Abstract
In response to physiological stimuli, skeletal muscle alters its myofiber composition to significantly affect muscle performance and metabolism. This process requires concerted regulation of myofiber-specific isoforms of sarcomeric and calcium regulatory proteins that couple action potentials to the generation of contractile force. Here, we identify Sox6 as a fast myofiber-enriched repressor of slow muscle gene expression in vivo. Mice lacking Sox6 specifically in skeletal muscle have an increased number of slow myofibers, elevated mitochondrial activity, and exhibit down-regulation of the fast myofiber gene program, resulting in enhanced muscular endurance. In addition, microarray profiling of Sox6 knockout muscle revealed extensive muscle fiber-type remodeling, and identified numerous genes that display distinctive fiber-type enrichment. Sox6 directly represses the transcription of slow myofiber-enriched genes by binding to conserved cis-regulatory elements. These results identify Sox6 as a robust regulator of muscle contractile phenotype and metabolism, and elucidate a mechanism by which functionally related muscle fiber-type specific gene isoforms are collectively controlled.
Keywords: calcium handling, myosin heavy-chain isoforms, slow-twitch fiber
The striated appearance of skeletal muscle arises from the tandem arrangement of individual sarcomeric contractile units. Sarcomere contraction occurs when thick filament myosins bind to actin thin filaments, and hydrolyze ATP to generate a power stroke (1). This process is tightly regulated by the calcium-sensitive troponin-tropomyosin complex, which alters its conformation to permit actin–myosin interactions in response to increased intracellular calcium (2). Multiple isoforms of contractile apparatus proteins are regulated in a fiber-type specific manner, and can influence overall muscle function (3). For example, differential expression of myosin heavy chain (Myh) isoforms alters force-velocity parameters of individual muscle fibers, and alternate combinations of troponin T, I, and C isoforms influence myofiber Ca2+ sensitivity (4, 5). The mechanism by which contractile and calcium regulatory genes are concordantly regulated to produce a unified effect on muscle contraction is poorly understood.
Skeletal muscle is composed of a heterogeneous population of slow and fast myofibers that display contrasting contractile and metabolic properties. Slow type I fibers exhibit oxidative metabolism, express slow isoforms of sarcomeric proteins, and are classically identified by their expression of type I myosin heavy chain (Myh7 or β-Myh). In contrast, fast type IIa, IIx/d, and IIb myofibers use glycolytic metabolism, express fast isoforms of contractile proteins, and are designated by their expression of fast myosin heavy chain isoforms (Myh2, Myh1, and Myh4, respectively) (6). Chronic muscle stimulation induces a reversible transition in myofiber phenotype from fast to slow, which occurs in a sequential manner through several fast myofiber intermediates (type IIb → type IIx/d → type IIa → type I) (7). Previous studies have implicated multiple transcriptional pathways in the regulation of basal-muscle fiber-type specific gene expression, stress-induced fiber-type remodeling, and myofiber metabolism (8–17).
Recently, we identified a microRNA (miRNA)-mediated transcriptional regulatory network that reinforces slow myofiber gene expression in skeletal muscle via a reciprocal negative-feedback loop (18). In this pathway, miRNAs miR-499 and miR-208b, intronically encoded within slow myosin heavy-chain genes, target a collection of transcriptional repressors, and promote a fast-to-slow myofiber-type switch. One transcriptional repressor integral to this regulatory pathway is Sox6, a member of the SoxD family of transcription factors that exhibits dual functions as a transcriptional activator or repressor (19, 20). Sox6-null mice exhibit failure to thrive and expire within 2 wk following birth (21); however, studies in these mice, as well as in zebrafish, have identified an important role for Sox6 in embryonic muscle development (22–24).
In the present study, we investigate the function of Sox6 in adult skeletal muscle. We show that Sox6 is required for fast myofiber maintenance in adult muscle, as conditional loss of Sox6 leads to conversion of muscle to a slow myofiber phenotype, resulting in significant changes in skeletal muscle mechanics. We demonstrate that Sox6 regulation of muscle phenotype occurs independent of changes in other regulatory factors implicated in fiber-type switching, such as PGC-1α and AMP-kinase (AMPK), and instead results from direct repression of a constellation of slow isoforms of sarcomeric and calcium regulatory proteins. Together, these data reveal an important role for Sox6 in the coordinated regulation of multiple properties of myofiber phenotype that profoundly effect muscle performance.
Results
Sox6 Is Enriched in Fast Myofibers.
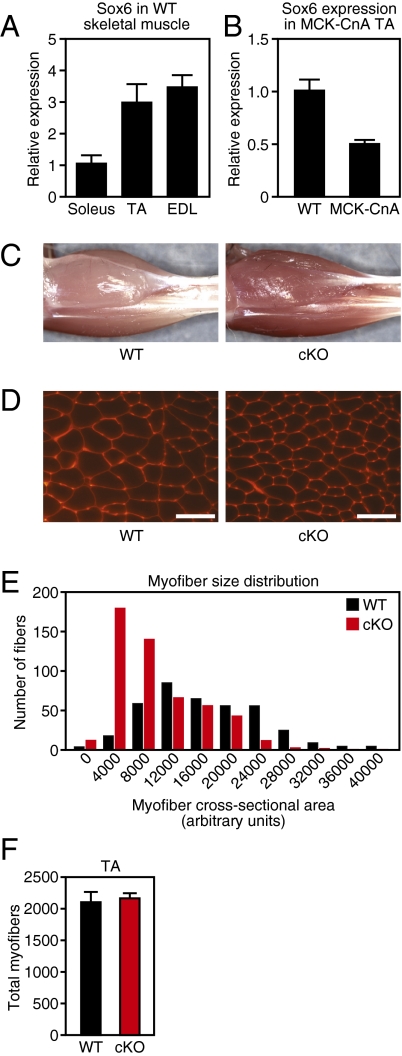
Previous work suggests a role for Sox6 in embryonic muscle differentiation, but the potential functions of Sox6 in adult muscle are unknown; therefore, we asked if Sox6 displays differential expression between muscle fiber types in adult mice. Comparison of slow myofiber-enriched soleus muscle to fast myofiber-enriched tibialis anterior (TA) and extensor digitorum longus (EDL) by qPCR revealed that Sox6 mRNA is expressed approximately threefold higher in fast TA and EDL muscle groups (Fig. 1A). To more thoroughly study the influence of fiber type on the expression of Sox6 within an individual muscle group, we utilized transgenic mice that express an active form of the phosphatase calcineurin (CnA) under control of muscle creatine kinase (MCK) regulatory elements, which drives slow myofiber formation in skeletal muscle (10). Expression of Sox6 in slow myofiber-enriched MCK-CnA TA muscle was reduced by ∼50% compared to WT TA by qPCR (Fig. 1B). Based on these results, we conclude that Sox6 is more highly expressed in fast myofibers.
Fig. 1.
Sox6 is enriched in fast muscle fibers and effects gross muscle fiber morphology. (A) Sox6 mRNA is enriched in muscle groups that predominantly contain fast myofibers (TA and EDL) compared with slow myofiber-enriched muscle (soleus), as measured by qPCR. (B) Sox6 expression levels are reduced by qPCR in slow fiber containing MCK-CnA transgenic TA muscle compared with WT fast TA muscle. (C) Gross examination of adult Sox6 cKO hindlimb muscles demonstrates increased redness and a reduction in muscle size compared with WT control. (D) Staining of myofiber cell membranes with wheat germ agglutinin indicates a reduction in myofiber size in Sox6 cKO TA muscle. (Scale bars, 200 μm.) (E) Quantification of myofiber size following wheat germ agglutinin staining reveals an increase in the proportion of smaller myofibers and a reduction in the proportion of larger myofibers in Sox6 cKO TA muscle. (F) Sox6 cKO and WT TA muscle contain a similar number of myofibers.
Loss of Sox6 Alters Myofiber Appearance and Morphology.
To study the function of Sox6 in skeletal muscle, we bred mice conditionally targeted at the Sox6 locus (25) with mice expressing Cre-recombinase in postnatal skeletal muscle under the control of MCK regulatory elements (26). Sox6f/f;MCK-Cre conditional knockout mice (henceforth referred to as Sox6 cKO) were born at Mendelian ratios and appeared identical to WT littermate controls. Analysis of Sox6 expression in 8-wk-old cKO mice by qPCR revealed a reduction in Sox6 transcript levels by 70% in TA and EDL muscles, and by 25% in the soleus muscle (Fig. S1A). By gross examination, hindlimb musculature of adult Sox6 cKO mice was markedly darker and more red in color than that of WT littermates (Fig. 1C). Furthermore, cKO muscle weighed less than WT controls (Fig. S1B). Examination of myofiber cross-sectional area by histochemical staining revealed that fiber size distribution was altered in Sox6 cKO TA muscle, with a shift toward a reduced cross-sectional area relative to WT (Fig. 1 D and E). Quantification of total myofiber number revealed that Sox6 cKO TA muscle contained a comparable number of individual fibers to WT controls (Fig. 1F), indicating that the decrease in Sox6 cKO muscle mass results solely from a reduction in myofiber size. Importantly, gross morphological changes in Sox6 cKO muscle were absent in young mice at stage P10 (Figs. S1C and S2 B, and C), suggesting that the observed phenotype in Sox6 cKO adult mice is not caused by adverse effects on normal muscle development or early fiber-type patterning.
Enhanced Muscle Performance in Sox6 cKO Mice.
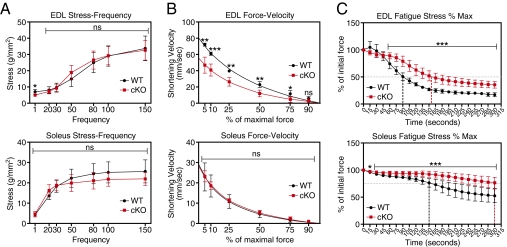
The altered appearance of Sox6 cKO muscle is consistent with changes observed in endurance trained muscle (27), leading us to ask if muscle from Sox6 cKO mice displayed altered mechanical performance. EDL and soleus muscles were isolated from WT and Sox6 cKO mice, and physiological performance was assessed ex vivo in a muscle bath. Sox6 cKO soleus and EDL muscle generated stress comparable to WT controls over a range of stimulation frequencies (Fig. 2A). Measurement of shortening velocity revealed that Sox6 cKO EDL, but not cKO soleus, had a reduced maximal shortening velocity compared with WT (Fig. 2B).
Fig. 2.
Altered physiological performance of Sox6 cKO muscle. (A) A stress-frequency plot of WT and Sox6 cKO EDL and soleus muscle reveals no difference in stress generation at various frequencies of stimulation. (B) Measurement of the shortening velocity in WT and Sox6 cKO EDL and soleus muscle reveals a reduced maximal shortening velocity (y-intercept) in Sox6 cKO EDL, but not soleus. Lines represent curve fit of datapoints. (C) Measurement of time-to-fatigue (i.e., to 50% of initial force; horizontal dashed line) for EDL muscle reveals an increase by 50% in Sox6 cKO EDL. Time-to-fatigue (i.e., 75% of initial force; horizontal dashed line) measurements for soleus muscle demonstrate an increase by 100%. These results indicate a significant improvement in Sox6 cKO muscle endurance. Vertical dashed lines indicate extrapolation of time-to-fatigue. WT (n = 4), Sox6 cKO (n = 6). *P < 0.05, **P < 0.01, ***P < 0.001. ns, not significant.
We assayed muscle endurance by calculating the time-to-fatigue: measured as the length of time for a muscle to fall to a defined percentage of its initial force under constant stimulating frequency. Remarkably, the time-to-fatigue for cKO EDL muscle (50% of initial force) was 50% greater than WT muscle, indicating a significant increase in cKO muscle endurance (Fig. 2C). This pronounced improvement in physiological performance was also evident in the cKO soleus, which displayed a 100% increase in the time-to-fatigue (75% of initial force) (Fig. 2C). Sox6 cKO EDL and soleus muscle also demonstrated improved recovery following fatigue (Fig. S2D).
Increased Slow Fibers in Sox6 cKO Mice.
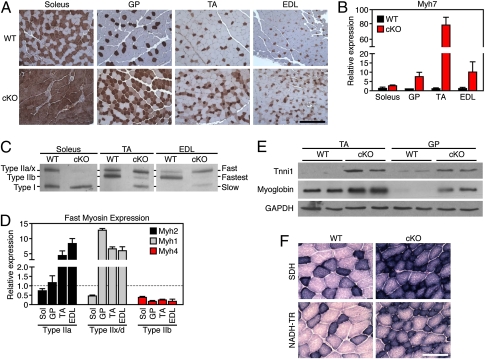
The reduction in maximal shortening velocity and increase in muscle endurance observed in Sox6 cKO skeletal muscle led us to investigate muscle fiber-type composition in these mice. Immunohistochemistry against type I slow myosin (β-Myh) revealed a dramatic increase in the number of slow myofibers in several muscle groups from Sox6 cKO mice (Fig. 3A). Quantification of the transcript encoding slow type I myosin, Myh7, by qPCR showed strong up-regulation in both fast and slow muscle groups of Sox6 cKO mice (Fig. 3B). Similarly, analysis of muscle myosin isoform content by gel electrophoresis demonstrated that WT soleus was composed of 50% type I myosin, but the Sox6 cKO soleus contained nearly 100% slow type I myosin (Fig. 3C). Sox6 cKO TA and EDL myosin electrophoretograms showed the presence of a band representing slow type I myosin; however, this band was absent in WT TA and EDL samples (Fig. 3C). In addition, Sox6 cKO TA and EDL lanes showed a shift from predominant expression of type IIb myosin, the fastest myosin isoform, toward expression of slower type IIa and IIx/d fast myosins. These changes in fast myofiber content were also evident by qPCR analysis of fast myosin isoform mRNA expression (Fig. 3D).
Fig. 3.
Increased slow myofibers and mitochondrial activity in Sox6 cKO mice. (A) Immunohistochemistry directed against type I slow myosin indicates an increase in slow myofibers in all muscle groups from Sox6 cKO mice at 8 wk of age. Slow myofibers are stained brown. (Scale bar, 400 μm.) (B) Measurement of Myh7, encoding type I slow myosin, by qPCR reveals significantly increased expression in Sox6 cKO muscle. (C) Separation of myosin isoforms by gel electrophoresis reveals increased type I myosin in soleus, TA, and EDL muscle from Sox6 cKO mice. In addition, a switch from type IIb to type IIa/x fibers is evident in cKO TA and EDL. (D) Sox6 cKO mice display altered expression of fast myosin isoforms compared with WT by qPCR. (E) Western blots for slow skeletal troponin (Tnni1) and myoglobin demonstrate robust up-regulation of these proteins in TA and gastrocnemius/plantaris (GP) muscle from Sox6 cKO mice. (F) Sox6 cKO TA muscle demonstrates increased SDH and NADH-TR enzymatic staining, consistent with an increase in mitochondrial activity in Sox6 cKO myofibers. (Scale bar, 200 μm.)
Next, we asked whether other slow myofiber-specific genes might be regulated in Sox6 cKO mice. By Western blot, we observed that the slow skeletal muscle-specific troponin isoform, Tnni1, and slow muscle-enriched myoglobin were both strongly up-regulated in cKO muscle (Fig. 3E).
We also compared the mitochondrial activity of muscle from Sox6 cKO mice to littermate controls. Sox6 cKO mice displayed higher levels of succinate dehydrogenase (SDH) and NADH dehydrogenase (NADH-TR) activity-dependant staining, indicating increased mitochondrial activity (Fig. 3F). The up-regulation of slow myofiber-specific gene expression, as well as the heightened mitochondrial activity in Sox6 cKO skeletal muscle, denotes a substantial increase in the number of slow type I myofibers.
Microarray Profiling of Sox6 cKO Mice Reveals Extensive Fiber-Type Switching and Novel Myofiber-Specific Gene Expression.
To more thoroughly analyze the changes that result from loss of Sox6 in skeletal muscle, we performed microarray analysis on mRNA isolated from WT and Sox6 cKO TA muscle. Of particular interest, microarray profiling revealed antithetical regulation of fiber-type specific isoforms of sarcomeric components and ion channels involved in excitation-contraction coupling (Table 1). Gene ontology analysis of transcripts regulated by more than twofold in Sox6 cKO TA demonstrated significant enrichment of genes known to function in skeletal and cardiac muscle contraction, as well as genes involved in various metabolic processes that differ between muscle fiber type (Fig. S3 C and D).
Table 1.
Opposing regulation of fast/slow myofiber programs in Sox6 cKO TA muscle
| Component | Fast isoform | Fold-change | Slow isoform | Fold-change |
| Myosin heavy chain | MYH4 | 0.16 | MYH7 | 4.18 |
| Myosin light chain | MYL1 | 0.51 | MYL2 | 31.53 |
| Na/K ATPase | ATP1B2 | 0.23 | ATP1B1 | 1.96 |
| Troponin T | TNNT3 | 0.72 | TNNT1 | 8.20 |
| Actinin | ACTN3 | 0.75 | ACTN2 | 1.74 |
| Actin | ACTA1 | 0.73 | ACTC1 | 10.87 |
| Sarcoplasmic calcium channel | ATP2A1 | 0.88 | ATP2A2 | 13.93 |
Given these robust changes in fiber-type specific gene expression, we next investigated if Sox6 cKO mice could be used as a tool to identify previously unexplored myofiber enriched genes. We generated a heat map depicting the most highly regulated genes in Sox6 cKO TA muscle (Fig. S3A), and identified 26 genes with no known association with muscle fiber type or muscle function. We next analyzed muscle fiber-type–associated expression of these 26 genes in WT mice by comparing their expression in soleus and TA muscle by qPCR (Fig. S3B). Cross referencing these datasets, we determined that Rspo3 (Thds2), Lrrn1, Lrba, DUPD1, and Psat1 displayed slow myofiber-enriched expression, while Zmynd17, Smox, Kcnc4, and Mod1 demonstrated elevated expression in fast myofibers (Fig. S3B). The transcripts Sv2b and Chrna1, encoding a neuron-specific synaptic vesicle glycoprotein and the nicotinic cholinergic receptor, were also up-regulated in slow muscle, potentially reflecting an increase in the number of neuromuscular junctions in Sox6 cKO TA and WT soleus muscle.
Sox6 Directly Represses Expression of Slow Contractile and Calcium Handling Genes.
To elucidate the mechanism by which Sox6 regulates myofiber-specific gene expression, we examined the possibility that Sox6 regulates factors known to promote slow myofiber gene expression. Pgc-1α, a transcriptional coactivator sufficient to drive slow fiber formation, was not changed in Sox6 cKO muscle as detected by Western blot (Fig. S4A). Additionally, levels of phosphoylated AMPK, a key metabolic regulator of muscle fiber transitions in response to endurance exercise (28), were unchanged in cKO muscle (Fig. S4B).
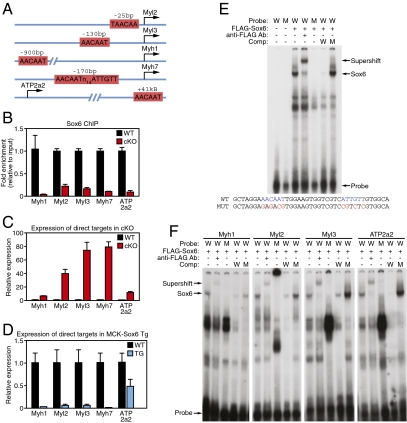
The strong increase in slow muscle-fiber gene expression in Sox6 cKO mice, as well as the enrichment of Sox6 mRNA in fast myofibers, led us to hypothesize that Sox6 acts as a direct repressor of slow myofiber gene expression. A search for the consensus Sox6 binding motif, AACAAT (29), near the loci of genes significantly regulated in cKO TA muscle by microarray identified conserved potential binding sites proximal to Myl2, Myl3, Myh1, Myh7, and ATP2a2 (Fig. 4A). These loci encode slow muscle-specific regulatory and essential myosin light chains (Myl2 and Myl3), a slow and intermediate isoform of myosin heavy chain (Myh7 and Myh1, respectively), and the slow fiber-specific isoform of the sarcoplasmic reticulum Ca2+ uptake channel SERCA (ATP2a2 or SERCA2). To determine if Sox6 is present at these loci in vivo, we performed ChIP assays from WT and Sox6 cKO TA muscle with an antibody directed against Sox6. Both qPCR and semiquantitative RT-PCR analysis of ChIP chromatin indicated that Sox6 bound to all five loci in WT TA muscle, but was largely absent in Sox6 cKO TA (Fig. 4B and Fig. S4C). ChIP analysis of FLAG-epitope tagged Sox6 in C2C12 myoblasts yielded similar results (Fig. S4D). In addition, gel-shift analysis of the Sox6 consensus binding sites at all five loci revealed that Sox6 specifically bound to these DNA sequences in vitro (Fig. 4 E and F). In support of Sox6 acting as a transcriptional repressor at these loci, transcript levels of Myl2, Myl3, Myh1, Myh7, and ATP2a2 were all increased in TA muscle from Sox6 cKO mice (Fig. 4C). Conversely, in transgenic mice that overexpress Sox6 specifically in skeletal muscle (18), Myl2, Myl3, Myh1, Myh7, and ATP2a2 mRNA transcripts were all strongly decreased (Fig. 4D). From these results, we conclude that Sox6 directly binds to loci encoding slow muscle fiber-enriched genes and effectively represses their transcription.
Fig. 4.
Sox6 is a direct repressor of slow myofiber genes. (A) Schematic representation of highly conserved Sox6 consensus binding motifs found near loci encoding slow fiber-enriched genes. (B) Sox6 ChIP reveals that all five loci are bound significantly more by Sox6 in WT TA muscle compared with Sox6 cKO TA negative-control tissue. (C) Direct Sox6 target genes are up-regulated in TA muscle from Sox6 cKO mice as measured by qPCR. (D) Sox6 target gene transcripts are reduced in hindlimb muscle from MCK-Sox6 transgenic mice as measured by qPCR. (E) Gel-shift analysis of the palindromic Sox6 binding site upstream of the Myh7 locus. Sox6 binds WT (W), but not mutant (M) consensus binding sequence. The Sox6 binding complex is supershifted with addition of α-FLAG antibody. (F) Gel-shift analysis of single Sox6 binding sites near loci for Myh1, Myl2, Myl3, and ATP2a2 reveals that Sox6 can bind to consensus binding sites in vitro.
Discussion
In this study, we demonstrate an essential role for the transcription factor Sox6 in postnatal muscle fiber-type differentiation, and consequently, muscle performance and metabolism. We show that mice lacking Sox6 in skeletal muscle display an increased number of slow muscle fibers and exhibit significantly improved endurance in response to mechanical workload. These changes result, at least in part, from a loss of transcriptional repression by Sox6 at loci encoding components of the contractile apparatus and calcium-storage machinery that are specific for slow skeletal muscle.
Our data suggest that Sox6 is a regulator of muscle fiber-type specification that acts in parallel to or downstream of PGC-1α and AMPK to induce slow myofiber formation. Additional experiments are necessary to elucidate the regulatory hierarchy that affects Sox6 function; however, our results are consistent with Sox6 as a target effector of the myomiR regulatory network, which is sufficient, and partially required for slow myofiber formation (18). It is possible that the β-MHC–encoded myomiR, miR-208b, serves as a nexus between slow myofiber stimuli, such as calcineurin activation by increased intracellular calcium, to dampen Sox6 activity and promote slow myofiber gene expression.
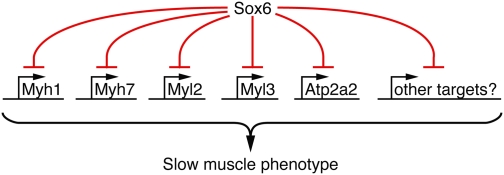
In addition to the regulation of opposing myofiber-specific gene profiles, muscle fiber-type remodeling requires regulation of a broad spectrum sarcomeric components that function in concert to effect contraction. Here, we find that Sox6 regulates Myh7, Myh1, Myl3, and Myl2, which together form the thick filament myosin motor, and the slow muscle-specific, ATP-dependent, sarcoplasmic reticulum Ca2+ uptake channel ATP2a2. Thus, Sox6 regulates both sarcomeric and calcium regulatory components that contribute to the contractile properties of slow myofibers (Fig. 5). Further studies are needed to investigate if other sarcomeric proteins that affect the dynamics of muscle contraction, such as troponins and actins, are also directly regulated by Sox6. Additionally, identification of Sox6 targets that promote oxidative metabolism in slow myofibers would further our understanding of how metabolic and contractile properties of muscle fibers are coregulated.
Fig. 5.
Our data suggest a model by which Sox6 represses multiple contractile genes that collectively contribute to slow muscle fiber contractile properties.
This study raises several important questions regarding the function of Sox6 in skeletal muscle. Previous in vivo loss-of-function studies of Sox6 have demonstrated significant functional overlap with the related SoxD family member, Sox5 (21). Sox5 is highly enriched in human cardiac and skeletal muscle tissue (30), indicating potentially overlapping roles for Sox5/6 in the regulation of myofiber phenotype. Furthermore, although we demonstrate transcriptional repression of slow myofiber genes by Sox6, the possibility that Sox6 also plays an opposing role as a direct transcriptional activator of fast myofiber-specific gene expression necessitates further inquiry.
Our results suggest a significant role for Sox6 in complex phenotypic remodeling of muscle fiber type. In the future, we aim to use the Sox6 cKO mouse as a tool for further exploring molecular mechanisms underlying fiber-type transitions, decoding the function of novel myofiber enriched genes, and understanding the connection between muscle fiber type and whole-body metabolism.
Methods
Immunohistochemistry.
Immunohistochemistry for β-Myh was performed as previously described (18).
MHC Electrophoresis.
Myosin was isolated from skeletal muscle and run on glycerol-SDS/PAGE gels, as previously described (31).
SDH and NADH-TR Staining.
Fresh-frozen sections were incubated in 0.2 M phosphate buffer (pH 7.6) containing sodium succinate and NBT for 60 min at 37 °C (SDH) or in 0.05 M Tris buffer (pH 7.6) containing NADH and NBT for 30 min at 37 °C (NADH-TR). Staining was then cleared with acetone and preserved with aqueous mounting medium.
Gel-Shift Assay.
Full-length Sox6 cDNA was cloned from skeletal muscle and placed into pcDNA3.1 vector containing N-terminal FLAG tag. TnT in vitro translation (Promega) was used to generate tagged Sox6 protein. Double-stranded gel-shift probes were radioactively labeled with ATP[γ-32P] by polynucleotide kinase (PNK) and incubated with Sox6 protein in the presence or absence of unlabeled competitor probes at room temperature. Mouse anti-FLAG antibody (Sigma) was used to supershift. Samples were resolved on a 5% native PAGE gel.
Western Blotting.
Western blotting was performed as previously described (18). For more information, see SI Methods.
Muscle Physiology.
Analysis of muscle contraction is described in SI Methods.
Quantification of Myofiber Size and Number.
Frozen sections were fixed in 100% ethanol and stained with Alexa594-conjugated wheat germ agglutinin (Invitrogen) at a concentration of 50 μg/mL. Fibers were measured and counted using ImageJ software.
Statistical Analysis.
All experiments represent an n = 3 for WT and Sox6 cKO samples unless otherwise stated. All graphs represent mean values ± SEM.
Mice.
MCK-calcineurin and MCK-Cre transgenic mice were previously described (10, 26). Sox6f/f mice were provided by Veronique Lefebvre (The Cleveland Clinic Foundation). All rights, title, and interest in the Sox6f/f mice are owned by The Cleveland Clinic Foundation. All animal procedures were previously approved by the Institutional Animal Care and Use Committee at the University of Texas Southwestern Medical Center and at Virginia Tech.
Supplementary Material
Acknowledgments
We thank Veronique Lefebvre for reagents; Doug Millay, David Patrick, Eric Small, Jason O'Rourke, and Ning Liu for helpful discussions; John Shelton for technical assistance; Nadia Rosenthal and Paul Rosenberg for insightful comments; and José Cabrera for assistance with graphics. This work is supported in part by grants from the National Institutes of Health, the Leducq Foundation, the Robert A. Welch Foundation (Grant I-0025), and the American Heart Association: Jon Holden DeHaan Foundation (to E.N.O.); and Kirschstein MD/PhD Fellowship 1F30HL103013 from the National Heart, Lung, and Blood Institute (to D.Q.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1107413108/-/DCSupplemental.
References
- 1.Spudich JA. The myosin swinging cross-bridge model. Nat Rev Mol Cell Biol. 2001;2:387–392. doi: 10.1038/35073086. [DOI] [PubMed] [Google Scholar]
- 2.Lehman W, Craig R, Vibert P. Ca(2+)-induced tropomyosin movement in Limulus thin filaments revealed by three-dimensional reconstruction. Nature. 1994;368:65–67. doi: 10.1038/368065a0. [DOI] [PubMed] [Google Scholar]
- 3.Bandman E. Contractile protein isoforms in muscle development. Dev Biol. 1992;154:273–283. doi: 10.1016/0012-1606(92)90067-q. [DOI] [PubMed] [Google Scholar]
- 4.Bottinelli R, Schiaffino S, Reggiani C. Force-velocity relations and myosin heavy chain isoform compositions of skinned fibres from rat skeletal muscle. J Physiol. 1991;437:655–672. doi: 10.1113/jphysiol.1991.sp018617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Geiger PC, Cody MJ, Sieck GC. Force-calcium relationship depends on myosin heavy chain and troponin isoforms in rat diaphragm muscle fibers. J Appl Physiol. 1999;87:1894–1900. doi: 10.1152/jappl.1999.87.5.1894. [DOI] [PubMed] [Google Scholar]
- 6.Bassel-Duby R, Olson EN. Signaling pathways in skeletal muscle remodeling. Annu Rev Biochem. 2006;75:19–37. doi: 10.1146/annurev.biochem.75.103004.142622. [DOI] [PubMed] [Google Scholar]
- 7.Pette D, Staron RS. Myosin isoforms, muscle fiber types, and transitions. Microsc Res Tech. 2000;50:500–509. doi: 10.1002/1097-0029(20000915)50:6<500::AID-JEMT7>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 8.Zechner C, et al. Total skeletal muscle PGC-1 deficiency uncouples mitochondrial derangements from fiber type determination and insulin sensitivity. Cell Metab. 2010;12:633–642. doi: 10.1016/j.cmet.2010.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu F, et al. Effects of thyroid hormone receptor gene disruption on myosin isoform expression in mouse skeletal muscles. Am J Physiol Regul Integr Comp Physiol. 2000;278:R1545–R1554. doi: 10.1152/ajpregu.2000.278.6.R1545. [DOI] [PubMed] [Google Scholar]
- 10.Naya FJ, et al. Stimulation of slow skeletal muscle fiber gene expression by calcineurin in vivo. J Biol Chem. 2000;275:4545–4548. doi: 10.1074/jbc.275.7.4545. [DOI] [PubMed] [Google Scholar]
- 11.Potthoff MJ, Olson EN, Bassel-Duby R. Skeletal muscle remodeling. Curr Opin Rheumatol. 2007;19:542–549. doi: 10.1097/BOR.0b013e3282efb761. [DOI] [PubMed] [Google Scholar]
- 12.Lin J, et al. Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres. Nature. 2002;418:797–801. doi: 10.1038/nature00904. [DOI] [PubMed] [Google Scholar]
- 13.Arany Z, et al. The transcriptional coactivator PGC-1beta drives the formation of oxidative type IIX fibers in skeletal muscle. Cell Metab. 2007;5:35–46. doi: 10.1016/j.cmet.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 14.Schuler M, et al. PGC1alpha expression is controlled in skeletal muscles by PPARbeta, whose ablation results in fiber-type switching, obesity, and type 2 diabetes. Cell Metab. 2006;4:407–414. doi: 10.1016/j.cmet.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 15.Allen DL, et al. Cardiac and skeletal muscle adaptations to voluntary wheel running in the mouse. J Appl Physiol. 2001;90:1900–1908. doi: 10.1152/jappl.2001.90.5.1900. [DOI] [PubMed] [Google Scholar]
- 16.Schiaffino S, Reggiani C. Molecular diversity of myofibrillar proteins: Gene regulation and functional significance. Physiol Rev. 1996;76:371–423. doi: 10.1152/physrev.1996.76.2.371. [DOI] [PubMed] [Google Scholar]
- 17.Narkar VA, et al. Exercise and PGC-1α-independent synchronization of type I muscle metabolism and vasculature by ERRγ. Cell Metab. 2011;13:283–293. doi: 10.1016/j.cmet.2011.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Rooij E, et al. A family of microRNAs encoded by myosin genes governs myosin expression and muscle performance. Dev Cell. 2009;17:662–673. doi: 10.1016/j.devcel.2009.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lefebvre V. Toward understanding the functions of the two highly related Sox5 and Sox6 genes. J Bone Miner Metab. 2002;20:121–130. doi: 10.1007/s007740200017. [DOI] [PubMed] [Google Scholar]
- 20.Lefebvre V, Li P, de Crombrugghe B. A new long form of Sox5 (L-Sox5), Sox6 and Sox9 are coexpressed in chondrogenesis and cooperatively activate the type II collagen gene. EMBO J. 1998;17:5718–5733. doi: 10.1093/emboj/17.19.5718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smits P, et al. The transcription factors L-Sox5 and Sox6 are essential for cartilage formation. Dev Cell. 2001;1:277–290. doi: 10.1016/s1534-5807(01)00003-x. [DOI] [PubMed] [Google Scholar]
- 22.Hagiwara N, Ma B, Ly A. Slow and fast fiber isoform gene expression is systematically altered in skeletal muscle of the Sox6 mutant, p100H. Dev Dyn. 2005;234:301–311. doi: 10.1002/dvdy.20535. [DOI] [PubMed] [Google Scholar]
- 23.Hagiwara N, Yeh M, Liu A. Sox6 is required for normal fiber type differentiation of fetal skeletal muscle in mice. Dev Dyn. 2007;236:2062–2076. doi: 10.1002/dvdy.21223. [DOI] [PubMed] [Google Scholar]
- 24.von Hofsten J, et al. Prdm1- and Sox6-mediated transcriptional repression specifies muscle fibre type in the zebrafish embryo. EMBO Rep. 2008;9:683–689. doi: 10.1038/embor.2008.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dumitriu B, Dy P, Smits P, Lefebvre V. Generation of mice harboring a Sox6 conditional null allele. Genesis. 2006;44:219–224. doi: 10.1002/dvg.20210. [DOI] [PubMed] [Google Scholar]
- 26.Brüning JC, et al. A muscle-specific insulin receptor knockout exhibits features of the metabolic syndrome of NIDDM without altering glucose tolerance. Mol Cell. 1998;2:559–569. doi: 10.1016/s1097-2765(00)80155-0. [DOI] [PubMed] [Google Scholar]
- 27.Holloszy JO, Booth FW. Biochemical adaptations to endurance exercise in muscle. Annu Rev Physiol. 1976;38:273–291. doi: 10.1146/annurev.ph.38.030176.001421. [DOI] [PubMed] [Google Scholar]
- 28.Röckl KS, et al. Skeletal muscle adaptation to exercise training: AMP-activated protein kinase mediates muscle fiber type shift. Diabetes. 2007;56:2062–2069. doi: 10.2337/db07-0255. [DOI] [PubMed] [Google Scholar]
- 29.Connor F, Wright E, Denny P, Koopman P, Ashworth A. The Sry-related HMG box-containing gene Sox6 is expressed in the adult testis and developing nervous system of the mouse. Nucleic Acids Res. 1995;23:3365–3372. doi: 10.1093/nar/23.17.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ikeda T, et al. Identification and characterization of the human long form of Sox5 (L-SOX5) gene. Gene. 2002;298:59–68. doi: 10.1016/s0378-1119(02)00927-7. [DOI] [PubMed] [Google Scholar]
- 31.Talmadge RJ, Roy RR. Electrophoretic separation of rat skeletal muscle myosin heavy-chain isoforms. J Appl Physiol. 1993;75:2337–2340. doi: 10.1152/jappl.1993.75.5.2337. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.