonic potential to prevent the system reorienting. The equilibration run was performed using NAMD2.6 (37) on a standard CPU cluster. All production runs using ACEMD (6) on the distributed computing project GPUGRID (7).
Free-Binding Production Simulations.
Five hundred different conformations were generated by sampling from a preproduction run of 50 ns. The ligand was kept at a minimum distance of 20 Å from the protein and allowed to diffuse in the xy plane only. All production runs were carried in the NVT ensemble. A longer time step of 4 fs was used thanks to the use of the hydrogen mass repartitioning scheme (38) implemented in ACEMD. Temperature, nonbonded forces cutoff, and PME grid size were kept the same as in the equilibration phase. In order to keep the protein orientation fixed and to avoid the benzamidine diffusing away, two restraint schemes were applied. First, an harmonic restraint of k = 1 kcal mol-1 Å-2 was applied on Cα atoms of the protein farther than 9 Å from the binding pocket. Second, a flat-bottomed harmonic restraint was applied to the C7 atom of benzamidine with box size b = (30,30,80) centered around the binding pocket and restraint potential of 0.1 kcal mol-1 Å-2. Five hundred production runs were submitted to GPUGRID. Upon data retrieval and analysis, 495 trajectories had reached the target of 100 ns of simulated time. The completed 495 trajectories (approximately 50 μs total aggregate data) constitute the total data used for trypsin-benzamidine binding analysis.
Markov State Models.
MSMs are useful to describe the dynamics of a system as a sequence of transitions between coarse-grained states (16). We obtained a dynamic description of the binding process, defining the states according to the ligand-protein geometry. Two- and three-dimensional reaction coordinates were defined from the Cartesian components of the position of the C7 atom in benzamidine, assuming its bound position as the origin of the axes. The values of the coordinates were discretized in a grid of states by rounding them to the closest multiple of a chosen bin size. The complete MSM model is then built and analyzed using the methods highlighted in SI Text S1 (16, 26, 39–41).
Supplementary Material
Acknowledgments.
We thank Dr. R. H. Henchman for his help revising the PMF derivation and also Matthew J. Harvey, Dr. S. Kashif Sadiq, and all the other colleagues who carefully revised the manuscript. We finally thank all the volunteers of GPUGRID who donated GPU computing time to the project. I.B. acknowledges support from the Fundació La Marató de TV3. T.G. acknowledges support from the Beatriu de Pinós programme of the Generalitat de Catalunya. Work was partially supported by the Virtual Physiological Human Network of Excellence. G.D.F. acknowledges support from the Ramón y Cajal scheme and support by the Spanish Ministry of Science and Innovation (Ref. FIS2008-01040).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1103547108/-/DCSupplemental.
References
- 1.Bleicher KH, Bohm H, Muller K, Alanine AI. Hit and lead generation: Beyond high-throughput screening. Nat Rev Drug Discov. 2003;2:369–378. doi: 10.1038/nrd1086. [DOI] [PubMed] [Google Scholar]
- 2.Jorgensen WL. The many roles of computation in drug discovery. Science. 2004;303:1813–1818. doi: 10.1126/science.1096361. [DOI] [PubMed] [Google Scholar]
- 3.Swinney DC. Biochemical mechanisms of drug action: What does it take for success? Nat Rev Drug Discov. 2004;3:801–808. doi: 10.1038/nrd1500. [DOI] [PubMed] [Google Scholar]
- 4.Swinney DC. The role of binding kinetics in therapeutically useful drug action. Curr Opin Drug Discov Devel. 2009;12:31–39. [PubMed] [Google Scholar]
- 5.Rich RL, Myszka DG. Why you should be using more SPR biosensor technology. Drug Discov Today. 2004;1:301–308. doi: 10.1016/j.ddtec.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 6.Harvey MJ, Giupponi G, De Fabritiis G. ACEMD: Accelerating biomolecular dynamics in the microsecond time scale. J Chem Theory Comput. 2009;5:1632–1639. doi: 10.1021/ct9000685. [DOI] [PubMed] [Google Scholar]
- 7.Buch I, Harvey MJ, Giorgino T, Anderson DP, De Fabritiis G. High-throughput all-atom molecular dynamics simulations using distributed computing. J Chem Inf Model. 2010;50:397–403. doi: 10.1021/ci900455r. [DOI] [PubMed] [Google Scholar]
- 8.Wu C, Biancalana M, Koide S, Shea J. Binding modes of thioflavin-T to the single-layer β-sheet of the peptide self-assembly mimics. J Mol Biol. 2009;394:627–633. doi: 10.1016/j.jmb.2009.09.056. [DOI] [PubMed] [Google Scholar]
- 9.Biancalana M, Makabe K, Koide A, Koide S. Molecular mechanism of thioflavin-T binding to the surface of β-rich peptide self-assemblies. J Mol Biol. 2009;385:1052–1063. doi: 10.1016/j.jmb.2008.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brannigan G, LeBard DN, Hénin J, Eckenhoff RG, Klein ML. Multiple binding sites for the general anesthetic isoflurane identified in the nicotinic acetylcholine receptor transmembrane domain. Proc Natl Acad Sci USA. 2010;107:14122–14127. doi: 10.1073/pnas.1008534107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vanni S, Neri M, Tavernelli I, Rothlisberger U. Predicting novel binding modes of agonists to β adrenergic receptors using All-Atom molecular dynamics simulations. PLoS Comput Biol. 2011;7:e1001053. doi: 10.1371/journal.pcbi.1001053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Selent J, Sanz F, Pastor M, Fabritiis GD. Induced effects of sodium ions on dopaminergic G-protein coupled receptors. PLoS Comput Biol. 2010;6:e1000884. doi: 10.1371/journal.pcbi.1000884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ahmad M, Gu W, Helms V. Mechanism of fast peptide recognition by SH3 domains. Angew Chem Int Ed Engl. 2008;47:7626–7630. doi: 10.1002/anie.200801856. [DOI] [PubMed] [Google Scholar]
- 14.Martí MA, Lebrero MCG, Roitberg AE, Estrin DA. Bond or cage effect: How nitrophorins transport and release nitric oxide. J Am Chem Soc. 2008;130:1611–1618. doi: 10.1021/ja075565a. [DOI] [PubMed] [Google Scholar]
- 15.Enkavi G, Tajkhorshid E. Simulation of spontaneous substrate binding revealing the binding pathway and mechanism and initial conformational response of GlpT. Biochemistry. 2010;49:1105–1114. doi: 10.1021/bi901412a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Noé F, Fischer S. Transition networks for modeling the kinetics of conformational change in macromolecules. Curr Opin Struct Biol. 2008;18:154–162. doi: 10.1016/j.sbi.2008.01.008. [DOI] [PubMed] [Google Scholar]
- 17.Warshel A, Sussman F, Hwang J. Evaluation of catalytic free energies in genetically modified proteins. J Mol Biol. 1988;201:139–159. doi: 10.1016/0022-2836(88)90445-7. [DOI] [PubMed] [Google Scholar]
- 18.Li X, He X, Wang B, Merz K. Conformational variability of benzamidinium-based inhibitors. J Am Chem Soc. 2009;131:7742–7754. doi: 10.1021/ja9010833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guvench O, Price DJ, Brooks CL. Receptor rigidity and ligand mobility in trypsin-ligand complexes. Proteins. 2005;58:407–417. doi: 10.1002/prot.20326. [DOI] [PubMed] [Google Scholar]
- 20.Essex JW, Severance DL, Tirado-Rives J, Jorgensen WL. Monte Carlo simulations for proteins: Binding affinities for trypsin-benzamidine complexes via free-energy perturbations. J Phys Chem B. 1997;101:9663–9669. [Google Scholar]
- 21.Resat H, Marrone TJ, McCammon J. Enzyme-inhibitor association thermodynamics: Explicit and continuum solvent studies. Biophys J. 1997;72:522–532. doi: 10.1016/s0006-3495(97)78692-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doudou S, Burton NA, Henchman RH. Standard free energy of binding from a one-dimensional potential of mean force. J Chem Theory Comput. 2009;5:909–918. doi: 10.1021/ct8002354. [DOI] [PubMed] [Google Scholar]
- 23.Gervasio FL, Laio A, Parrinello M. Flexible docking in solution using metadynamics. J Am Chem Soc. 2005;127:2600–2607. doi: 10.1021/ja0445950. [DOI] [PubMed] [Google Scholar]
- 24.Marquart M, Walter J, Deisenhofer J, Bode W, Huber R. The geometry of the reactive site and of the peptide groups in trypsin, trypsinogen and its complexes with inhibitors. Acta Crystallogr B. 1983;39:480–490. [Google Scholar]
- 25.Mares-Guia M, Shaw E. Studies on the active center of trypsin. J Biol Chem. 1965;240:1579–1585. [PubMed] [Google Scholar]
- 26.Singhal N, Snow CD, Pande VS. Using path sampling to build better Markovian state models: Predicting the folding rate and mechanism of a tryptophan zipper beta hairpin. J Chem Phys. 2004;121:415–425. doi: 10.1063/1.1738647. [DOI] [PubMed] [Google Scholar]
- 27.Huang D, Caflisch A. The free energy landscape of small molecule unbinding. PLoS Comput Biol. 2011;7(2) doi: 10.1371/journal.pcbi.1002002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guillain F, Thusius D. Use of proflavine as an indicator in temperature-jump studies of the binding of a competitive inhibitor to trypsin. J Am Chem Soc. 1970;92:5534–5536. doi: 10.1021/ja00721a051. [DOI] [PubMed] [Google Scholar]
- 29.Prinz J, et al. Markov models of molecular kinetics: Generation and validation. J Chem Phys. 2011;134:174105. doi: 10.1063/1.3565032. [DOI] [PubMed] [Google Scholar]
- 30.Bowman GR, Ensign DL, Pande VS. Enhanced modeling via network theory: Adaptive sampling of markov state models. J Chem Theory Comput. 2010;6:787–794. doi: 10.1021/ct900620b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Voelz VA, Bowman GR, Beauchamp K, Pande VS. Molecular simulation of ab initio protein folding for a millisecond folder NTL9(1-39) J Am Chem Soc. 2010;132:1526–1528. doi: 10.1021/ja9090353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deng Y, Roux B. Computations of standard binding free energies with molecular dynamics simulations. J Phys Chem B. 2009;113:2234–2246. doi: 10.1021/jp807701h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singh N, Warshel A. Absolute binding free energy calculations: On the accuracy of computational scoring of protein-ligand interactions. Proteins. 2010;78:1705–1723. doi: 10.1002/prot.22687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hornak V, et al. Comparison of multiple amber force fields and development of improved protein backbone parameters. Proteins. 2006;65:712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA. Development and testing of a general amber force field. J Comput Chem. 2004;25:1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 36.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 37.Phillips JC, et al. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Feenstra KA, Hess B, Berendsen HJC. Improving efficiency of large time-scale molecular dynamics simulations of hydrogen-rich systems. J Comput Chem. 1999;20:786–798. doi: 10.1002/(SICI)1096-987X(199906)20:8<786::AID-JCC5>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 39.Noé F, Schütte C, Vanden-Eijnden E, Reich L, Weikl TR. Constructing the equilibrium ensemble of folding pathways from short off-equilibrium simulations. Proc Natl Acad Sci USA. 2009;106:19011–19016. doi: 10.1073/pnas.0905466106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Noé F, Horenko I, Schütte C, Smith JC. Hierarchical analysis of conformational dynamics in biomolecules: Transition networks of metastable states. J Chem Phys. 2007;126:155102. doi: 10.1063/1.2714539. [DOI] [PubMed] [Google Scholar]
- 41.Bowman GR, Beauchamp KA, Boxer G, Pande VS. Progress and challenges in the automated construction of markov state models for full protein systems. J Chem Phys. 2009;131:124101. doi: 10.1063/1.3216567. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.