In the field of protein folding, the membrane is uncharted territory. The energetic factors that govern the folding and stability of a protein in the hydrophobic lipid bilayer are sparsely understood. To probe these thermodynamic influences, several energy scales have been determined that measure the energy to transfer an amino acid into a membrane-like environment: the Wimley-White (WW) scale (1, 2), which measures residue partitioning between water and octanol, and the translocon scale, which measures the partitioning of residues on an α-helical segment from the translocon machinery into the membrane (3). Although these hydrophobicity scales provide energetic benchmarks, they do not convey the actual equilibrium change in free energy (ΔGo) of transferring a residue from the water into a real membrane core, in the context of a membrane protein structure.
In PNAS, Moon and Fleming (4) overcome this obstacle by developing a membrane protein system that reversibly and spontaneously folds between the aqueous solution and the lipid bilayer, enabling the measurement of the free energy of putting a side chain into the bilayer core. To do this, the authors use a unique host–guest system: the outer membrane bacterial phospholipase OmpLA as the host and amino acid hitchhiking onto the OmpLA backbone as the guest. By measuring tryptophan fluorescence and phospholipase activity, they determine the population of unfolded vs. native protein as a function of denaturant concentration. From these results they are able to measure the energetic impact of each residue on the free energy of folding and derive the Moon-Fleming whole-protein hydrophobicity scale.
This sounds like a “classic” protein folding experiment, but it is anything but. It has been notoriously difficult to do these types of studies with membrane proteins because the folding pathways are more complex and are less understood. At some point, a membrane protein must partition into the lipid membrane on its way to a folded, functional structure. In both prokaryotic and eukaryotic cells this is often assisted by the translocon, a protein pore that chaperones the folding and insertion process (5, 6). However, it is not clear how this happens—whether proteins partition one transmembrane segment at a time and then come together to fold in the membrane or instead fold modularly in the translocon and then partition into the lipid core. In any case, it has been difficult to reproduce this in an experimental setting, and most models of membrane protein stability and folding are either constrained to single α-helices that spontaneously insert from solution (7, 8) or partition from the translocon pore (3).
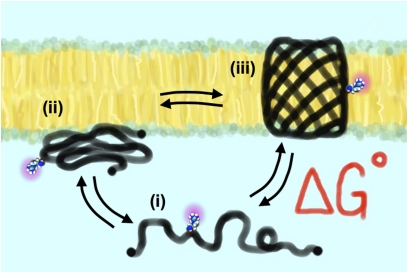
It turns out that OmpLA folds in a very different way. Folding of β-barrel membrane proteins does not involve the translocon (9). Instead, there is equilibrium between the unfolded aqueous state, a prefolded state that spontaneously inserts into the membrane, and the final membrane imbedded structure (Fig. 1). From many years of work, the Fleming group and others (10–13) have detailed the energetic factors influencing this process. In their current study, Moon and Fleming work out the final trick for completely reversible folding into the lipid bilayer: low pH. Using this, they sculpt OmpLA into a spontaneous, reversible folding model for a functionally relevant membrane protein structure.
Fig. 1.
Reversible, spontaneous OmpLA folding system. At low pH and in the presence of denaturants, the β-barrel protein is in equilibrium between (i) a soluble unfolded state, (ii) a prefolded intermediate that spontaneously inserts into the lipid bilayer, and (iii) the final folded functional state of the protein. The free energy of the folding, ΔG°, is measured by quantifying the unfolded and folded populations. Moon and Fleming (4) use OmpLA as a host system and measure the change in free energy each residue confers when constrained to the center of the lipid bilayer, providing a whole-protein hydrophobicity energy scale for the 20 natural amino acids, relative to alanine.
With this advancement, their hydrophobicity scale is the closest in reflecting the actual energy of taking a residue from the water and putting it into the membrane core. How does the Moon-Fleming scale compare with previous hydrophobicity scales? In general it is similar to the WW water-octanol scale, except that it shows transfer energies that are larger at both extremes; there is a higher energetic cost in putting polar and charged residues into the membrane, whereas hydrophobic residues are more favorable (see figure S5 in ref. 4 for a direct comparison). The differences between the two scales could arise from the fact that OmpLA constrains the amino acid at the center of the membrane, whereas partitioning of residues into octanol is isotropic. The authors examine this further by introducing mutations of leucine or arginine at varying depths within the bilayer, and they demonstrate the critical role of membrane “solvent” anisotropy in the energetic contribution to protein stability.
Finally, the authors go on to study a controversial question: what is the cost of putting a charged arginine into the lipophilic core? Beyond being a general curiosity in the protein-folding field, this is also an important question for understanding the mechanism of voltage gating in ion channels. Excitable Na+, Ca2+, and K+ channels all contain a voltage-sensing domain (VSD) comprising four α-helices, with one of these segments, S4, containing up to eight positive charges (14). Electrostatic theory dictates that putting charges into the lipid bilayer is energetically prohibitive, and so there is an ongoing debate regarding how the many VSD charges are stabilized inside the membrane core. The Moon-Fleming whole-protein energy scale shows that this energy is a modest +2 kcal/mole for the arginine side chain alone (+4 kcal/mole stability difference relative to alanine). It is interesting to note that lysine is measured as the most destabilizing residue on the scale, which may help explain why arginines are preferred over lysines in VSD sequences. Furthermore, when arginine is already at the center of the membrane, addition of another arginine in close vicinity shifts the stability by only +1 kcal/mole. Numerous computer simulations have shown that water pathways form to allow for “snorkeling” of the side chain (15–17), and this may lead to the observed ease of bringing a second arginine residue into the membrane.
With this work the authors have made a strong declaration: we choose to go to the membrane, not because it is easy but because it is hard. They have solved the hard problem of developing a tractable system to study how proteins are stabilized in a hydrophobic environment. They are further benefited by the fact that the OmpLA β-barrel structure is robust and will hopefully withstand the significant perturbations that are required for studying interesting questions (e.g., different lipid environments, hydrophobic mismatch, and pH dependency of membrane insertion). Even though this is just one model of a membrane protein system, the measurements themselves are true thermodynamic values, and in this respect provide us with unique benchmarks for membrane protein energetics in general.
Footnotes
The author declares no conflict of interest.
See companion article on page 10174.
References
- 1.White SH, Wimley WC. Hydrophobic interactions of peptides with membrane interfaces. Biochim Biophys Acta. 1998;1376:339–352. doi: 10.1016/s0304-4157(98)00021-5. [DOI] [PubMed] [Google Scholar]
- 2.White SH, Wimley WC. Membrane protein folding and stability: Physical principles. Annu Rev Biophys Biomol Struct. 1999;28:319–365. doi: 10.1146/annurev.biophys.28.1.319. [DOI] [PubMed] [Google Scholar]
- 3.Hessa T, et al. Recognition of transmembrane helices by the endoplasmic reticulum translocon. Nature. 2005;433:377–381. doi: 10.1038/nature03216. [DOI] [PubMed] [Google Scholar]
- 4.Moon CP, Fleming KG. Side-chain hydrophobicity scale derived from transmembrane protein folding into lipid bilayers. Proc Natl Acad Sci USA. 2011;108:10174–10177. doi: 10.1073/pnas.1103979108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.White SH, von Heijne G. The machinery of membrane protein assembly. Curr Opin Struct Biol. 2004;14:397–404. doi: 10.1016/j.sbi.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 6.Rapoport TA, Goder V, Heinrich SU, Matlack KE. Membrane-protein integration and the role of the translocation channel. Trends Cell Biol. 2004;14:568–575. doi: 10.1016/j.tcb.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 7.Hunt JF, Rath P, Rothschild KJ, Engelman DM. Spontaneous, pH-dependent membrane insertion of a transbilayer alpha-helix. Biochemistry. 1997;36:15177–15192. doi: 10.1021/bi970147b. [DOI] [PubMed] [Google Scholar]
- 8.Wimley WC, White SH. Designing transmembrane alpha-helices that insert spontaneously. Biochemistry. 2000;39:4432–4442. doi: 10.1021/bi992746j. [DOI] [PubMed] [Google Scholar]
- 9.Tamm LK, Arora A, Kleinschmidt JH. Structure and assembly of beta-barrel membrane proteins. J Biol Chem. 2001;276:32399–32402. doi: 10.1074/jbc.R100021200. [DOI] [PubMed] [Google Scholar]
- 10.Buchanan SK. Beta-barrel proteins from bacterial outer membranes: Structure, function and refolding. Curr Opin Struct Biol. 1999;9:455–461. doi: 10.1016/S0959-440X(99)80064-5. [DOI] [PubMed] [Google Scholar]
- 11.Stanley AM, Fleming KG. The process of folding proteins into membranes: Challenges and progress. Arch Biochem Biophys. 2008;469:46–66. doi: 10.1016/j.abb.2007.09.024. [DOI] [PubMed] [Google Scholar]
- 12.Burgess NK, Dao TP, Stanley AM, Fleming KG. Beta-barrel proteins that reside in the Escherichia coli outer membrane in vivo demonstrate varied folding behavior in vitro. J Biol Chem. 2008;283:26748–26758. doi: 10.1074/jbc.M802754200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dekker N, Merck K, Tommassen J, Verheij HM. In vitro folding of Escherichia coli outer-membrane phospholipase A. Eur J Biochem. 1995;232:214–219. doi: 10.1111/j.1432-1033.1995.tb20801.x. [DOI] [PubMed] [Google Scholar]
- 14.Hille B. Ion Channels of Excitable Membranes. Sunderland, MA: Sinauer Associates; 2001. pp. 605–608. [Google Scholar]
- 15.Dorairaj S, Allen TW. On the thermodynamic stability of a charged arginine side chain in a transmembrane helix. Proc Natl Acad Sci USA. 2007;104:4943–4948. doi: 10.1073/pnas.0610470104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.MacCallum JL, Bennett WF, Tieleman DP. Partitioning of amino acid side chains into lipid bilayers: Results from computer simulations and comparison to experiment. J Gen Physiol. 2007;129:371–377. doi: 10.1085/jgp.200709745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schow EV, et al. Arginine in membranes: The connection between molecular dynamics simulations and translocon-mediated insertion experiments. J Membr Biol. 2011;239:35–48. doi: 10.1007/s00232-010-9330-x. [DOI] [PMC free article] [PubMed] [Google Scholar]