Abstract
Altering the expression level of proteins that are subunits of complexes has been proposed to be particularly detrimental because the resulting stoichiometric imbalance among components would lead to misassembly of the complex. Here we show that assembly of the phage HK97 connector complex is severely inhibited by the overexpression of one of its component proteins, gp6. However, this effect is a result of the unusual mechanism by which the oligomerization and assembly of gp6 are controlled. Alteration of this mechanism by single amino acid substitutions leads to a reversal of the response to gp6 overexpression. Surprisingly, the binding partner of gp6 within the phage particle is expressed at a 500-fold higher concentration despite their identical stoichiometry within the complex. Our data emphasize that a generalized prediction of the effects of changes in the expression level of protein complex subunits is very difficult because these effects are dependent upon assembly mechanism.
Keywords: macromolecular assembly, protein overexpression
Most biological processes are mediated by multicomponent protein complexes. Although tremendous effort has been devoted to defining the composition of protein complexes in a variety of organisms (1–4), our understanding of the regulation of the assembly of these complexes and the role of gene expression levels in governing this process is limited. It has been argued that the concentrations of proteins found in the same complex must be “balanced” to prevent accumulation of off-pathway intermediates, which might disrupt assembly (5–7). Thus, genes encoding proteins found in complexes are expected to more frequently display “dosage sensitivity,” meaning that increasing or decreasing their expression levels will reduce cell viability. In apparent confirmation of this hypothesis, changes in the expression level of proteins that are members of complexes have been found to be more deleterious than proteins that are not (6). In addition, analysis of codon biases has suggested that pairs of genes that encode interacting proteins are coordinately expressed (5). However, more recent large-scale studies have failed to find increased dosage sensitivity among genes encoding members of protein complexes, and no correlation was observed between genes displaying haploinsufficiency and overexpression phenotypes (8, 9). To reconcile these conflicting conclusions, the topology of proteins within complexes, not just membership in the complex, has been suggested to be a key determinant in dosage sensitivity (10).
In studies of the relationship between gene expression level and complex assembly, the interactions between components within a complex are usually assumed to be controlled primarily by their intracellular concentrations; thus, these concentrations would need to be precisely balanced to maximize the efficiency of assembly (6, 10, 11). Some of the earliest evidence supporting this idea came from genetic studies on phage morphogenesis, which showed that reductions in assembly efficiency caused by a limiting concentration of one assembly subunit could be ameliorated by reducing the concentration of a second subunit in the complex (12, 13). However, a number of subsequent studies have shown that phage assembly is most often regulated by conformational changes, such that individual pathway components can be present at high concentration and remain unassembled (14–18). For example, gpU, the tail terminator protein of phage λ is expressed during phage infection at a level 40% higher than the tail tube protein (19), gpV, yet 32-fold more copies of gpV are found within the phage particle. Although it is found as a hexamer within the assembled phage, gpU on its own can be maintained in a monomeric state at concentrations up to 1 mM (20). Oligomerization of gpU during phage assembly occurs only in the presence of the appropriate assembly precursor. Because phage particles are complicated structures comprised of multiple oligomeric species coming together through well-ordered pathways, investigations of these systems, which are tractable to both detailed in vitro and in vivo analysis, continue to provide important insights into the mechanism and regulation of macromolecular complex assembly.
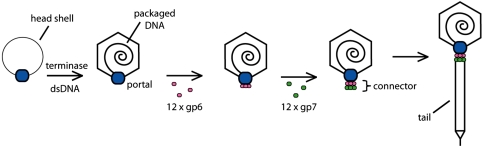
The focus of this study is the head–tail connector of bacteriophage HK97. Phage HK97 possesses an icosahedral head, which contains its dsDNA genome, joined to a long noncontractile tail by a ring-shaped complex known as the connector. Assembly of the HK97 phage particle proceeds through formation of an empty head precursor, known as the prohead, into which the genome is packaged. Once packaging is completed, the components of the connector, gp6 and gp7, assemble onto the portal protein, which is a dodecameric ring positioned at a unique vertex of the head (Fig. 1). The addition of gp6 and gp7 stabilizes the DNA within the head (21). This connector complex then serves as the attachment point for the tail, which is assembled in an independent pathway. Upon infection, the DNA leaves the head through the connector and proceeds down the tail tube to be injected into the host cell. Although the structure of the assembled HK97 connector has not been determined, the cryoEM structure of the connector complex of a related phage, SPP1, indicates that both gp6 and gp7 assemble into the phage particle as dodecameric rings with gp6 forming the interface between the portal protein and gp7 (21–23) (Fig. 1).
Fig. 1.
Schematic of phage HK97 connector assembly. DNA is packaged into an empty head precursor through the dodecameric ring formed by the portal protein in a reaction mediated by the phage-encoded terminase enzyme. Once the head is filled with DNA, gp6 is assembled onto the portal protein as a dodecameric ring. Subsequently, gp7 is assembled onto gp6 completing connector assembly and providing the site of attachment for tails, which are assembled in an independent pathway.
We recently showed that purified gp6 can form stable ring-shaped structures even at relatively low protein concentrations (21). This was a surprising result in light of many previous observations, as mentioned above, that other phage particle components remain monomeric in solution at high concentrations and do not oligomerize into the form found within phage particles until they contact the correct assembly intermediate. This phenomenon is thought to be a key factor in controlling assembly by preventing premature self-association and misassociation with other components before the accumulation of the correct assembly precursor, even if some components are present in excess. Thus, a consequence of the oligomerization of gp6 in the absence of other assembly intermediates is that the rings formed might participate in off-pathway interactions leading to deleterious misassembly. The possibility of misassembly of gp6 is reinforced by the observation of a 13-membered ring in the crystal structure of gp6 rather than the dodecameric ring expected to be found in assembled phage particles (21).
In this study, we show that overexpression of gp6 from a plasmid vector during HK97 infection resulted in more than a 100-fold decrease in phage titre. This dramatic dosage sensitivity of gp6 contrasts sharply with other phage morphogenetic proteins previously studied and led us to address the hypothesis that this phenomenon was caused by premature gp6 oligomerization. We have found that the assembly of gp6 is controlled during the HK97 life cycle by severely restricting its expression level. However, the requirement for this stringent regulation can be bypassed by single amino acid substitutions that abrogate premature oligomerization. Our data demonstrate that the dosage sensitivity of a gene is uniquely dependent on the assembly mechanism of the encoded protein.
Results
Overexpression of WT gp6 Inhibits HK97 Assembly.
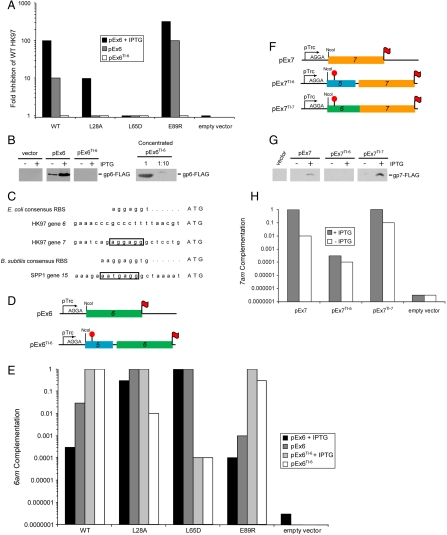
In constructing and testing expression plasmid constructs for structural studies on gp6, we observed an unexpected inhibitory effect of overexpression of gp6 on the growth of WT HK97 phage. As seen in Fig. 2A, the plating efficiency of WT HK97 on cells carrying a gp6-overexpressing plasmid (pEx6) was reduced by more than 100-fold compared to cells carrying an empty vector. Under conditions where gp6 expression was lowered by omission of IPTG (the inducer of transcription), only a 10-fold inhibitory effect was observed (Fig. 2A), indicating that this inhibition was sensitive to the expression level of gp6, which was approximately 5-fold lower under these conditions (Fig. 2B). Transmission electron micrographs of lysates of an HK97 infection in the presence of gp6 overexpression revealed an overwhelming accumulation of protein ring complexes not seen in the empty vector control lysate (Fig. S1 A and B). These rings were the same size and shape as the gp6 rings observed in our previous studies (21).
Fig. 2.
Analysis of the expression of HK97 connector proteins. (A) Inhibition of WT HK97 phage plating efficiency resulting from expression of gp6 from the indicated plasmids. (B) Plasmid-based expression levels of gp6. Lysates made from cells carrying the indicated plasmids were separated by SDS-PAGE and immunoblotted with an anti-FLAG antibody (Left). gp6 produced from pEx6TI-6 was concentrated by Ni-NTA chromatography [2,000-fold relative to samples analyzed (Left)] and dilutions of this preparation were analyzed by SDS-PAGE and immunoblotting (Right). (C) The nucleotide sequences upstream of HK97 genes 6 and 7, and SPP1 gene 15. Ribosome binding sites are boxed. Consensus ribosome binding sites for the hosts of these phages [E. coli (34) and B. subtilis (35)] are also shown. (D) Schematic representations of gp6 constructs used in this work. pEx6 contains gene 6 (green) under the translational control of the strong ribosome binding site encoded within the parent expression vector. pEx6TI-6 contains 150 bp upstream of gene 6, which includes a portion of gene 5 (blue) cloned out of frame with respect to the plasmid encoded translation start (red “stop” sign). Translation of gene 6 on this vector is mediated by its native translation initiation site. Both vectors express gp6 with a C-terminal FLAG epitope (red flag). (E) Complementation of an HK97 6am phage by WT and mutant forms of gp6 expressed from the indicated constructs. Values were normalized to the complementation mediated by WT gp6 expressed from pEx6TI-6. (F) Schematic representations of gp7 constructs used in this work. For pEx7, gene 7 (orange) is expressed under the translational control of the plasmid-based translation initiation site as in pEx6. pEx7TI-6 is constructed similarly to pEx6TI-6 such that translation of gene 7 is mediated by the native translation initiation site of gene 6. In pEx7TI-7, the translation of gene 7 is mediated by its native translation initiation site. (G) Expression level of gp7 in cells carrying pEx7, pEx7TI-6, and pEx7TI-7. These experiments were carried out as described in panel 2B. (H) Complementation of an HK97 7am phage by gp7 expressed from the indicated construct. Values were normalized to the complementation mediated by gp7 expressed from pEx7TI-7 in the presence of IPTG.
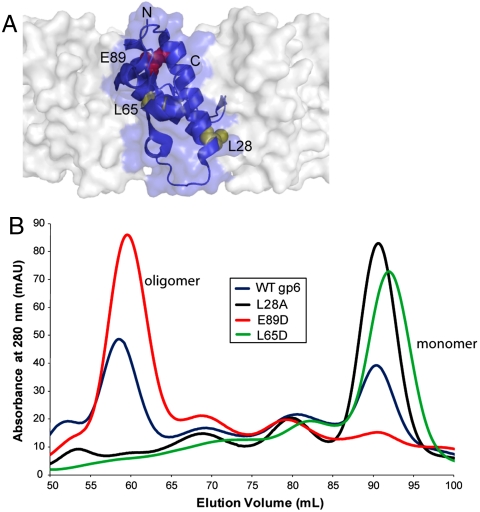
To elucidate the relationship between formation of the gp6 oligomer and the overexpression inhibition phenotype, we constructed gp6 mutants bearing amino acid substitutions at positions in the intersubunit interface within the 13-mer gp6 structure (Fig. 3A). Two of these mutants, L28A and L65D, clearly displayed weakened intersubunit interactions; their gel filtration elution profiles demonstrated only peaks at the monomer position (Fig. 3B). By contrast, the WT protein eluted as a mixture of multimeric and monomeric species. A third mutant, E89R, appeared to possess a strengthened interface as it eluted almost exclusively at the volume expected for a multimer. Consistent with their reduced levels of oligomerization, the L65D mutant displayed no overexpression inhibition phenotype, and the L28A mutant moderately inhibited phage growth only in the presence of IPTG. Conversely, the E89R mutant protein, which oligomerizes more readily than WT, was 10-fold more inhibitory in the absence of IPTG (Fig. 2A). Also consistent with these results, we observed by electron microscopy the same ring-shaped complexes in lysates from the E89R-expressing cells as were present in WT samples, whereas no rings were detected in cells expressing the L28A protein (Fig. S1 C and D). These data clearly demonstrate that the overexpression inhibition phenotype is correlated with the level of gp6 oligomerization.
Fig. 3.
Analysis of amino acid substitutions that alter gp6 oligomerization. (A) The positions of the amino acids within the intersubunit interface of gp6 (PDB ID code 3JVO) that were substituted to alter oligomerization. One monomer is shown in ribbon format, whereas neighboring subunits in the gp6 ring are depicted in surface representation. This figure was generated using PyMol (http://pymol.sourceforge.net/). (B) Gel filtration elution profiles of gp6 mutants altering oligomerization. Purified preparations of WT gp6 (blue), and the L28A (black), E89R (red), and L65D (green) mutants were each analyzed by gel filtration chromatography on a Superdex-200 column. The proteins were loaded at concentrations between 20 and 80 μM. The elution volumes were consistent with two species, a monomer and an oligomer with a stoichiometry of approximately 13.
The Native Translation Initiation Site of Gene 6 Is Very Weak, But Is Sufficient for Phage Growth.
Because high expression of gp6 severely reduced phage viability, we surmised that gp6 might be expressed at a low level during a WT phage infection. Supporting this hypothesis, gene 6 lacks an obvious ribosome binding site in the expected position within 6–10 base pairs of its initiation codon (24) (Fig. 2C); thus, it may be translated very poorly in its native context. To assess the gp6 expression level when its translation is driven by its own translation start site rather than the strong site present in pEx6, we created the construct, pEx6TI-6 (Fig. 2D). In this vector, transcription is driven from the same promoter as the overexpression plasmid pEx6, but inclusion of the 150 bp upstream of the 6 start codon ensured that translation would initiate from the same site as would be utilized within the context of the phage genome (Fig. 2D). Although the expression of gp6 from pEx6 could be easily detected in whole cell lysates by immunoblotting in the absence of IPTG, the expression of gp6 from pEx6TI-6 was undetectable even when IPTG was included (Fig. 2B). Only when we concentrated the gp6 expressed from pEx6TI-6 through Ni-affinity purification were we able to detect this protein and determine that the expression level of gp6 from this plasmid was at least 2,000-fold lower than the expression from pEx6 (Fig. 2B).
A Low Expression Level Is Sufficient for the in Vivo Function of WT gp6, But Insufficient for Oligomerization Mutants.
To determine whether the low level of gp6 expressed from pEx6TI-6 was sufficient for phage propagation, we assessed the ability of plasmid-expressed gp6 to complement a mutant phage bearing a nonsense (amber) mutation in gene 6 [HK97 6am phage, (21)]. Remarkably, pEx6TI-6 was able to complement the HK97 6am mutant greater than 10-fold more effectively than pEx6 in spite of its extremely low gp6 expression level (Fig. 2E). By contrast, the L28A and L65D mutants, which do not oligomerize in vitro (Fig. 3B), complemented considerably less efficiently than WT when expressed from the pEx6TI-6 plasmid (Fig. 2E). In particular, the activity of the L65D protein was reduced by 10,000-fold, whereas the L28A mutant displayed an intermediate activity that could be increased to the WT level by addition of IPTG. Strikingly, the high level of expression mediated by pEx6 allowed the L65D mutant to overcome its oligomerization defect and fully complement. The E89R mutant was similar to WT in requiring the low expression mediated by pEx6TI-6 for optimal activity, and low expression of E89R eliminated its overexpression inhibition phenotype (Fig. 2A). These data demonstrate that the weakened intersubunit interactions of the L28A and L65D mutants can be compensated for by an increased intracellular concentration that serves to drive assembly of these proteins onto the portal. The higher activity of the L28A mutant as compared to the L65D mutant is likely due to the less severe nature of this substitution allowing stronger intersubunit interactions in vivo. On the other hand, the very strong intersubunit interactions of WT and E89R gp6 allow them to assemble efficiently even at very low concentrations.
Gene 7 Requires a High Level of Expression for Optimal in Vivo Activity
Because HK97 gp7 interacts directly with gp6 in the connector complex and is incorporated with the same stoichiometry, one might expect, based on the “balance hypothesis,” the expression level of gp7 to be the same as that of gp6. Because both genes 6 and 7 in the HK97 genome are transcribed from the same promoter (24), differences in their expression levels during phage infection would likely be mediated through modulation of translation efficiency as has been observed for the morphogenetic proteins of phage λ, a relative of HK97 (25). To assess the strength of the gene 7 translation initiation site, we constructed a plasmid called pEx7TI-7, which is analogous to pEx6TI-6 except that it contains gene 7 and the 150 bp upstream of its initiation codon (Fig. 2F). In sharp contrast to pEx6TI-6, pEx7TI-7 mediated expression of a high amount of gp7 that even exceeded the expression level of gp7 mediated by the strong translation start site of the parent gp7 expression vector, pEx7 (Fig. 2 F and G). Quantitative analyses of immunoblots of cells bearing pEx6TI-6 and pEx7TI-7 indicated that the expression levels of gp6 and gp7 vary by more than 500-fold when translated from their native ribosome binding sites despite their assembly into the same complex with the same stoichiometry.
To evaluate the relationship between gp7 expression and biological activity, we assessed the ability of the gp7-expressing plasmids to complement an HK97 7am mutant (see SI Text). It can be seen that both pEx7 and pEx7TI-7 could complement the HK97 7am phage and that the efficiency of complementation increased significantly upon addition of IPTG (Fig. 2H). These data indicate that in the absence of IPTG, the expression level of gp7 limits the phage yield. Consistent with this finding, a plasmid in which the translation of gene 7 was driven by the weak translation initiation site of gene 6 (pEx7TI-6) complemented very poorly even in the presence of IPTG (Figs. 2 F–H). These data demonstrate that the expression level requirements of gp6 and gp7 for optimal assembly are drastically different. Furthermore, even when gp7 was expressed in excess of its requirement for optimal function (i.e., when expressed from pEx7TI-7 in the presence of IPTG), we nevertheless observed no inhibitory effect on the plating efficiency of WT HK97.
Discussion
Our data clearly show that the dosage sensitivity of a protein found within a complex is profoundly influenced by the manner in which its incorporation into that complex is regulated. WT gp6 has a very strong oligomerization interface that causes it to oligomerize aberrantly when expressed at a high level. This aberrant oligomerization likely inhibits phage assembly by sequestering gp6 in nonproductive complexes that are unable to bind appropriately to upstream or downstream components of the pathway. For this reason, in the context of the phage genome, gene 6 is translated very poorly such that its intracellular concentration is maintained at a very low level during the phage growth cycle. Because of its very low concentration, gp6 likely polymerizes only upon contact with the dodecameric portal ring after the head is filled with DNA; thus, avoiding formation of dead-end oligomeric intermediates. The strong intersubunit binding surface of WT gp6 allows this reaction to occur at high enough efficiency for optimal phage yield, even under conditions of low protein concentration. However, when this binding surface is disrupted by amino acid substitutions, a much higher protein concentration is required to drive the weakened oligomerization reaction. Because these mutants are no longer able to oligomerize efficiently on their own, high level expression can be tolerated with no resulting inhibition of phage growth. In this way, we see that a single amino acid substitution can reverse both the mechanism by which assembly is controlled and the effect of high dosage. While WT gp6 displays dosage sensitivity when expressed at high levels, the oligomerization deficient mutants display dosage sensitivity when expressed at low levels.
It could be argued that the gp6 weakened oligomerization mutants characterized here are not representative of naturally occurring proteins and that the normal functioning of proteins in the gp6 family requires strong oligomerization as is seen for gp6. However, gp15 of SPP1, a homologue of gp6 (21), does not form an oligomer on its own and was solved as a monomer by NMR spectroscopy despite its dodecameric structure in the assembled connector complex (22). Comparison of the DNA sequence upstream of gene 15 to the consensus ribosome binding site for Bacillus subtilis, the host organism for SPP1, indicates that it possesses a strong translation initiation site (Fig. 2C) and would likely be well expressed during an SPP1 infection. Thus, the assembly mechanism of SPP1 gp15 resembles that of the L28A or L65D mutants of gp6 for which a high level of expression is required for optimal function. These data demonstrate that the assembly of homologous proteins performing precisely the same function may be controlled by distinct mechanisms, which can lead to widely varying expression level requirements for function. This comparison also highlights the divergent evolutionary strategies that can be employed by homologous proteins to achieve the same goal of controlling complex assembly. In the case of SPP1, assembly is controlled utilizing a weak oligomerization interface that prevents premature aberrant assembly, whereas in HK97 aberrant assembly is prevented through low gene expression.
Gp7 is the binding partner of gp6 within the connector complex, and it is present with equivalent stoichiometry. Nevertheless, our plasmid-based assays using the native translation start sites of genes 6 and 7 indicated that gp7 is present within phage-infected cells at a concentration at least 500-fold higher than that of gp6 (Fig. 2 B and G). Despite this relatively high expression level, plasmid-based expression of gp7 did not inhibit HK97 growth. Moreover, the addition of IPTG increased plating efficiency, indicating that the concentration of gp7 was actually limiting the yield of phage particles even though the intracellular concentration of gp6 was vastly lower under these conditions. These results emphasize that though two proteins may be found within the same complex with the same stoichiometry, their expression levels can vary tremendously. Although the oligomerization properties of gp7 in isolation have not been assessed due to its poor solubility in vitro, it is likely maintained in a monomeric state in vivo until the appropriate assembly intermediate is contacted. This has been shown to be the case for the homologues of gp7 in bacteriophage λ and SPP1, gpFII and gp16, respectively (17, 22). The monomeric states of these proteins could be maintained at protein concentrations high enough to determine their solution structures by NMR, yet they multimerize in vivo upon incorporation into the phage particle. The incorporation and oligomerization of both of these proteins appear to be controlled by disorder-to-order transitions occurring upon assembly (22, 26).
Previous discussions of the dosage sensitivity of proteins that are involved in complex formation in eukaryotic cells have generally assumed that the number of on- and off-pathway complexes formed is determined solely by the intracellular concentrations of the components comprising those complexes. However, in phage assembly systems, pathways appear to be predominantly regulated by allosteric transitions that occur when monomeric proteins contact the correct assembly intermediate. The monomeric solution structures of many such proteins have been solved at high protein concentration by NMR spectroscopy even though all oligomerize upon assembly (17, 18, 20–22, 27–29). These results imply that, in contrast to gp6, these proteins in isolation possess very weak oligomerization interfaces that become strong only after interaction with the proper precursor in the assembly reaction. Because these complex subunits are maintained in a monomeric state until the proper assembly intermediate is encountered, the impact of imbalances in the expression levels of these components is minimized. Assuming that complex assembly in other systems is often regulated in a similar manner to phages, it is unsurprising that the overexpression of protein complex subunits in eukaryotic cells is often tolerated without adverse effects (8) and 50% reductions of gene expression in yeast diploid cells rarely cause phenotypic effects (30).
Overall, our data demonstrate that the inhibitory effect of overexpression or underexpression of a protein on complex assembly is intimately tied to its propensity to interact with itself and its binding partners when present in its unassembled state. Because this property will naturally vary among proteins, even among homologues such as gp6 and gp15, and can be drastically altered by single amino acid substitutions, prediction of the dosage sensitivity of any set of proteins is extremely difficult. In eukaryotic cells, where many proteins are members of several distinct complexes (31, 32), each potentially with their own copy number requirements, allosteric mechanisms for the regulation of complex assembly similar to those seen in phages will likely be more important than modulation of expression level. Thus, knowledge of the mechanisms controlling the assembly of multiprotein complexes is crucial for comprehending the relationship between protein complex membership and overexpression inhibition or haploinsufficiency phenotypes.
Methods
In Vivo Analysis of gp6 and gp7 Activity.
Complementation assays were performed as described (21). For assays that included IPTG, cells were preincubated with IPTG (final concentration in top agar was 0.1 mM) for 15 min at room temperature before plating. Lysates of amber mutant and WT HK97 were prepared by standard procedures. Results were quantified by spotting serial dilutions of the test phage and scoring for activity based on the formation of a zone of clearance caused by lysis of the bacterial lawn by phage.
Protein Purification and Analysis.
All proteins were expressed, purified, and analyzed by gel filtration as previously described (21).
Analysis of gp6 and gp7 Expression Levels.
Escherichia coli 594 cells transformed with expression plasmids grown in the presence or absence of IPTG were pelleted and resuspended in 1/5 volume of SDS-PAGE sample buffer. To assess the low level of gp6 expression from pEx6TI-6, the total gp6 produced from a 1-L culture of cells containing this plasmid was purified by Ni-affinity chromatography and resuspended in 0.1 mL of sample buffer, thus providing a 10,000-fold increase in gp6 concentration. Samples were separated by SDS-PAGE and immunoblotted with anti-FLAG antibody using standard procedures (33). Bands were visualized using a Typhoon 9400 system, and intensities were quantitated using ImageQuant v. 5.2 software. The expression levels of gp6 and gp7 from their native translation sites were quantitated by comparing band intensities of appropriately diluted samples of gp6 and gp7 expressed from pEx6TI-6 and pEx7TI-7 to purified gp6 protein standards of known concentration. Control experiments were performed with purified gp6 and gp7 to determine that these proteins were detected at equivalent levels by the anti-FLAG antibody.
Supplementary Material
Acknowledgments.
The authors thank Lindsay A. Baker and John L. Rubinstein for aid in generating EM micrographs, Dennis Yu for technical assistance, and Paul Sadowski and Andrew Spence for critical reading of the manuscript. This work was supported by operating grants from the Canadian Institutes of Health Research (MOP-77680 to A.R.D. and MOP-6279 to K.L.M.). L.C. was supported by a Postgraduate Scholarship (Doctoral) from the Natural Sciences and Engineering Research Council of Canada.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1100759108/-/DCSupplemental.
References
- 1.Butland G, et al. Interaction network containing conserved and essential protein complexes in Escherichia coli. Nature. 2005;433:531–537. doi: 10.1038/nature03239. [DOI] [PubMed] [Google Scholar]
- 2.Li S, et al. A map of the interactome network of the metazoan C. elegans. Science. 2004;303:540–543. doi: 10.1126/science.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ito T, et al. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci USA. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stelzl U, et al. A human protein-protein interaction network: A resource for annotating the proteome. Cell. 2005;122:957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- 5.Fraser HB, Hirsh AE, Wall DP, Eisen MB. Coevolution of gene expression among interacting proteins. Proc Natl Acad Sci USA. 2004;101:9033–9038. doi: 10.1073/pnas.0402591101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Papp B, Pal C, Hurst LD. Dosage sensitivity and the evolution of gene families in yeast. Nature. 2003;424:194–197. doi: 10.1038/nature01771. [DOI] [PubMed] [Google Scholar]
- 7.Carmi S, Levanon EY, Eisenberg E. Efficiency of complex production in changing environment. BMC Syst Biol. 2009;3:3. doi: 10.1186/1752-0509-3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sopko R, et al. Mapping pathways and phenotypes by systematic gene overexpression. Mol Cell. 2006;21:319–330. doi: 10.1016/j.molcel.2005.12.011. [DOI] [PubMed] [Google Scholar]
- 9.Deutschbauer AM, et al. Mechanisms of haploinsufficiency revealed by genome-wide profiling in yeast. Genetics. 2005;169:1915–1925. doi: 10.1534/genetics.104.036871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oberdorf R, Kortemme T. Complex topology rather than complex membership is a determinant of protein dosage sensitivity. Mol Syst Biol. 2009;5:253. doi: 10.1038/msb.2009.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bray D, Lay S. Computer-based analysis of the binding steps in protein complex formation. Proc Natl Acad Sci USA. 1997;94:13493–13498. doi: 10.1073/pnas.94.25.13493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sternberg N. A genetic analysis of bacteriophage lambda head assembly. Virology. 1976;71:568–582. doi: 10.1016/0042-6822(76)90382-2. [DOI] [PubMed] [Google Scholar]
- 13.Floor E. Interaction of morphogenetic genes of bacteriophage T4. J Mol Biol. 1970;47:293–306. doi: 10.1016/0022-2836(70)90303-7. [DOI] [PubMed] [Google Scholar]
- 14.Maxwell KL, Davidson AR, Murialdo H, Gold M. Thermodynamic and functional characterization of protein W from bacteriophage lambda. The three C-terminal residues are critical for activity. J Biol Chem. 2000;275:18879–18886. doi: 10.1074/jbc.M001178200. [DOI] [PubMed] [Google Scholar]
- 15.Yang F, et al. Novel fold and capsid-binding properties of the lambda-phage display platform protein gpD. Nat Struct Biol. 2000;7(3):230–237. doi: 10.1038/73347. [DOI] [PubMed] [Google Scholar]
- 16.Katsura I, Tsugita A. Purification and characterization of the major protein and the terminator protein of the bacteriophage lambda tail. Virology. 1977;76:129–145. doi: 10.1016/0042-6822(77)90290-2. [DOI] [PubMed] [Google Scholar]
- 17.Maxwell KL, Yee AA, Arrowsmith CH, Gold M, Davidson AR. The solution structure of the bacteriophage lambda head-tail joining protein, gpFII. J Mol Biol. 2002;318:1395–1404. doi: 10.1016/s0022-2836(02)00276-0. [DOI] [PubMed] [Google Scholar]
- 18.Maxwell KL, et al. The solution structure of bacteriophage lambda protein W, a small morphogenetic protein possessing a novel fold. J Mol Biol. 2001;308:9–14. doi: 10.1006/jmbi.2001.4582. [DOI] [PubMed] [Google Scholar]
- 19.Murialdo H, Siminovitch L. The morphogenesis of phage lambda. V. Form-determining function of the genes required for the assembly of the head. Virology. 1972;48:824–835. doi: 10.1016/0042-6822(72)90163-8. [DOI] [PubMed] [Google Scholar]
- 20.Edmonds L, et al. The NMR structure of the gpU tail-terminator protein from bacteriophage lambda: identification of sites contributing to Mg(II)-mediated oligomerization and biological function. J Mol Biol. 2007;365:175–186. doi: 10.1016/j.jmb.2006.09.068. [DOI] [PubMed] [Google Scholar]
- 21.Cardarelli L, et al. The crystal structure of bacteriophage HK97 gp6: Defining a large family of head-tail connector proteins. J Mol Biol. 2010;395:754–768. doi: 10.1016/j.jmb.2009.10.067. [DOI] [PubMed] [Google Scholar]
- 22.Lhuillier S, et al. Structure of bacteriophage SPP1 head-to-tail connection reveals mechanism for viral DNA gating. Proc Natl Acad Sci USA. 2009;106:8507–8512. doi: 10.1073/pnas.0812407106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Orlova EV, et al. Structure of a viral DNA gatekeeper at 10 A resolution by cryo-electron microscopy. EMBO J. 2003;22:1255–1262. doi: 10.1093/emboj/cdg123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Juhala RJ, et al. Genomic sequences of bacteriophages HK97 and HK022: Pervasive genetic mosaicism in the lambdoid bacteriophages. J Mol Biol. 2000;299:27–51. doi: 10.1006/jmbi.2000.3729. [DOI] [PubMed] [Google Scholar]
- 25.Sampson LL, Hendrix RW, Huang WM, Casjens SR. Translation initiation controls the relative rates of expression of the bacteriophage lambda late genes. Proc Natl Acad Sci USA. 1988;85:5439–5443. doi: 10.1073/pnas.85.15.5439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cardarelli L, et al. Phages have adapted the same protein fold to fulfill multiple functions in virion assembly. Proc Natl Acad Sci USA. 2010;107:14384–14389. doi: 10.1073/pnas.1005822107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iwai H, Forrer P, Pluckthun A, Guntert P. NMR solution structure of the monomeric form of the bacteriophage lambda capsid stabilizing protein gpD. J Biomol NMR. 2005;31:351–356. doi: 10.1007/s10858-005-0945-7. [DOI] [PubMed] [Google Scholar]
- 28.Pell LG, Kanelis V, Donaldson LW, Howell PL, Davidson AR. The phage lambda major tail protein structure reveals a common evolution for long-tailed phages and the type VI bacterial secretion system. Proc Natl Acad Sci USA. 2009;106:4160–4165. doi: 10.1073/pnas.0900044106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kanamaru S, et al. Structure of the cell-puncturing device of bacteriophage T4. Nature. 2002;415:553–557. doi: 10.1038/415553a. [DOI] [PubMed] [Google Scholar]
- 30.Springer M, Weissman JS, Kirschner MW. A general lack of compensation for gene dosage in yeast. Mol Syst Biol. 2010;6:368. doi: 10.1038/msb.2010.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gavin AC, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- 32.Krogan NJ, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 33.Pell LG, et al. The solution structure of the C-terminal Ig-like domain of the bacteriophage lambda tail tube protein. J Mol Biol. 2010;403:468–479. doi: 10.1016/j.jmb.2010.08.044. [DOI] [PubMed] [Google Scholar]
- 34.Shine J, Dalgarno L. Determinant of cistron specificity in bacterial ribosomes. Nature. 1975;254:34–38. doi: 10.1038/254034a0. [DOI] [PubMed] [Google Scholar]
- 35.Moran CP, Jr, et al. Nucleotide sequences that signal the initiation of transcription and translation in Bacillus subtilis. Mol Gen Genet. 1982;186:339–346. doi: 10.1007/BF00729452. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.