Abstract
MicroRNA 21 (miR-21) is overexpressed in virtually all types of carcinomas and various types of hematological malignancies. To determine whether miR-21 promotes tumor development in vivo, we knocked out the miR-21 allele in mice. In response to the 7,12-dimethylbenz[a]anthracene (DMBA)/12-O-tetradecanoylphorbol-13-acetate mouse skin carcinogenesis protocol, miR-21-null mice showed a significant reduction in papilloma formation compared with wild-type mice. We revealed that cellular apoptosis was elevated and cell proliferation was decreased in mice deficient of miR-21 compared to wild-type animals. In addition, we found that a large number of validated or predicted miR-21 target genes were up-regulated in miR-21-null keratinocytes, which are precursor cells to skin papillomas. Specifically, up-regulation of Spry1, Pten, and Pdcd4 when miR-21 was ablated coincided with reduced phosphorylation of ERK, AKT, and JNK, three major downstream effectors of Ras activation that plays a predominant role in DMBA-initiated skin carcinogenesis. These results provide in vivo evidence that miR-21 exerts its oncogenic function through negatively regulating its target genes.
Keywords: microRNA, chemical-induced carcinogenesis, Ras effector pathways
The multistage murine skin carcinogenesis model has been a paradigm for studying epithelial tumors for over 60 years. In this model, mice are first treated with a single dose of a genotoxic carcinogen (initiator, such as 7,12-dimethylbenz[a]anthracene [DMBA]), followed by multiple applications of a nongenotoxic tumor promoter, usually a phorbol ester such as 12-O-tetradecanoylphorbol-13-acetate (TPA). This gradually results in the generation of benign squamous papillomas that develop progressively into dysplastic foci and finally become invasive squamous carcinomas. Both the carcinogen and the promoter are required for completion of the carcinogenic process in this model. This chemical-induced skin carcinogenesis recapitulates many aspects of the development of human carcinoma, particularly squamous cell carcinoma. Numerous protein-coding genes (1) play critical roles in this process, yet the role of noncoding RNAs has been overlooked.
MicroRNAs (miRNAs) are short 20–25 nucleotide RNA molecules that negatively regulate gene expression by targeting the 3′ UTRs of mRNAs. Over 1,000 human miRNAs (2–4) have been identified. Computational methods have indicated that up to 92% of human genes may be regulated by miRNAs (5). Experimentally, however, the physiological function of only a small number of these miRNAs has been revealed. The pathological involvement of miRNAs in skin carcinogenesis has yet to be explored. MicroRNA 21 (miR-21) is overexpressed in carcinomas of lung, prostate, breast, pancreas, colon, head and neck, stomach, esophagus, liver, etc., compared to normal adjacent tissues, and it is arguably the only miRNA that is up-regulated in all types of carcinomas (6, 7). In addition, it is also up-regulated in blood cancers such as leukemia (8), lymphoma (9), and multiple myeloma (10), supporting miR-21 as a ubiquitous oncogene.
Using in vitro assays, the oncogenic properties of miR-21 have been extensively studied with molecular and cellular assays in various cell lines (11–15). Recently, two reports from the laboratories of Slack (16) and Olson (17) revealed that overexpression of miR-21 leads to a pre-B malignant lymphoid-like phenotype (16) and enhances Kras-mediated lung tumorigenesis (17), whereas genetic deletion of miR-21 partially protects against tumorigenesis (17). However, in the miR-21 knockout model, it was found that deletion of the miR-21 locus has little impact on the expression of miR-21 target genes in lung (17) or in heart (18); i.e., few miR-21 targets are up-regulated in miR-21-null mice. In addition, reduced cellular apoptosis is observed in both miR-21 transgenic models (16, 17), yet deletion of miR-21 does not cause significant change in cellular apoptosis in lung tissues and tumors (17). The authors speculate that compensation by other biological inputs to normalize target regulation in the absence of miR-21, rather than increased levels of miR-21 targets, results in suppressed lung tumor development in mice without miR-21 (17).
In this study, we report that miR-21 deficiency leads to up-regulation of its target genes in epidermal keratinocytes and reduces tumorigenesis in a murine skin carcinogenesis model. Data generated from this work allow us to reconcile target efficacy with the phenotype, i.e., reduced tumorigenesis of miR-21-null mice, providing added evidence to support that miR-21 is an oncogene that suppresses the expression of its target genes to exert its oncogenic function.
Results
Gene Targeting of miR-21 in Mice.
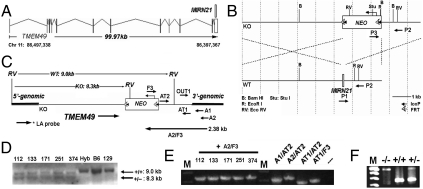
The mouse miR-21 allele is located in the 3′UTR of a protein-coding gene TMEM49 (Fig. 1A). To delete the miR-21 allele in mouse ES cells, we generated a vector that deletes the precursor to miR-21 by replacing it with a neomycin (NEO)-resistance expression cassette (Fig. 1B). Correctly targeted ES colonies were selected by the presence of NEO, the shortened long arm, and the deletion-specific PCR products (Fig. 1 C–E, SI Text, and Table S1). One ES clone was used to successfully generate germ line transmission by blastocyst injection. Mice heterozygous or homozygous for the miR-21 deletion allele (Fig. 1F) were fertile and appeared phenotypically normal, similar to a previous report (17). Also we found the expression of TMEM49 is unaffected by miR-21 loss and the inserted NEO cassette as we did not observe any significant difference in the levels of TMEM49 in tissues from skin, heart, lung, and pancreas. There were no significant differences in major organs between the wild-type and miR-21-null mice of up to one and half years old.
Fig. 1.
Conventional knockout of the miR-21 allele. (A) The mouse gene structure of TMEM49 ∼ MIRN21. miR-21 is located in the 3′UTR region of the TMEM49 transcript. (B) The construct for miR-21 gene targeting. The neomycin gene (NEO) replaced the miR-21 gene in the TMEM49 ∼ MIRN21 transcriptional unit. (C) Strategy for confirmation of five miR-21-null positives from more than 400 ES colonies. (D) Southern blotting to confirm the long homology arm (LA, 5.4 kb) with a LA probe as in C. (E) PCR to confirm the short homology arm (SA, 2.0 kb) with A2 and F3 primers (shown in C). M (Top to Bottom): 3.0 and 2.0 kb. (F) Duplex PCR with P1, P2, P3 (shown in B) to confirm the miR-21-null mice. M ((Top to Bottom)): 500, 400, 300, and 200 bp. The founder heterozygous mice are from ES colony #374.
miR-21 Deficiency Reduces Chemical-Induced Skin Carcinogenesis.
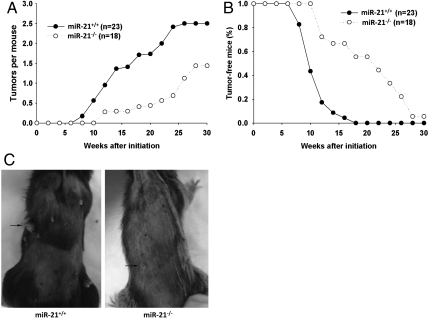
We subjected 18 miR-21-null mice along with 23 of their wild-type littermates to a murine skin carcinogenesis (DMBA/TPA) protocol with an initial one dose of DMBA and multiple doses of TPA twice weekly afterward. TPA painting was stopped at the end of the 29th week when there was no increase of papillomas in both groups of mice for two weeks. Overall, there were fewer papillomas in miR-21-null mice compared to their wild-type littermates (Fig. 2 A and C). Additionally, the percentage of mice with papillomas in miR-21-null mice was also significantly less than the control during the treatment; 100% tumor penetration was reached at week 17 for wild-type mice, yet there was still one miR-21-null animal tumor-free at the end of the regimen (Fig. 2B). These results suggest that miR-21 ablation negatively impacts chemical-induced epidermal carcinogenesis.
Fig. 2.
Fewer papillomas developed in miR-21-null mice upon the DMBA/TPA regimen. (A) Papilloma multiplicity of miR-21-null and wild-type mice. P = 0.004 with Mann–Whitney U test. (B) Papilloma incidence, i.e., the percentage of mice with at least one papilloma. P = 0.008 with Fisher’s exact test. (C) A representative photo of papilloma formation in wild-type and miR-21-null mice. Arrows point to one papilloma in each animal.
Reduced Cell Proliferation and Elevated Cellular Apoptosis in Keratinocytes and in the Epidermis of miR-21-Null Mice.
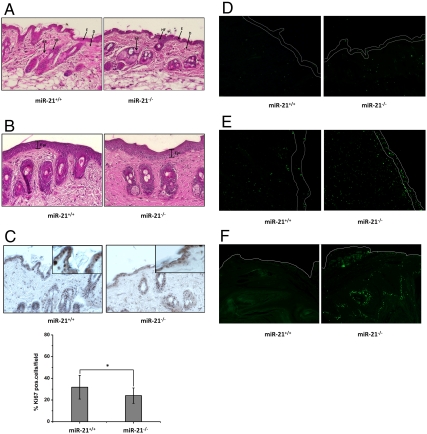
We first analyzed the naive dorsal skin of wild-type and miR-21-null mice. They exhibited normal overall epidermal morphology and thickness. Sebaceous glands and hair follicles also appeared to be normal (Fig. 3A). When treated with three doses of TPA (without DMBA), which is known to induce a robust proliferative response leading to substantial epidermal hyperplasia, both wild-type and miR-21-null skin developed comparable epidermal thickening (Fig. 3B), suggesting miR-21 is not required for TPA-induced epidermal hyperplasia. When skin sections were staining for Ki-67, a cellular marker for proliferation, there was a small yet statistically significant reduction (approximately 8%) of Ki-67-positive cells in the epidermis of miR-21-null mice compared to the control (Fig. 3C), supporting a role of miR-21 in cell proliferation.
Fig. 3.
Comparison of skin epidermis of wild-type and miR-21-null mice. Magnification, 20×. (A) H&E staining of naive skin. E, epidermis; D, dermis; HF, hair follicle; SB, sebaceous gland. (B) Hematoxylin and eosin (H&E) staining of skin treated with three doses of TPA. Thickness of the epidermis (Epi) is indicated. (C) Ki-67 staining of naive skin. (Insets) Area of higher magnification (40×). Bar graph below shows the percentage of cells that are Ki-67 positive in wild-type and miR-21-null epidermis. Values represent the means ± SD of three separate experiments. *P < 0.05. (D–F) TUNEL analysis. White curves indicate the edges of the epidermis. Green fluorescence indicates apoptosis-positive cells in naive skin (D), skin treated with three doses of TPA (E), and papillomas (F).
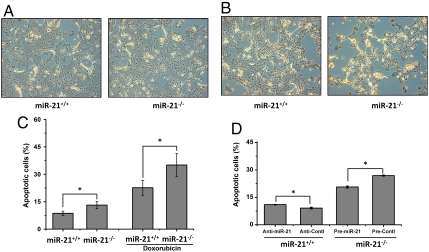
We next examined cellular apoptosis using the TdT-mediated dUTP nick end (TUNEL) assay. We found there are more apoptotic cells in miR-21-null epidermis with or without TPA treatment (Fig. 3 D and E). In addition, miR-21 loss had also increased the amount of apoptotic cells in the papillomas, suggesting that apoptosis contributes to the reduced tumorigenesis in miR-21-null mice (Fig. 3F). To validate the role of miR-21 in apoptosis, we isolated epidermal keratinocytes from skin tissues of newborn mice. miR-21-null keratinocytes displayed more apoptosis than their wild-type counterparts (Fig. 4 A and C). When induced by doxorubicin, more apoptosis was observed in cells without miR-21 (Fig. 4 B and C). Next, we transfected wild-type keratinocytes with inhibitors to miR-21 (anti-miR-21) and miR-21-null cells with precursors to miR-21 (pre-miR-21) and analyzed cellular apoptosis using FACS. We found that the apoptosis rate of wild-type cells was increased when miR-21 was inhibited, whereas reintroduction of miR-21 into miR-21-null cells reduced cellular apoptosis (Fig. 4D). These results support that miR-21 is antiapoptotic both in vivo and in vitro.
Fig. 4.
Apoptosis analyses of isolated keratinocytes. (A and B) TUNEL analysis in wild-type and miR-21-null cells (A) and in cells treated with doxorubicin (B). (C) Bar graph shows the percentage of cells that are apoptosis-positive in wild-type and miR-21-null keratinocytes using TUNEL. (D) FACS analyses of wild-type cells transfected with anti-miR-21 and miR-21-null cells with pre-miR-21. Anti-Contl, Anti-miR™ miRNA Inhibitors—Negative Control No. 1; Pre-Contl, Pre-miR™ miRNA Precursor Molecules—Negative Control No. 1. Values represent the means ± SD of three separate experiments. *P < 0.05.
Increased Expression of miR-21 Target Genes in miR-21-Null Keratinocytes.
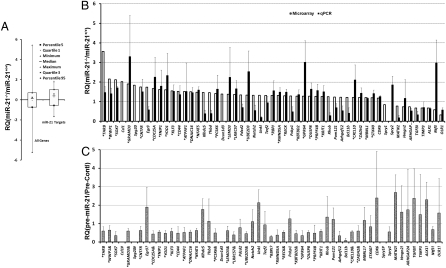
As miRNAs generally exert their function through suppression of target genes, we profiled global expression of mRNAs in keratinocytes isolated from wild-type and miR-21-null mice. We grouped genes that are differentially expressed in wild-type and miR-21-null keratinocytes and drew a Whisker-box plot to visualize the percentile values of all genes expressed in keratinocytes versus those targeted by miR-21 as predicted by TargetScan (http://targetscan.org). Overall there were 153 putative or validated miR-21 targets that were detectable in either sample. These target genes as a group were highly expressed in miR-21-null keratinocytes compared to all genes (Fig. 5A, P = 4.76E-05). To validate this finding, we used Taqman® quantitative real-time PCR (qPCR) to analyze a total of 55 selected genes, including putative miR-21 targets that are differentially expressed in wild-type or miR-21-null keratinocytes as determined by expression profiling and some validated miR-21 targets such as Pdcd4 that were not detected by microarray. Beyond Ccl1 and Spry1, all qPCR reagents were able to amplify quality products from cDNAs of two groups of keratinocytes. We found that there was approximately 65% (36 out of 55) concordance between microarray data and the qPCR data. Among them, 30 validated or predicted miR-21 targets are up-regulated in miR-21-null cells compared to the control as determined by both microarray and qPCR (Fig. 5B and Table S2). To further corroborate that these target genes are modulated by miR-21 directly, rather than some unknown compensatory indirect mechanisms, we transfected miR-21-null keratinocytes with pre-miR-21 and examined the expression of miR-21 target genes. We found that except three genes (MBNL1, STK40, and CDK6), the expression of all 30 putative or validated miR-21 targets that showed concordance between microarray data and the qPCR data was reversed (down-regulated) when miR-21 was reintroduced (Fig. 5C). This result suggests that up-regulation of most miR-21 targets in miR-21-null cells is directly due to the loss of miR-21.
Fig. 5.
Expression of miR-21 targets in wild-type and miR-21-null keratinocytes. (A) The expression levels of miR-21 targets in wild-type and miR-21-null cells as determined by the microarray assay. A Whisker-box plot is presented with 5th, 95th percentile values as well as minimum, maximum, median, and 25th and 75th percentiles. The average values of all genes is 0.071, compared to 0.35 for miR-21 targets (P = 4.8E-05). RQ, Relative quantity of mRNA levels (Y axis) is reflected by the log 2 value of signal intensity (log 2(SDmiR-21-null-SDwild type). (B) The expression of miR-21 targets as determined by the qPCR assay compared to that by the microarray assay. (C) The expression of miR-21 targets in miR-21-null cells transfected with pre-miR-21. For B and C, RQ of mRNA levels (Y axis) is reflected by the log 2 value of signal intensity (log 2(SDmiR-21-null-SDwild type) normalized to that of Gapdh. Gene name in capital denotes that there is concordance between microarray and qPCR. “*” prefix, genes are down-regulated in miR-21-null cells when introduced with pre-miR-21. “&” postfix, UTR was confirmed to be down-regulated in a reporter assay; “#”, UTR down-regulated by no qPCR data; “?” UTR not down-regulated; “!”, UTR down-regulated but expression repression by miR-21 not supported by microarray or qPCR.
Elevated Expression of miR-21 Target Genes and Inhibition of Ras Effector Pathways in miR-21-Null Cells.
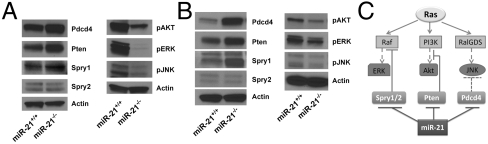
The development of papillomas induced by DMBA-TPA is dependent on mutant activation of H-Ras, which further activates the phosphorylation of ERK, AKT, and JNK. It is reported that the Raf kinase is inhibited by both Spry1 and Spry2 (19). Pten converts PIP3 into PIP2 to inhibit the PI3K-Akt cascade (20). Pdcd4 inhibits JNK activation by down-regulating MAP4K1, an upstream kinase of JNK (21). Thus, we hypothesize that up-regulation of these miR-21 target genes, namely Spry1 (22), Spry2 (23), Pten (24, 25), and Pdcd4, in keratinocytes contributed to reduced tumorigenesis in miR-21-null mice subjected to multiple-stage skin carcinogenesis. We first examined the expression of these miR-21 targets using Western blotting analyses and found the protein levels of Spry1, Pten, and Pdcd4 were increased in miR-21-null keratinocytes, whereas that of Spry2 remained unchanged (Fig. 6A, Left). We next determined the phosphorylation status of these three kinases in keratinocytes and found that all of them were phosphorylated at a lower level in miR-21-null cells compared to the control (Fig. 6A, Right). To further validate that miR-21 targets are up-regulated and Ras effector pathways are inhibited by loss of miR-21, we isolated mouse embryonic fibroblasts (MEFs) from wild-type and miR-21-null embryos. Western blotting analyses confirmed that the expression of Spry1, Pten, and Pdcd4 was up-regulated and the phosphorylation of ERK, AKT, and JNK was reduced in MEFs (Fig. 6B). Collectively, these data provide a strong link between elevated expression of miR-21 to attenuate Ras signaling and reduced skin tumorigenesis.
Fig. 6.
miR-21 target gene expression and Ras effector pathways in keratinocytes and in MEFs. (A) The expression of miR-21 targets (Pdcd4, Pten, Spry1, and Spry2) and the phosphorylation of ERK, AKT, and JNK in keratinocytes. (B) Similar to A but in MEFs. (C) A proposed model showing that miR-21 suppresses multiple targets that are inhibitory to Ras signaling.
Discussion
It is well established that Ras activation is the initiating event of multistage murine skin carcinogenesis as over 90% of DMBA/TPA-induced papillomas harbor A182 → T mutation in the H-RAS gene (26). There are three Ras effectors that play major roles in cell transformation: Raf, PI3K, and Ral-GEFs. Ras activates Raf and subsequently the ERK MAPKs through MEK. The second effector of Ras is phosphoinositide 3-kinase (PI3K), which catalyzes the conversion of PIP2 to PIP3. PIP3 recruits Akt and PDK1 to the plasma membrane where Akt is activated. A family of GDP-GTP exchange factors (GEFs) for the Ral small GTPases (RalGDS, for instance) represents the third family of Ras effectors (27). RalGDS mediates tumor cell survival through the activation of the JNK pathway (28). All three effectors are key components in Ras-dependent DMBA/TPA-induced murine carcinogenesis (28–30). In this study, we found that respective ERK, AKT, and JNK inhibitors such as Spry1, Pten, and Pdcd4 are up-regulated in miR-21-null keratinocytes. As these genes are known to be repressed by miR-21, these results provide direct evidence that miR-21 target genes are elevated to attenuate three major Ras effector pathways to inhibit tumorigenesis in miR-21-null animals (Fig. 6C). We are cognizant that other miR-21 targets may participate in pathways mediated by three major Ras effectors or a handful of other distinct Ras effectors such as Tiam1, Rassf1, and PLCϵ (27) and that Ras effector pathways are nonlinear cascades with extensive feedback inhibition and interconnections. These data nonetheless demonstrate the critical role of miR-21 in Ras signaling.
Beyond these three miR-21 target genes, we also found that a large number of other miR-21 putative and validated targets are up-regulated upon loss of the miR-21 allele. This is in line with a report on miR-223 (31), but in contrast with results from lung and heart tissues of miR-21-null mice in another study (17). Two findings are noteworthy in our analyses of miR-21 target expression. First, up-regulation of miR-21 target genes in miR-21-null cells is moderate, in accord with previous reports (31, 32). Second, we found a 65% concordance between microarray data and the qPCR data on miR-21 target expression. We believe that this is largely due to the nature of moderate differential expression in our data. When we use a cutoff of 50% differential expression, there is a better concordance between microarray and qPCR (10 out of 11 miR-21 targets). This suggests that additional experimental validation is needed before a conclusive declaration that these genes are miR-21 targets. We have tested the specificity of the 3′UTR from 27 of these putative or experimentally validated miR-21 targets and found that a reporter with the vast majority (23 out of 27) of the respective 3′UTRs is down-regulated > 20% when miR-21 is overexpressed (Fig. S1). This result indicates that most of the target genes we identified in this study are likely bona fide miR-21 targets. It is intriguing that Spry2 is up-regulated in miR-21-null lung tissues (17), but not in miR-21-null keratinocytes and MEFs. On the other hand, Spry1 and Pdcd4 are up-regulated in miR-21-null keratinocytes and MEFs, but not in lung and heart tissues (17, 18). It implies that tissue and/or cell-type specificity play a role in miR-21 targeting efficacy. It is also possible that the difference in target efficacy is due to the use of isolated cells in our study rather than whole tissue lysates that are employed in the lung tumor study (17). We note that Pdcd4 is less significantly down-regulated in miR-21-null samples when whole skin lysate is used for Western blotting analyses compared to lysate from torn epidermis (mainly keratinocytes) (Fig. S2).
Finally, we summarize and compare two transgenic and two knockout models for miR-21 (Table 1). The major difference of two transgenic models is that miR-21 overexpression driven by the Nestin promoter but not that by the CAG promoter leads to pre-B cell malignancies. This could be due to the gene dosage (> 10-fold vs. 4- to 6-fold miR-21 overexpression). There are three disparities between the miR-21 knockout model with Kras activation from Olson and colleagues (the Olson model) (17) and the present study with skin carcinogenesis. First, in the Olson model, deletion of the miR-21 locus has little impact on the expression of miR-21 target genes in the lung; in our model, we found statistically significant yet moderate up-regulation of scores of miR-21 targets. Second, in the Olson model, miR-21 loss does not cause significant change in cellular apoptosis and proliferation in lung tissues and tumors; in our model, increased apoptosis was observed in miR-21-null cells and skin tissues, along with slightly reduced proliferation, underscoring the role of miR-21 in suppressing apoptosis and promoting cell proliferation. Third, our study provides evidence to support that miR-21 promotes all three major Ras effector pathways, whereas the activation of the Raf-ERK pathway is not affected in the Olson model. Concisely, the phenotype of miR-21 knockout mice under chemical induction (reduced tumorigenesis accompanied with elevated expression of miR-21 targets, increased apoptosis, and attenuated Ras signaling) in this study is roughly an opposite to that of miR-21 transgenic animals.
Table 1.
Comparison of four genetic models for miR-21
| miR-21 genetics | Technology | Promoter | Genetic background* | Phenotype | Tumor driver gene | Changes in cell biology | miR-21 Targets | Reference |
| Transgenic | tet-off-mediated conditional overexpression | nestin promoter (active in multiple organs of adult animals) | unknown | pre-B lymphoma | miR-21 overexpression (> 10-fold) | increased cell proliferation; decreased apoptosis | ND† | 16 |
| Transgenic | global overexpression | cytomegalovirus immediate early enhancer-chicken beta-actin hybrid promoter (CAG, global) | B6C3F1 | no tumors in major organs | miR-21 overexpression (4- to 6-fold) alone did not induce tumorigenesis | ND† | Spry1, Spry2, Btg2 | 17 |
| B6C3F1x (B6x129) | when crossed with KrasLA2 mice, enhances lung tumorigenesis | mutant Kras | increased cell proliferation; decreased apoptosis; enhanced Ras signaling | Spry1, Spry2, Btg2, Pdcd4, and others | ||||
| Gene targeting | conventional knockout | NA† | B6x129 | without any gross phenotype; suppresses Kras-driven lung tumorigenesis | mutant Kras | no change in proliferation, apoptosis, or Ras signaling | Spry2 | |
| Gene targeting | conventional knockout | NA† | B6.129 | without any gross phenotype; suppresses chemical-induced skin carcinogenesis; some immune defect when fed with high-fat diet | mutant Hras | slight reduction in proliferation; increased apoptosis; attenuated Ras signaling | Spry1, Pten, Pdcd4, and others | this study |
Bold indicates the difference between two knockout models.
*B6, C57Bl/6; 129, 129/SvEv; B6C3F1, F1 of c57Bl6 x C3H/HeJ.
†ND, not determined; NA, not applicable.
miR-21 is up-regulated in virtually all types of carcinomas, and it is reported to be the most differentially up-regulated miRNA in 31 types of solid tumors (7). Illuminating its role in epidermal tumor development and the target efficacy using a mouse knockout model in the present work is critical to provide previously undescribed information on multistage carcinogenesis and to yield crucial insights regarding whether we can target miR-21 or its target genes in cancer prevention and cancer therapy. Beyond tumorigenesis, miR-21 is implicated as a negative regulator of inflammation as one of its targets Pdcd4 promotes NF-κB activation to produce proinflammation cytokine IL-6 (33). We found there is no difference in basal IL-6 levels between wild-type and miR-21 knockout mice. However, when mice were subjected to high-fat diet, IL-6 production was higher in miR-21-null mice, suggesting that miR-21 loss results in a deficiency in response to low-grade chronic inflammation (high fat). This physiological function of miR-21 is currently under investigation.
Materials and Methods
Details of gene targeting of miR-21 in mice are provided in SI Text. Female littermate animals were used in this study because they display less aggressive behavior, and repeated wounds due to fighting or irritation of the skin can act to promote skin tumor formation. The back skin of weight-matched 7-wk-old mice was shaved with surgical clippers. Two days later the mice were examined to determine whether they had entered the resting phase of hair follicles, or the telogen growth cycle, as indicated by the lack of hair growth. Mice showing no hair growth were treated with 100 nmoles of DMBA in 200 μL of acetone, painted on the skin in a chemical fume hood appropriate for handling chemical carcinogens. The technician applying the tumor initiator and promoter was blind to the genotypes of the animals. Starting 10 d after DMBA administration, twice weekly doses of 5 nmoles TPA in 200 μL of acetone were painted on the shaved area. Higher doses of DMBA/TPA were chosen (100 nmoles of DMBA and 5 nmoles TPA) because C57Bl/6(B6) × 129/SvEv (129) is a resistant strain to DMBA/TPA protocol (34–36). TPA painting was discontinued when the number of papillomas per mouse did not change over 2 wk. All experimental procedures involving animals in this study were reviewed and approved by the Institutional Animal Care and Use Committee at the University of Louisville. All other experimental details are provided in SI Text.
Supplementary Material
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1103735108/-/DCSupplemental.
References
- 1.Yuspa SH. The pathogenesis of squamous cell cancer: Lessons learned from studies of skin carcinogenesis—Thirty-third G. H. A. Clowes Memorial Award Lecture. Cancer Res. 1994;54:1178–1189. [PubMed] [Google Scholar]
- 2.Bentwich I, et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet. 2005;37:766–770. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- 3.Lagos-Quintana M, Rauhut R, Meyer J, Borkhardt A, Tuschl T. New microRNAs from mouse and human. RNA. 2003;9:175–179. doi: 10.1261/rna.2146903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miranda KC, et al. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203–1217. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 6.Liu M-F, Jiang S, Lu Z, Young KH, Li Y. Physiological and pathological functions of mammalian microRNAs. In: McQueen CA, editor. Comprehensive Toxicology. 2nd Ed. London, UK: Elsevier Science; 2010. (Cellular and Molecular Toxicology). [Google Scholar]
- 7.Volinia S, et al. Reprogramming of miRNA networks in cancer and leukemia. Genome Research. 2010;20:589–599. doi: 10.1101/gr.098046.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fulci V, et al. Quantitative technologies establish a novel microRNA profile of chronic lymphocytic leukemia. Blood. 2007;109:4944–4951. doi: 10.1182/blood-2006-12-062398. [DOI] [PubMed] [Google Scholar]
- 9.Lawrie CH, et al. Microrna expression distinguishes between germinal center B cell-like and activated B cell-like subtypes of diffuse large B cell lymphoma. Int J Cancer. 2007;121:1156–1161. doi: 10.1002/ijc.22800. [DOI] [PubMed] [Google Scholar]
- 10.Pichiorri F, et al. MicroRNAs regulate critical genes associated with multiple myeloma pathogenesis. Proc Natl Acad Sci USA. 2008;105:12885–12890. doi: 10.1073/pnas.0806202105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Asangani I, et al. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27:2128–2136. doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- 12.Frezzetti D, et al. Upregulation of miR-21 by Ras in vivo and its role in tumor growth. Oncogene. 2010;30:275–286. doi: 10.1038/onc.2010.416. [DOI] [PubMed] [Google Scholar]
- 13.Si ML, et al. miR-21-mediated tumor growth. Oncogene. 2007;26:2799–2803. doi: 10.1038/sj.onc.1210083. [DOI] [PubMed] [Google Scholar]
- 14.Talotta F, et al. An autoregulatory loop mediated by miR-21 and PDCD4 controls the AP-1 activity in RAS transformation. Oncogene. 2009;28:73–84. doi: 10.1038/onc.2008.370. [DOI] [PubMed] [Google Scholar]
- 15.Lu Z, et al. MicroRNA-21 promotes cell transformation by targeting the programmed cell death 4 gene. Oncogene. 2008;27:4373–4379. doi: 10.1038/onc.2008.72. [DOI] [PubMed] [Google Scholar]
- 16.Medina PP, Nolde M, Slack FJ. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature. 2010;467:86–90. doi: 10.1038/nature09284. [DOI] [PubMed] [Google Scholar]
- 17.Hatley ME, et al. Modulation of K-Ras-dependent lung tumorigenesis by microRNA-21. Cancer Cells. 2010;18(3):282–293. doi: 10.1016/j.ccr.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Patrick DM, et al. Stress-dependent cardiac remodeling occurs in the absence of microRNA-21 in mice. J Clin Invest. 2010;120:3912–3916. doi: 10.1172/JCI43604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gross I, Bassit B, Benezra M, Licht JD. Mammalian sprouty proteins inhibit cell growth and differentiation by preventing Ras activation. J Biol Chem. 2001;276:46460–46468. doi: 10.1074/jbc.M108234200. [DOI] [PubMed] [Google Scholar]
- 20.Stambolic V, et al. Negative regulation of PKB/Akt-dependent cell survival by the tumor suppressor PTEN. Cell. 1998;95:29–39. doi: 10.1016/s0092-8674(00)81780-8. [DOI] [PubMed] [Google Scholar]
- 21.Yang HS, et al. Tumorigenesis suppressor Pdcd4 down-regulates mitogen-activated protein kinase kinase kinase kinase 1 expression to suppress colon carcinoma cell invasion. Mol Cell Biol. 2006;26:1297–1306. doi: 10.1128/MCB.26.4.1297-1306.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thum T, et al. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature. 2008;456:980–984. doi: 10.1038/nature07511. [DOI] [PubMed] [Google Scholar]
- 23.Sayed D, et al. MicroRNA-21 targets sprouty2 and promotes cellular outgrowths. Mol Biol Cell. 2008;19:3272–3282. doi: 10.1091/mbc.E08-02-0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meng F, et al. Involvement of human micro-RNA in growth and response to chemotherapy in human cholangiocarcinoma cell lines. Gastroenterology. 2006;130:2113–2129. doi: 10.1053/j.gastro.2006.02.057. [DOI] [PubMed] [Google Scholar]
- 25.Meng F, et al. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–658. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quintanilla M, Brown K, Ramsden M, Balmain A. Carcinogen-specific mutation and amplification of Ha-ras during mouse skin carcinogenesis. Nature. 1986;322:78–80. doi: 10.1038/322078a0. [DOI] [PubMed] [Google Scholar]
- 27.Malumbres M, Barbacid M. RAS oncogenes: The first 30 years. Nat Rev Cancer. 2003;3(6):459–465. doi: 10.1038/nrc1097. [DOI] [PubMed] [Google Scholar]
- 28.Gonzalez-Garcia A, et al. RalGDS is required for tumor formation in a model of skin carcinogenesis. Cancer Cells. 2005;7:219–226. doi: 10.1016/j.ccr.2005.01.029. [DOI] [PubMed] [Google Scholar]
- 29.Ehrenreiter K, et al. Raf-1 addiction in Ras-induced skin carcinogenesis. Cancer Cells. 2009;16:149–160. doi: 10.1016/j.ccr.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 30.Gupta S, et al. Binding of ras to phosphoinositide 3-kinase p110alpha is required for ras-driven tumorigenesis in mice. Cell. 2007;129:957–968. doi: 10.1016/j.cell.2007.03.051. [DOI] [PubMed] [Google Scholar]
- 31.Baek D, et al. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sheedy FJ, et al. Negative regulation of TLR4 via targeting of the proinflammatory tumor suppressor PDCD4 by the microRNA miR-21. Nat Immunol. 2010;11:141–147. doi: 10.1038/ni.1828. [DOI] [PubMed] [Google Scholar]
- 34.Reiners JJ, Jr, Singh KP. Susceptibility of 129/SvEv mice in two-stage carcinogenesis protocols to 12-O-tetradecanoylphorbol-13-acetate promotion. Carcinogenesis. 1997;18:593–597. doi: 10.1093/carcin/18.3.593. [DOI] [PubMed] [Google Scholar]
- 35.Jansen AP, Camalier CE, Colburn NH. Epidermal expression of the translation inhibitor programmed cell death 4 suppresses tumorigenesis. Cancer Res. 2005;65:6034–6041. doi: 10.1158/0008-5472.CAN-04-2119. [DOI] [PubMed] [Google Scholar]
- 36.Segrelles C, et al. Deregulated activity of Akt in epithelial basal cells induces spontaneous tumors and heightened sensitivity to skin carcinogenesis. Cancer Res. 2007;67:10879–10888. doi: 10.1158/0008-5472.CAN-07-2564. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.