Abstract
Structural vaccinology is an emerging strategy for the rational design of vaccine candidates. We successfully applied structural vaccinology to design a fully synthetic protein with multivalent protection activity. In Group B Streptococcus, cell-surface pili have aroused great interest because of their direct roles in virulence and importance as protective antigens. The backbone subunit of type 2a pilus (BP-2a) is present in six immunogenically different but structurally similar variants. We determined the 3D structure of one of the variants, and experimentally demonstrated that protective antibodies specifically recognize one of the four domains that comprise the protein. We therefore constructed a synthetic protein constituted by the protective domain of each one of the six variants and showed that the chimeric protein protects mice against the challenge with all of the type 2a pilus-carrying strains. This work demonstrates the power of structural vaccinology and will facilitate the development of an optimized, broadly protective pilus-based vaccine against Group B Streptococcus by combining the uniquely generated chimeric protein with protective pilin subunits from two other previously identified pilus types. In addition, this work describes a template procedure that can be followed to develop vaccines against other bacterial pathogens.
Keywords: backbone protein, isopeptide bonds, homology modeling
Structural vaccinology is a branch of structural biology that is emerging as a promising platform for the identification of effective protective antigens. This technology stems from the observation that epitopes inducing protective immune responses are restricted to specific domains within an immonogenic protein (1). Thus, once the domains are identified and expressed in a recombinant form, they can be used as potent immunogens devoid of other regions that are irrelevant from a vaccine standpoint. One of the advantages of working with protein domains is that they are typically structurally ordered, and can be easily handled in vitro, allowing the construction of synthetic fusion proteins hosting two or more immunogenic domains.
Streptococcus agalactiae, also known as Group B Streptococcus (GBS), is a Gram-positive pathogen that causes life-threatening pneumonia, sepsis, and meningitis in newborn and young infants (2). This microorganism is classified into 10 capsular polysaccharide serotypes, each antigenically and structurally unique. Although major efforts have been made in the development of multivalent capsular conjugate vaccines, currently there is no vaccine against GBS (3). To overcome serotype-specific immunity and the increasing number of nontypeable isolates, vaccines based on conserved protective proteins are highly desirable (4).
The most promising protein vaccine candidates selected so far are the structural subunits of pili that are long filamentous structures protruding from the bacterial surface (5–8), which play a key role in bacterial virulence and pathogenesis (9–13). Extensive analysis of a large panel of GBS isolates has revealed the presence of three pilus islands, PI-1, PI-2a, and PI-2b. All strains characterized so far have at least one, but more frequently two, of the three islands. Each island encodes a pilus composed of three structural proteins, two of which induce protective antibodies (8): the shaft-forming subunit or backbone protein (BP) and the major ancillary protein (AP1), which exhibits adhesin functions (9, 10). Moreover, DNA sequence analysis has shown that the three subunits in strains carrying the same island are highly conserved, with the exception of BP-2a, which is grouped into six main different immunologically variants (named 515, CJB111, DK21, H36B, 2603, and CJB110, based on their reference strain) (8). As a result, immunization with BP-2a induces variant-specific protection, but immunization with BP and AP1 from both PI-1 and PI-2b, and immunization with AP1 from PI-2a (AP-2a), induce pilus island-specific protection (8). However, although both BP and AP1 are significantly protective in animal models, BPs tend to perform better than AP1 (5, 8), a difference that is particularly highlighted in the in vitro opsonophagocytic assay (Fig. S1). This difference is likely to be linked to the relative abundance of the two subunits in the pilus, with BP forming the bulk of the pilus structure (7). Thus, considering their capacity to elicit high bactericidal antibody titers, the “ideal” vaccine should include all pilus BPs, a formulation however, that is demanding from a manufacturing standpoint because of the variability of BP-2a.
In an attempt to develop an easily producible and efficacious BP-based vaccine, we successfully applied structural vaccinology to the BP-2a protein. We determined the 3D structure of one of the six main BP-2a variants (BP-2a-515). Subsequently, we expressed the single domains into which the protein is structurally organized, in Escherichia coli, and defined the minimal domain carrying the protective epitopes. Finally, as the other five BP-2a variants have a similar structural organization, despite their primary structure differences, we fused together the protective domains of each of the six variants in a synthetic protein and demonstrated that this chimera is able to induce strong protection and opsonophagocytic activity against strains carrying all BP-2a variants.
Results
X-Ray Crystal Structure of BP-2a-515 Pilus Subunit.
With the aim to identify the domains carrying the protective epitopes, the crystal structure of BP-2a-515 was solved and refined at 1.75 Å resolution via molecular replacement. Data collection and refinement statistics are shown in Table S1. The crystal asymmetric unit contains a dimer of two independent chains (A: residues 192–640, and B: residues 190–641), each made up of three distinct domains that adopt a modified IgG-fold (14): D2 (residues 190–332), D3 (residues 333–455), and D4 (residues 456–641) (Fig. 1A). The observed BP-2a-515 dimer is not expected to occur in solution and it is a likely consequence of crystal packing, as indicated by the Protein Interfaces, Surfaces and Assemblies (PISA) Service (15) at the European Bioinformatics Institute (http://www.ebi.ac.uk/msd-srv/prot_int/pistart.html), which gave a PISA complexation significance score (CSS) of 0 (CSS scores range from 0 to 1, with increasing values reflecting a corresponding increase in complexation relevance), and by the fact that there are no strong interactions at the association interface (555 Å2). Moreover, as the dimer is “closed” (i.e., the monomers are arranged in a head-to-head fashion), the BP2a-515 dimer-association mode is not relevant for pili filament formation.
Fig. 1.
Structural analysis of BP-2a-515. (A) Ribbon representation of the crystal structure of BP-2a-515 (residues 190–640), illustrating the N and C termini, domains D2, D3, and D4, two potassium ions (blue spheres), and the three intramolecular isopeptide bonds (spheres). (B) Superimposition of BP-2a-515 (purple) with RrgB from Streptococcus pneumoniae (blue), highlighting the structural similarity between the two proteins. (C) Structural details of the D2, D3, and D4 domains in the regions involved in isopeptide bond formation. All images were generated using Pymol Version 1.1r1 (www.pymol.org).
Although crystallization was carried out using the full-length protein, ∼190 aa from the N terminus (D1 domain) were absent in the crystal, an observation also reported for its homolog, the pneumococcal RrgB pilus protein (14). The D1 domain is likely to be cleaved off during crystallization and, in fact, there is no available space in the crystal asymmetric unit that indicates that it could be present but conformationally disordered.
Three potassium cations (derived from potassium-sodium tartrate in the crystallization solution) are bound at strategic positions in the structure and act to stabilize flexible portions of the protein (Fig. 1A). Superimposition of the Cα atoms of BP-2a-515 chain B and RrgB, using the pairwise structural alignment Cα match program (http://bioinfo3d.cs.tau.ac.il/c_alpha_match/), indicates significant structural homology, with a rmsd value of 1.37 Å over 280/452 residues (Fig. 1B). The major structural differences between the two proteins regard the spatial location of the D3 domain, the movement of two α-helices in the D4 domain that are connected to the β-sandwich by two β-strands not present in RrgB, and flexible regions (Fig. 1B). Similarly to RrgB, BP-2a-515 contains three stabilizing isopeptide bonds between: Lys199 and Asn325 (D2 domain), Lys355 and Asn437 (D3 domain), and Lys463 and Asn636 (D4 domain) (Fig. 1C). The surrounding area around these bonds is largely hydrophobic, comprising several aromatic residues, in agreement with observations made for the isopeptide bonds in several pilus proteins.
The presence of the three intramolecular isopeptide bonds was experimentally confirmed by mass spectrometry (MS) using a recently developed protocol (SI Materials and Methods) (Fig. S2). BP-2a-515 was digested with the Lys-C endoprotease to generate peptides with a C-terminal lysine, and the digestion products were analyzed by MS, before and after incubation with O-methylisourea. Because the compound modifies the C-terminal lysine to homoarginine with a consequent mass increase of 42 Da, digestion products constituted by two peptides that are covalently linked by isopeptide bonds will show a mass shift after O-methylisourea treatment of either 42 or 84 Da, depending on whether the two C-terminal lysines are partially or completely derivatized. As shown in Fig. S2, ions of m/z 4040.85, 2145.18, and 1762.05 were assigned to linked Lys-C peptides bearing the isopeptide bonds observed in the crystal structure in domains D2, D3, and D4, respectively. Noteworthy, no isopeptide bond was identified in the N-terminal part corresponding to domain D1 of the full-length recombinant protein. Each of the four domains appears to fold independently, as demonstrated by expressing and purifying each domain, selecting the N and C termini based on the domain boundaries defined in the crystal structure of BP-2a-515 (Fig. 1A). All four domains were expressed in soluble form in E. coli, and MS analysis of tryptic digests of D2, D3, and D4 revealed that the domains carried the same isopeptide bonds found in the full-length protein. This finding suggested that the overall structural organization of the independently expressed domains was sufficiently preserved to bring the lysine and asparagine residues at a suitable reaction distance.
Domain D3 of the BP-2a 515 Allele Is the Most Important for Protection.
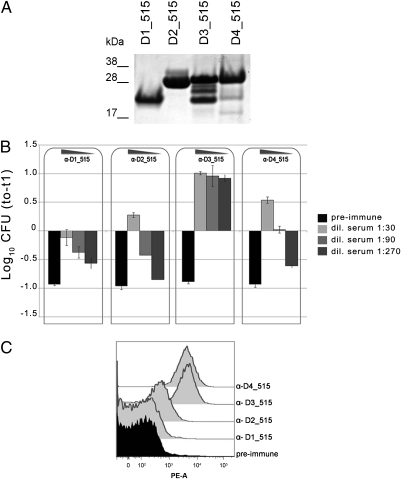
We then asked whether protective epitopes in BP-2a-515 were specifically concentrated in one of the four distinct domains. As said above, the four domains could be well expressed in E. coli as soluble His-tagged fusions, with D3 being the only domain undergoing partial degradation during expression/purification (Fig. 2A). The four purified domains were independently used to immunize CD1 mice and the protection activity of each was tested in an active maternal mouse immunization/neonatal pup challenge model and in an in vitro opsonophagocitosis assay. For the challenge, we used the strain 515 expressing the homologous BP-2a-515 variant. As shown in Table 1, domains D3 and D4, and not domains D1 and D2, conferred in vivo protection to a level comparable to that of full length protein. In terms of bactericidal activities, the D3 domain promoted the most significant complement-dependent bacterial killing (Fig. 2B), indicating that this region is the main target of protective antibodies. Interestingly, when the bactericidal activity of anti–full-length antibodies was compared with that elicited by the D3 domain, it appeared to be slightly superior (Figs. S1 and S4), in line with the observation that some bactericidal activity was also induced by D4 and D2 domains. In addition, flow cytometry analysis of whole GBS cells using domain-specific sera confirmed that the D3 and D4 domains were easily accessible to antibodies, but weak and no bacterial recognition was obtained with anti-D2 and anti-D1 antibodies, respectively (Fig. 2C).
Fig. 2.
Domain D3 of BP-2a-515 is essential for protection. (A) SDS/PAGE of the four single domains of BP-2a-515, expressed in and purified from E. coli, as His-tagged fusion proteins. (B) Opsonophagocitosis activity of sera from mice immunized with each domain. 104 CFUs of GBS 515 strain were incubated for 1 h with differentiated HL60 cells, baby rabbit complement, and mouse sera (1:30, 1:90, and 1:270 dilutions) collected from mice before (preimmune sera, black bars) and after immunization with each domain (gray scale bars). Error bars indicate SD from two independent experiments. (C) FACS analysis on 515 strain with mouse sera raised against each domain of BP-2a-515. Fixed bacteria were stained with antisera and then labeled with R-Phycoerythrin conjugated goat anti-mouse secondary antibodies. Black histogram indicates staining of bacteria with preimmune serum.
Table 1.
Protection conferred by single domains of BP-2a-515 allele against GBS strain 515, assessed by active maternal mouse immunization/neonatal pup challenge model
| Antigen | Protected/treated | Protection (%) | Statistical significance (P value)* |
| D1-515 | 19/59 | 24 | 0.0098 |
| D2-515 | 7/25 | 20 | 0.0687 |
| D3-515 | 21/28 | 72 | P < 0.0001 |
| D4-515 | 42/60 | 67 | P < 0.0001 |
| Full-length BP-2a-515 | 44/60 | 70 | P < 0.0001 |
| BP-2a-515K199A/K355A/K463A | 28/38 | 71 | P < 0.0001 |
| PBS | 4/39 |
Groups of female mice received three doses (on days 1, 21, and 35) of either 20 μg antigen or buffer (PBS) combined with Freund's adjuvant. Mice were then mated, and their offspring were challenged with a GBS dose calculated to induce death in 90% of the pups. *P value, by Fisher's exact test.
D3 Domain from All BP-2a Variants Has Similar Structural and Protective Features.
Having shown that in the BP-2a-515 variant protective epitopes are mostly concentrated in the D3 domain, we investigated whether similar structural and protective properties occur in the other BP-2a variants. To this aim we performed a structural analysis of the other variants by comparative homology modeling using, as a template, the crystal structure of BP-2a-515. In particular, we modeled the BP variants from strains H36B and CJB111, which are the most evolutionary distant and closest variants, among the other five variants of the 515 allele, respectively. Even for BP-2a from H36B, a protein which shares 42.2% sequence identity (62.9% similarity) with the 515 variant, the model adopted a similar modular domain organization, comprising three IgG-like fold domains, each containing an internal Lys–Asn isopeptide bond within relatively conserved surroundings. The only relevant difference observed in the H36B model structure was the presence of two insertion loops (spanning residue 211–222 and residue 373–376), which are not present in the 515 variant structure (Fig. S3). Conversely, as expected for a protein that shares 76% sequence identity, the homolog model built for BP-2a from the CJB111 strain was essentially superimposable with the 515 variant.
The modeling analyses strengthened the idea that the overall topological organization of the six BP-2a variants is remarkably conserved, suggesting that the same domain boundaries used for the 515 variant could be used to study/validate domain functions in the other variants. Therefore, we investigated whether the D3 domain of BP-2a variants from the CJB111 and H36B strains were also the major players in eliciting protection. We divided both proteins into four partly overlapping fragments, each carrying one of the four domains, and evaluated the levels of protection induced by each fragment in the active maternal immunization mouse model. As observed for the 515 variant, the most significant levels of protection were obtained upon immunization with fragments carrying the D3 domain (Tables S2 and S3).
In conclusion, the data described above indicate that all six BP-2a variants share a similar four-domain organization and that the D3 domain of the three variants analyzed carries most of the epitopes that induce protective antibody responses.
Fusion Protein Carrying Protective D3 Domains from the Six Different BP-2a Alleles Confers Cross-Protection in Mice.
The discovery that protective epitopes in BP-2a reside in a single domain, which can be easily cloned and expressed in recombinant form, led us to design a synthetic protein and test whether it could induce broad protection against all GBS strains carrying PI-2a pili. To this aim, we generated a chimeric protein (6xD3) by fusing the D3 domains of BPs from six PI-2a reference GBS strains: 515, CJB111, H36B, DK21, CJB110, and 2603 (8). The domains were intercalated by Gly-Ser-Gly-Ser spacers to provide enough flexibility to the structure, thus preventing possible unfavorable interactions (Fig. 3A). The chimeric protein was expressed in, and purified from, E. coli fused to a N-terminal His-tag. Interestingly, although the single D3 domain appeared to be partially degraded during the expression in E. coli, the synthetic protein was purified as a single stable species (Fig. 3B). The protein was then analyzed in the animal model to evaluate its ability to confer broad protection. As shown in Table 2, the 6xD3 chimera was able to elicit protective immunity in mice challenged with six different GBS strains, each expressing a different variant of BP-2a. Protection was clearly mediated by the elicitation of opsonophagocytosis antibodies, as confirmed by the in vitro, complement-dependent, phagocytic killing of the same GBS strains used in the mouse-challenge model (Fig. 3C). Interestingly, the killing activity observed with anti-6xD3 serum was always higher than that obtained with antibodies against the major ancillary protein from the same pilus type (AP1-2a) (Fig. 3C). In accordance with these data, the anti-6xD3 mouse serum was also able to recognize the pilus-like structures on the bacterial surface as revealed by FACS analysis on whole bacterial cells (Fig. 3D).
Fig. 3.
Opsonophagocytic activity and FACS analysis of sera from mice immunized with the fusion protein 6xD3. (A) Schematic representation of the chimeric protein 6xD3. The amino acid boundaries of each domain as deduced from the 3D structures, the Histidine tag and the stop codon are indicated. Domains are intercalated by Gly-Ser-Gly-Ser spacers. (B) SDS/PAGE of the chimera 6xD3 expressed in, and purified from E. coli as an N-terminally His-tagged protein. Total protein extracts from E. coli BL21 (DE3) transformed with the pET vector carrying 6xD3 gene, before induction with IPTG (lane 1) after induction (lane 2), from the soluble fraction (lane 3), and purified 6xD3 protein after affinity chromatography and gel filtration of the soluble fraction (lane 4). Gels were Coomassie Blue G-250 stained. (C) Opsonophagocitosis activity of sera from mice immunized with the fusion protein 6xD3 (gray bars), preimmune sera from the same animals (black bars), and sera from mice immunized with AP1-2a (light-gray bars). Six different strains expressing different BP-2a variants were used in the assay, strain CJB111 (CJB111 allele); strain 515 (515 allele); 5401 (H36B allele); 3050 (2603 allele); DK21 (DK21 allele); and strain CDC89 (CJB110 allele). Error bars indicate SD of two independent experiments. (D) Flow cytometry analysis on whole fixed bacteria corresponding to six strains, as in C, probed with mouse antiserum anti chimera 6xD3 and labeled with R-Phycoerythrin conjugated goat anti-mouse secondary antibodies.
Table 2.
Protection by active maternal mouse immunization/neonatal pup challenge model conferred by fusion protein 6xD3 against a panel of GBS strains expressing different BP-2a allelic variants
| GBS challenge strain | BP-2a allele | Antigen 6xD3 (protected/treated) | PBS (protected/treated) | Protection (%) |
| 515 | 515 | 50/68 | 13/50 | 65* |
| CJB111 | CJB111 | 38/48 | 7/30 | 73* |
| 3050 | 2603 | 53/70 | 12/40 | 66* |
| 5401 | H36B | 22/30 | 11/40 | 63* |
| DK21 | DK21 | 29/38 | 6/29 | 70* |
| CDC89 | CJB110 | 26/40 | 6/26 | 55* |
Groups of female mice received three doses (on days 1, 21, and 35) of either 20 μg antigen or buffer (PBS) combined with Freund's adjuvant. Mice were then mated, and their offspring were challenged with a GBS dose calculated to induce death in 90% of the pups. *P value, P < 0.0001 by Fisher's exact test.
Discussion
The data presented in this work convey three main messages of major scientific relevance.
First, this article provides the crystal structure of a pilus subunit from Streptococci, thus contributing to the understanding of the structural organization of these organelles playing key roles in virulence and pathogenesis. Crystal structures of four backbone subunits, Streptococcus pyogenes Spy0128 (16), Bacillus cereus BcpA (17), Corynebacterium diphtheriae Spa (18), and Streptococcus pneumoniae RrgB (14), are available and have revealed unique structural-functional properties. Despite variation in size and relatively low sequence similarity, backbone pilins show a very similar tertiary structure, organized in IgG-like domains, typical of bacterial cell-surface adhesins known as MSCRAMMs (Microbial Surface Components Recognizing Adhesive Matrix Molecules). Furthermore, they carry intramolecular isopeptide bonds, which represent the Gram-positive counterparts of disulfide bonds found in Gram-negative bacterial pili. Isopeptide bonds are the result of an autocatalytic reaction that renders pili extremely resistant to proteolysis and high temperatures (19, 20).
The high-resolution crystal structure of GBS pilus type 2a backbone subunit (BP-2a-515), described in this work, provides further evidence of the overall structural conservation of Gram-positive pilin subunits. Similarly to other backbone proteins, BP-2a-515 is organized into four major structural domains, three of which (D2, D3, and D4) house the isopeptide bonds. Our data show that the four domains are endowed with self-assembly properties, demonstrated by the observation that each of them can be expressed in soluble form in E. coli. Moreover, formation of the isopeptide bonds occurs equally well in the independently expressed D2, D3 and D4 domains. Interestingly, we have also recently observed that, although isopeptide bonds are important for the stability of pilus subunits, they are not required for determining the overall final domain folding. This finding was indirectly demonstrated by replacing the lysine residues of BP-2a-515 involved in isopeptide bonds with alanine residues, and analyzing if this mutant could still elicit opsonophagocytic antibodies and protection in the animal model. Indeed, the mutant that expressed well in E. coli as a soluble protein showed the same immunological behavior as the native protein (Table 1 and Fig. S4). Therefore, it is likely that all pilus subunits originate from a common heat-labile ancestor, which subsequently acquires temperature (and proteolysis) resistance by inserting highly stable domains into the most exposed regions.
The second important message regards the potential of structural biology in developing efficacious vaccines. An increasing number of elegant publications underlies the relevance of this emerging technology in vaccinology. For example, the immunogenic epitopes of the influenza virus hemoagglutinin (HA) have been mapped on the 3D structure of the viral protein. From these studies, it appears that most of the neutralizing antibodies target regions of the protein that are not functionally relevant. This process allows the virus to readily escape antibody binding to HA by continuously mutating the antibody binding sites. Recently, Ekiert et al. selected high affinity neutralizing monoclonal antibodies from convalescent patients that target a well-conserved region of HA involved in membrane fusion (21). This region is naturally poorly immunogenic but future protein engineering strategies applied to HA could lead to a durable and cross-protective universal vaccine against influenza A. In human CMV, Macagno et al. showed that high-affinity neutralizing antibodies recognize conformational epitopes shared by more than one protein of the gH/gL/UL128-131A protein complex (22). This fascinating observation anticipates the possible failure in eliciting protective immunity using single proteins as immunogens, and paves the way to the development of efficacious anti-CMV vaccines, based on the three-protein complex with preserved quaternary structural organization.
Data reported in this work show how structural biology can support vaccine development by solving a problem related to manufacturing. The backbone subunit of GBS pilus 2a is a highly protective antigen. However, it is variable and, to protect against all GBS strains carrying this pilus, the combination of al least six major protein variants would be necessary, which poses an obvious challenge from a manufacturing viewpoint. The discovery that most of the protective epitopes in BP-2a are localized in a single protein domain that maintains its conformation when expressed in E. coli allowed us to construct a single fusion protein including the protective domains from all six BP-2a variants. The fusion protein protected mice against challenge with any of the pilus 2a-carrying strains, and protection was mediated by the elicitation of opsonophagocytic antibodies.
Finally, in this work we provide data that pave the way to the development of an optimized broadly protective GBS vaccine. GBS infections still represent a serious threat for newborns despite the intrapartum antibiotic prophylaxis of GBS-colonized pregnant women adopted by most industrialized countries. Because children delivered from women with high titers of anti-GBS opsonophagocytic antibodies are protected from GBS infections (23, 24), vaccination of women in child-bearing age or pregnant women appears to be the ideal long-term solution to newborn infections. We have shown that all GBS isolates carry pili, whose subunits induce protection in mice (8). Moreover, these subunits are highly conserved, with the exception of the backbone protein of pilus 2a, which is separated into six main alleles (8). Although a three-pilin combination, including the backbone proteins from PI-1 (BP-1) and PI-2b (BP-2b) and the ancillary protein of pilus 2a (AP1-2a), induced protective responses in mice challenged with different GBS serotypes (8), the ideal combination should include all backbone proteins for their ability to induce the highest opsonophagocytic titers. This is a solution that, in the absence of the data presented in this work, would have requested the design, production and testing of eight recombinant proteins (BP -1, BP-2b, and the six alleles of BP-2a). The possibility to combine the protective activities of the six BP-2a alleles in a single synthetic construct renders practically feasible the development of a protein-based vaccine, constituted by the three most potent pilus antigens described so far (i.e., BP-1, BP-2b, and 6xD3). Although not shown in this work, we predict that this optimized pilus-based vaccine could be effective against most of the circulating GBS strains.
Materials and Methods
Protein Crystallization.
Crystallization trials were set up in 96-well microbatch plates (Greiner) using the Orxy 8.0 crystallization robot (Douglas Instruments). Crystals of BP-2a-515 grew after 1 to 2 wk at 20 °C in a 0.5-μL drop consisting of 0.3 μL protein (180 mg/mL) in 10 mM Hepes pH 7.0 and 0.2 μL crystallization solution [25% (wt/vol) PEG 4000, 0.1 M Hepes pH 7.0 and 90 mM potassium sodium tartrate tetrahydrate], layered with silicon oil and paraffin, mixed at a ratio of 1:1. Crystals were cryoprotected in the crystallization solution containing increased precipitant concentration [40% (wt/vol) PEG4000]. Crystals belong to the orthorhombic P212121 space group with an estimated solvent content of 53%.
Structure Solution and Refinement.
Diffraction data from a single crystal of BP-2a-515 were collected at a resolution of 1.75 Å at the European Synchrotron Radiation Facility (Grenoble, France; beam line ID23-1). Data were processed using programs available from the CCP4 Program Suite (25) and the crystal structure was solved by molecular replacement using the structure of RrgB from S. pneumoniae (14) (PDB 2X9W), as a search model. The structure was refined to give satisfactory refinement and geometric parameter (Table S1). Additional details are provided in SI Materials and Methods. Atomic coordinates and structure factors for residues 190 to 640 of BP-2a-515 have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org) under accession PDB code 2XTL (26). The nucleotide sequence accession number is ZP_00789556.1.
Cloning and Protein Purification.
The full-length BP-2a proteins, corresponding to 515 and CJB111 alleles, were produced as previously reported (8), and the full-length H36B allele was cloned in pET24b+ (Novagen) using strain H36B as source of DNA. DNA encoding the single domains (D1, D2, D3, and D4) of the 515, CJB111, and H36B alleles were PCR amplified, cloned into the pENTR⁄TEV⁄D-TOPO vector (Invitrogen) and then into pET54 or pET59 vectors (Novagen) using the GATEWAY cloning system (Invitrogen), with the exception of the D3 domain of H36B allele, which was cloned into the pSpeedET vector by the PIPE (Polymerase Incomplete Primer Extension) method (27). The oligos used are listed in Table S4. The gene coding for the fusion protein 6xD3 was synthetically constructed by GENEART. The 6xD3 gene was subcloned into pET15, and adapted in-house using PIPE cloning in the E. coli HK100 strain. Proteins were expressed in E. coli BL21 (DE3) (Novagen) cells as His- or TRX-tagged fusion proteins and purified by affinity chromatography and gel filtration. Additional details are provided in SI Materials and Methods.
Mouse Immunization and Flow Cytometry Analysis.
Antisera specific for each protein were produced by immunizing CD1 mice with the purified recombinant proteins, as previously described (5). Protein-specific immune responses (total Ig) in pooled sera were monitored by ELISA. FACS analysis were performed as described elsewhere (7). Additional details are provided in SI Materials and Methods.
Opsonophagocytosis Assay.
The opsonophagocytosis assay was performed using GBS strains as target cells and HL-60 cell line (ATCC; CCL-240), differentiated into granulocyte-like cells, by adding 100 mM N, N dimethylformamide (Sigma) to the growth medium for 4 d. Midexponential bacterial cells were incubated at 37 °C for 1 h in the presence of phagocytic cells, 10% baby rabbit complement (Cedarlane), and heat-inactivated mouse antisera. Negative controls consisted of reactions either with preimmune sera, or without HL-60, or with heat-inactivated complement. The amount of opsonophagocytic killing was determined by subtracting the log of the number of colonies surviving the 1-h assay from the log of the number of CFU at the zero time point.
Mouse Active Maternal Immunization Model.
A mouse maternal immunization/pup challenge model of GBS infection was used to verify the protective efficacy of the antigens, as previously described (5). In brief, groups of four to eight CD-1 female mice (age, 6–8 wk) were immunized on days 1, 21, and 35 with 20 μg of antigen or buffer (PBS) formulated in Freund's adjuvant. Mice were then mated, and their offspring were challenged intraperitoneally with a GBS dose calculated to induce dead in 90% of the pups. Protection values were calculated as [(% dead in control – % dead in vaccine)/% dead in control] × 100. Mice were monitored on a daily basis and killed when they exhibited defined humane endpoints that had been pre-established for the study in agreement with Novartis Animal Welfare Policies. All animal studies were performed according to guidelines of the Istituto Superiore di Sanità (Italy). Statistical analysis was performed using Fisher's exact test.
Supplementary Material
Acknowledgments
We thank Marco Tortoli and the Animal Care Facility at Novartis Siena for expert technical assistance and Antonietta Maiorino for her expert secretarial assistance and the careful editing of the manuscript. Part of this work was funded by the European Community FP6 project, “Assessment of Structural Requirements in Complement-Mediated Bactericidal Events: Towards a Global Approach to the Selection of New Vaccine Candidates” (BacAbs) (Contract LSH-2005-037325), and by a Ministero dell'Istruzione, dell'Università e della Ricerca Prin-2008 (Project 2008K37RHP). L.J.G. is a recipient of Assegno di Ricerca (2009) from the University of Milan.
Footnotes
The authors declare no conflict of interest.
See Commentary on page 10029.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1106590108/-/DCSupplemental.
References
- 1.Dormitzer PR, Ulmer JB, Rappuoli R. Structure-based antigen design: A strategy for next generation vaccines. Trends Biotechnol. 2008;26:659–667. doi: 10.1016/j.tibtech.2008.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johri AK, et al. Group B Streptococcus: Global incidence and vaccine development. Nat Rev Microbiol. 2006;4:932–942. doi: 10.1038/nrmicro1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker CJ, Edwards MS. Group B streptococcal conjugate vaccines. Arch Dis Child. 2003;88:375–378. doi: 10.1136/adc.88.5.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Edwards MS, Rench MA, Palazzi DL, Baker CJ. Group B streptococcal colonization and serotype-specific immunity in healthy elderly persons. Clin Infect Dis. 2005;40:352–357. doi: 10.1086/426820. [DOI] [PubMed] [Google Scholar]
- 5.Maione D, et al. Identification of a universal Group B Streptococcus vaccine by multiple genome screen. Science. 2005;309:148–150. doi: 10.1126/science.1109869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lauer P, et al. Genome analysis reveals pili in Group B Streptococcus. Science. 2005;309:105. doi: 10.1126/science.1111563. [DOI] [PubMed] [Google Scholar]
- 7.Rosini R, et al. Identification of novel genomic islands coding for antigenic pilus-like structures in Streptococcus agalactiae. Mol Microbiol. 2006;61:126–141. doi: 10.1111/j.1365-2958.2006.05225.x. [DOI] [PubMed] [Google Scholar]
- 8.Margarit I, et al. Preventing bacterial infections with pilus-based vaccines: The group B streptococcus paradigm. J Infect Dis. 2009;199:108–115. doi: 10.1086/595564. [DOI] [PubMed] [Google Scholar]
- 9.Dramsi S, et al. Assembly and role of pili in group B streptococci. Mol Microbiol. 2006;60:1401–1413. doi: 10.1111/j.1365-2958.2006.05190.x. [DOI] [PubMed] [Google Scholar]
- 10.Pezzicoli A, et al. Pilus backbone contributes to group B Streptococcus paracellular translocation through epithelial cells. J Infect Dis. 2008;198:890–898. doi: 10.1086/591182. [DOI] [PubMed] [Google Scholar]
- 11.Rinaudo CD, et al. Specific involvement of pilus type 2a in biofilm formation in group B Streptococcus. PLoS ONE. 2010;5:e9216. doi: 10.1371/journal.pone.0009216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mandlik A, Swierczynski A, Das A, Ton-That H. Pili in Gram-positive bacteria: Assembly, involvement in colonization and biofilm development. Trends Microbiol. 2008;16:33–40. doi: 10.1016/j.tim.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Konto-Ghiorghi Y, et al. Dual role for pilus in adherence to epithelial cells and biofilm formation in Streptococcus agalactiae. PLoS Pathog. 2009;5:e1000422. doi: 10.1371/journal.ppat.1000422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spraggon G, et al. Supramolecular organization of the repetitive backbone unit of the Streptococcus pneumoniae pilus. PLoS ONE. 2010;5:e10919. doi: 10.1371/journal.pone.0010919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 16.Kang HJ, Coulibaly F, Clow F, Proft T, Baker EN. Stabilizing isopeptide bonds revealed in Gram-positive bacterial pilus structure. Science. 2007;318:1625–1628. doi: 10.1126/science.1145806. [DOI] [PubMed] [Google Scholar]
- 17.Budzik JM, et al. Amide bonds assemble pili on the surface of bacilli. Proc Natl Acad Sci USA. 2008;105:10215–10220. doi: 10.1073/pnas.0803565105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kang HJ, Paterson NG, Gaspar AH, Ton-That H, Baker EN. The Corynebacterium diphtheriae shaft pilin SpaA is built of tandem Ig-like modules with stabilizing isopeptide and disulfide bonds. Proc Natl Acad Sci USA. 2009;106:16967–16971. doi: 10.1073/pnas.0906826106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alegre-Cebollada J, Badilla CL, Fernández JM. Isopeptide bonds block the mechanical extension of pili in pathogenic Streptococcus pyogenes. J Biol Chem. 2010;285:11235–11242. doi: 10.1074/jbc.M110.102962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Budzik JM, et al. Intramolecular amide bonds stabilize pili on the surface of bacilli. Proc Natl Acad Sci USA. 2009;106:19992–19997. doi: 10.1073/pnas.0910887106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ekiert DC, et al. Antibody recognition of a highly conserved influenza virus epitope. Science. 2009;324:246–251. doi: 10.1126/science.1171491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Macagno A, et al. Isolation of human monoclonal antibodies that potently neutralize human cytomegalovirus infection by targeting different epitopes on the gH/gL/UL128-131A complex. J Virol. 2010;84:1005–1013. doi: 10.1128/JVI.01809-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baker CJ, Kasper DL. Correlation of maternal antibody deficiency with susceptibility to neonatal group B streptococcal infection. N Engl J Med. 1976;294:753–756. doi: 10.1056/NEJM197604012941404. [DOI] [PubMed] [Google Scholar]
- 24.Lin FY, et al. Level of maternal IgG anti-group B streptococcus type III antibody correlated with protection of neonates against early-onset disease caused by this pathogen. J Infect Dis. 2004;190:928–934. doi: 10.1086/422756. [DOI] [PubMed] [Google Scholar]
- 25.Evans PR. Proceedings of the CCP4 Study Weekend on Data Collection and Processing. United Kingdom: CLRC Daresbury Laboratory, Warrington; 1993. pp. 114–122. [Google Scholar]
- 26.Berman HM, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klock HE, Lesley SA. The Polymerase Incomplete Primer Extension (PIPE) method applied to high-throughput cloning and site-directed mutagenesis. Methods Mol Biol. 2009;498:91–103. doi: 10.1007/978-1-59745-196-3_6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.