Abstract
Adhesion of circulating monocytes to vascular endothelial cells (ECs) is a critical event leading to vascular inflammation and, hence, development of atherosclerosis. MicroRNAs (miRs) are a class of endogenous, highly conserved, noncoding small RNAs that play important roles in regulating gene expression and cellular function, as well as pathogenesis of atherosclerosis. Here, we showed that oscillatory shear stress (OSS) induces the expression of miR-21 at the transcriptional level in cultured human umbilical vein ECs via an increased binding of c-Jun, which is a component of transcription factor activator protein-1 (AP-1), to the promoter region of miR-21. OSS induction of miR-21 inhibited the translation, but not transcription, of peroxisome proliferators-activated receptor-α (PPARα) by 3′-UTR targeting. Overexpression of miR-21 up-regulated AP-1 activation, which was attenuated by exogenous expression of PPARα. OSS and overexpression of miR-21 enhanced the expression of adhesion molecules vascular cell adhesion molecule-1 and monocyte chemotactic protein-1 and the consequential adhesion of monocytes to ECs. Overexpression of PPARα significantly attenuated the AP-1–mediated miR-21 expression. These results demonstrate a unique mechanism by which OSS induces AP-1–dependent miR-21 expression, which directly targets PPARα to inhibit its expression, thereby allowing activation of AP-1 and the promotion of monocyte adhesion. Our findings suggest the presence of a positive feedback loop that enables the sustained induction of miR-21, thus contributing to the proinflammatory responses of vascular endothelium under OSS.
Keywords: noncoding RNA, endothelial inflammatory response, feedback control, hemodynamics, atherogenesis
Vascular endothelial cells (ECs) are constantly exposed to hemodynamic shear stress, which regulates EC function and influences the development of vascular pathologies, including atherosclerosis (1). The pulsatile shear stress (PSS) in the straight part of the arteries, with its significant forward direction, is atheroprotective. Atherosclerosis occurs preferentially at arterial branches and curvatures, where the shear stress is low and oscillates back and forth (2, 3). The oscillatory shear stress (OSS) at these loci up-regulates proinflammatory molecules in ECs and, hence, enhances the adhesion of circulating monocytes to ECs (1). Increased expressions of proatherogenic genes, including vascular cell adhesion molecule-1 (VCAM-1) and monocyte chemotactic protein-1 (MCP-1), have been observed at the inner curvature of aortic arch and the orifices of arch branches (2, 4) and in cultured ECs subjected to prolonged OSS (5–7). Activator protein-1 (AP-1), which is composed of c-Jun/c-Jun and c-Jun/c-Fos protein dimers and has been shown to regulate VCAM-1 and MCP-1 transcriptions (8, 9), can be activated at both the phorsphorylation and de novo protein synthesis levels in ECs subjected to OSS (10). The metabolic complications of atherosclerosis have directed attention toward peroxisome proliferators-activated receptors (PPARs) as antiinflammatory molecules involved in the vascular wall (11, 12). Activation of PPARα with specific agonist represses AP-1 signaling in response to proinflammatory stimuli in ECs (13, 14). It was not clear whether PPARα plays a role in modulating OSS-induced endothelial inflammation.
MicroRNAs (miRs), the noncoding single-stranded RNA molecules of ≈22 nucleotides, bind to target sites in 3′-untranslated regions (3′-UTRs) of mRNAs to cause their degradation or translation repression (15). MiRs have distinct expression profiles in the cardiovascular system and play crucial roles in the pathogenesis of vascular diseases characterized by inflammatory status (16–20). The molecular mechanisms by which miRs epigenetically modulate OSS-induced EC responses, however, remain unclear. MiR-21 is considered an onco-miR that has been implicated in a variety of disorders and found to play important roles in cardiovascular diseases (21, 22). It is highly expressed in cardiovascular cells and is aberrantly expressed during many processes of cardiovascular pathologies, e.g., vascular neointimal lesion formation and acute myocardial infarction (23–25). Here, we report that OSS induces a sustained miR-21 expression to inhibit PPARα translation, thus leading to the activation of AP-1 and increases of VCAM-1 and MCP-1 expression, as well as monocyte adhesion to ECs. Expression of miR-21 is autoamplified through a positive feedback loop, including the miR-21/PPARα/c-Jun signaling cascade. This regulatory circuit integrates transcription factors, miR-21, and its direct target PPARα into connected molecular pathways that are responsible for the OSS-induced inflammatory responses in ECs.
Results
OSS Induces Functional miR-21 Expression to Cause Inflammatory Responses in ECs.
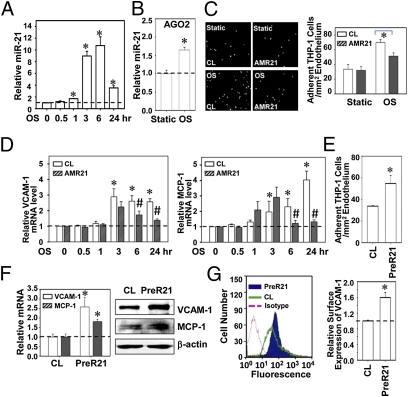
ECs were exposed to OSS at 0.5 ± 4 dynes/cm2 or PSS at 12 ± 4 dynes/cm2 for 0.5, 1, 3, 6, and 24 h, and their miR-21 expression was determined. OSS induced significant increases in miR-21 expression over static control (1.70 ± 0.03 fold at 1 h, P < 0.01), with a peak increase at 6 h (10.74 ± 1.50 fold, P < 0.01) (Fig. 1A), and sustained for 24 h (3.53 ± 0.37 fold, P < 0.01). In contrast, PSS induced a transient down-regulation of miR-21, which returned to basal level after 24 h (Fig. S1A). The divergent regulations of miR-21 by OSS and PSS were confirmed by FISH staining (Fig. S1B). A significant enrichment of miR-21 was observed in the Argonaut-2 (AGO2)-containing miR-inducing silencing complexes (miRISCs) after 24 h of shearing (Fig. 1B), indicating that OSS induced the expression of functional miR-21. The protein level of AGO2 in neither the total cell lysate nor the immunocomplexes was affected by shearing (Fig. S1C).
Fig. 1.
OSS induction of functional miR-21 leads to inflammatory responses in ECs. (A) ECs were kept as static controls or exposed to OSS (0.5 ± 4 dynes/cm2) for 0.5, 1, 3, 6, and 24 h, and miR-21 expression was analyzed by quantitative RT-PCR. (B) AGO2 pull-down assay was performed at 24 h time point, and miR-21 expression in the immunocomplexes was determined by quantitative RT-PCR. *P < 0.05 vs. static control cells. (C) ECs were transfected with AMR21 or the negative control molecules and kept as static controls or exposed to OSS for 24 h, followed by incubating with fluorescent-labeled THP-1 cells (5 × 105 cells per mL). Representative images of THP-1 adhesion are shown in Left, and statistic results were shown in Right, expressed as cells per mm2. *P < 0.05 vs. negative controls. (D) ECs were transfected with AMR21 or the negative control molecules and kept as static controls or exposed to OSS, and expressions of VCAM-1 and MCP-1 were analyzed by quantitative RT-PCR. *P < 0.05 vs. static control. #P < 0.05 vs. negative control at matched time points. (E, F, and G) ECs were transfected with PreR21 or the negative control molecules and THP-1 adhesion assay was performed 48 h after transfection (E); mRNA and protein (F) levels of VCAM-1 and MCP-1 were analyzed. (G) ECs were transfected with PreR21 or the negative control molecules, and VCAM-1 expression was measured by flow cytometry (Left), and statistical results are shown in bar graph (Right). Data are shown as mean ± SEM from three independent experiments. Results in H are representative of triplicate experiments with similar results. *P < 0.05 vs. negative control.
To investigate the role of OSS-induced miR-21 in inflammatory responses, ECs were transfected with anti–miR-21 inhibitor (AMR21), pre-miR-21–mimic (PreR21), and the respective negative control molecules. The association of miR-21 with miRISCs was reduced by AMR21 and induced by PreR21 (Fig. S2A). To further validate the function of miR-21, we cotransfected AMR21 or PreR21 and a luciferase reporter construct containing an insert of miR-21 binding sites into HeLa cells; the luciferase reporter activity was desuppressed by AMR21 and repressed by PreR21 (Fig. S2B). HeLa cells were used because of their much higher transfection efficiency than ECs.
Application of OSS to ECs for 24 h induced a significant increase in their adhesiveness for THP-1 cells, which was attenuated by AMR21, confirming the proinflammatory role of miR-21 in OSS-induced EC inflammatory response (Fig. 1C). The OSS inductions of EC VCAM-1 and MCP-1 were also suppressed by AMR21 (Fig. 1D). Exogenous overexpression of miR-21 per se was sufficient to induce THP-1 cell adhesion to ECs (Fig. 1E) and the EC expressions of VCAM-1 and MCP-1 at mRNA and protein (Fig. 1F) levels. PreR21 also increased EC surface expression of VCAM-1 (Fig. 1G). These results indicate that miR-21 plays important roles in mediating OSS-induced proinflammatory responses in ECs.
MiR-21 Negatively Regulates PPARα Expression by Targeting Its 3′-UTR.
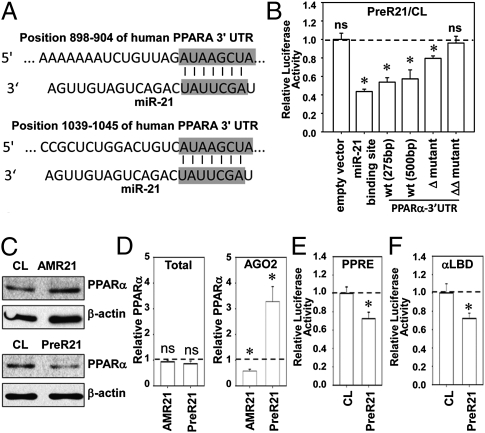
Bioinformatic databases TargetScan, EIMMo, and Pic Tar predicted that human PPARA mRNA is a potential target of miR-21. PPARα is an antiinflammatory molecule that contains two miR-21 binding sites in the 3′-UTRs of its mRNA (Fig. 2A). To test whether miR-21 directly regulates PPARα, we constructed a series of luciferase reporter constructs containing a short (275 bp) or long (500 bp) fragment of predicted miR-21 recognition sequences in the wild-type 3′-UTRs of PPARα inserted into pMir-Report. A construct harboring a direct-match miR-21 binding site was used as positive controls. PreR21 decreased luciferase activities of the reporter constructs with 275 bp or 500 bp of 3′-UTR to 0.54 ± 0.05 and 0.57 ± 0.10 fold, respectively, in comparison with the negative control molecules (Fig. 2B). Deletion of one predicted miR-21 binding site reduced miR-21 inhibition to 0.79 ± 0.03 fold, whereas double deletions totally abolished the inhibitory effects (Fig. 2B). These data indicate that miR-21 can regulate PPARα level by targeting its 3′-UTRs.
Fig. 2.
MiR-21 targets 3′-UTRs of PPARα to regulate its expression and activation. (A) The two conserved miR-21 binding sites locate in 3′-UTRs of PPARA. (B) HeLa cells were cotransfected with PreR21 or the negative control molecules and a series of luciferase reporter plasmids: empty vector only, synthetic consensus miR-21 binding sequences, wild-type PPARα 3′-UTR [wt (275 bp) and wt (500 bp)], and mutants of PPARα 3′-UTR (Δ mutant and ΔΔ mutant, with the miR-21 binding sites single or double deleted) to assess the miR-21 targeting PPARα 3′-UTR. (C) PPARα protein expression in ECs transfected with AMR21, PreR21, or the negative control molecules. Images are representatives of triplicate experiments with similar results. (D) AGO2 pull-down assay was performed 48 h after transfection with AMR21 or PreR21. The transcript levels of PPARα in total cell lysate (Left) or in AGO immunocomplexes (Right) were determined by quantitative RT-PCR. (E) PreR21 attenuates PPARα transcriptional activities determined by luciferase assays with PPRE×3-TK-Luc. (F) PreR21 decreases PPARα binding activities determined by luciferase assays of GAL-hPPAR-α-LBD and MH100 × 4-TK-Luc cotransfection. Bar graphs show mean ± SEM from three independent experiments. *P < 0.05 vs. cells transfected with negative control molecules.
Transfecting ECs with AMR21 increased PPARα protein expression; conversely, PreR21 decreased PPARα expression (Fig. 2C). There was no detectable alteration in PPARα mRNA levels by AMR21 or PreR21. However, in the precipitated miRISCs, the relative PPARα enrichment was reduced to 0.58 ± 0.07 fold by AMR21 and increased to 3.29 ± 0.58 fold by PreR21 (Fig. 2D). These results indicate that miR-21 regulates PPARα expression by inhibiting the translation, but not destabilization, of its mRNA.
MiR-21 Impairs PPARα Binding to PPAR-Response Elements and Activities.
PPARs regulate gene expression by binding to specific PPAR-response elements (PPREs) (26). PPARα binding activity was determined by cotransfection of PreR21 and a luciferase reporter driven by three copies of PPRE. PPARα activation was further tested by the cotransfection of a GAL4 reporter (MH100 × 4-TK-Luc), a GAL-hPPARα-LBD (ligand binding domain) vector, and PreR21. The results showed that miR-21 impaired both the DNA binding activity and ligand-dependent binding activity of PPARα (Fig. 2 E and F).
OSS Represses PPARα Expression to Induce Monocyte Adhesion.
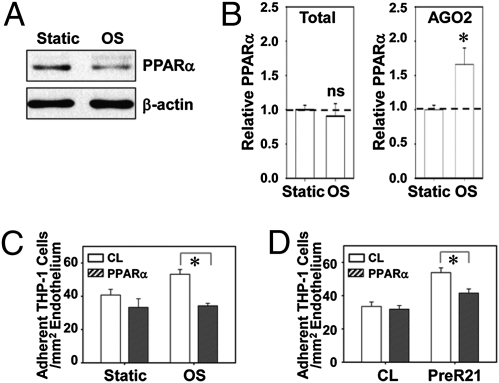
The protein level of PPARα in ECs was repressed by OSS (Fig. 3A). The mRNA level of PPARα in cells remained unchanged under OSS, whereas the PPARα mRNA in miRISCs was increased to 1.66 ± 0.24 fold by OSS (Fig. 3B), indicating that OSS promotes miR-21–mediated translational repression of PPARα. Overexpression of PPARα by infecting cells with adenovirus bearing the full length of coding regions of human PPARA gene suppressed both OSS- and PreR21-induced THP-1 cell adhesion to ECs (Fig. 3 C and D), indicating the inhibitory role of PPARα in miR-21–mediated EC inflammatory responses induced by OSS.
Fig. 3.
OSS represses PPARα expression to induce monocyte adhesion. (A and B) ECs were kept as static controls or exposed to OSS for 24 h; PPARα protein level was determined by Western blot analysis (A). (B) PPARα transcript levels were determined by quantitative RT-PCR in total cell lysate (Left) and in AGO immunocomplexes (Right). (C) ECs were infected with Ad-PPARα or control virus. Twenty-four hours after infection, ECs were kept as static controls or exposed to OSS for 24 h, and then subjected to THP-1 adhesion assay. (D) ECs were transfected with PreR21 or the negative control molecules, infected with Ad-PPARα or control virus, and then subjected to THP-1 adhesion assay. Data in B–D are shown as mean ± SEM from three independent experiments. Results in A are representative of triplicate experiments with similar results. *P < 0.05 vs. control virus.
AP-1 Is Involved in miR-21–Mediated EC Inflammatory Response.
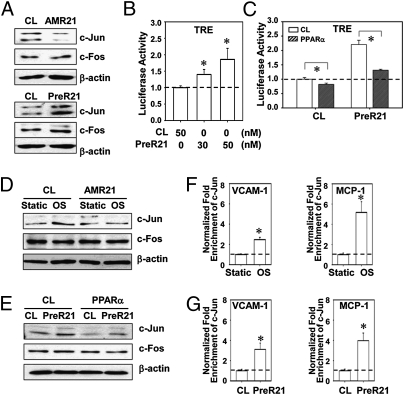
We tested whether AP-1 is a downstream target of the OSS/miR-21/PPARα cascade. The protein level of AP-1 component c-Jun (but not c-Fos) in ECs was reduced by AMR21 and was elevated by PreR21 (Fig. 4A). Overexpression of miR-21 also enhanced AP-1 activation, as demonstrated by a dose-dependent increase of luciferase levels in HeLa cells cotransfected with PreR21 and a luciferase reporter driven by four copies of AP-1 sites (TRE×4-luc) (Fig. 4B). The miR-21 activation of AP-1 was counteracted by overexpression of PPARα (Fig. 4C). The involvement of c-Jun in the OSS/miR-21/PPARα cascade was confirmed by the finding that inhibition of miR-21 attenuated OSS-induced synthesis of c-Jun, but not c-Fos (Fig. 4D); overexpression of PPARα attenuated the miR-21 induction of c-Jun (Fig. 4E).
Fig. 4.
AP-1 is involved in miR-21–mediated EC inflammation. (A) c-Jun and c-fos protein levels in ECs transfected with AMR21, PreR21, or the respective negative control molecules. (B) TRE×4-Luc activities in HeLa cells cotransfected with PreR21 or negative control molecules. *P < 0.05 vs. negative control molecules. (C) TRE×4-Luc activities in HeLa cells cotransfected with PreR21 or negative control molecules followed by Ad-PPARα or control viral infection. *P < 0.05 vs. control virus. (D) c-Jun and c-fos protein levels in ECs transfected with AMR21 or the negative control molecules under static or 24 h-OSS. (E) c-Jun and c-fos protein levels in ECs transfected with PreR21 or the negative control molecules, followed by Ad-PPARα or control viral infections. (F) ChIP assays for c-Jun association to promoter regions of VCAM-1 (Left) and MCP-1 (Right) in ECs under static or 24-h OSS. *P < 0.05 vs. static control. (G) ChIP assays for c-Jun association to promoter regions of VCAM-1 (Left) and MCP-1 (Right) in ECs transfected with PreR21 or negative control molecules, under static control or 24-h OSS. Data in B, C, F, and G are mean ± SEM from three independent experiments. Results in A, D, and E are representative of triplicate experiments with similar results. *P < 0.05 vs. negative control molecules.
Chromatin immunoprecipitation (ChIP) assay showed that OSS induced c-Jun binding to VCAM-1 and MCP-1 promoter regions to 2.47 ± 0.23 and 5.17 ± 1.05 folds, respectively, in ECs (Fig. 4F). Transfection with PreR21 induced enrichments of c-Jun in these two promoter regions to 3.11 ± 0.62 and 3.98 ± 0.75 folds, respectively (Fig. 4G). These data are in keeping with the thesis that miR-21 modulates OSS-induced EC inflammatory responses via AP-1 activation (Fig. 1).
PPARα Negatively Regulates miR-21 Transcription Through AP-1.
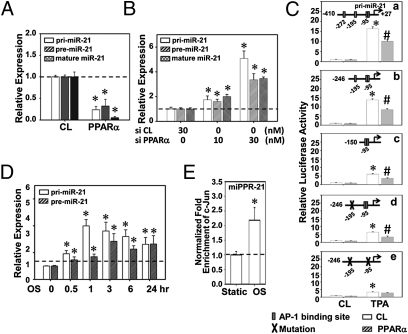
We next determined the accumulations of primary transcripts of miR-21 gene (pri-miR-21), the hairpin intermediate (pre-miR-21), and mature miR-21 molecules in ECs infected with Ad-PPARα or transfected with PPARα-specific siRNA. Overexpression of PPARα decreased the amounts of pri-miR-21, pre-miR-21, and mature miR-21 (Fig. 5A), whereas knockdown of PPARα increased the expression of these molecules (Fig. 5B), indicating that PPARα negatively regulates miR-21 biosynthesis at the transcriptional level.
Fig. 5.
OSS-induced miR-21 biosynthesis is mediated by PPARα/AP-1 at the transcriptional level. (A) ECs were infected with Ad-PPARα or control virus; expressions of pri-miR-21, pre-miR-21, and mature miR-21 were measured by quantitative RT-PCR. (B) ECs were transfected with PPARα-specific or control siRNA; expressions of pri-miR-21, pre-miR-21, and mature miR-21 were measured by quantitative RT-PCR. Data are shown as mean ± SEM from three independent experiments. *P < 0.05 vs. control virus or control siRNA. (C) HeLa cells were transfected with a series of luciferase reporter vectors bearing the full-length or truncated promoter fragments of miR-21 with the wild-type putative AP-1 binding sites or mutated binding sites (shown at the top of each graph). Twenty-four hours after transfection, cells were infected with Ad-PPARα or control virus, treated with TPA (50 nM) or PBS control for 4 h, and then subjected to luciferase activity assay. *P < 0.05 vs. control virus and without TPA treatment. #P < 0.05 vs. control virus and with TPA treatment. (D) ECs were kept as static controls or exposed to OSS for 0.5, 1, 3, 6, and 24 h, and expressions of pri-miR-21 and pre-miR-21 were analyzed by quantitative RT-PCR. *P < 0.05 vs. the respective static controls. (E) ECs were kept as static control or exposed to OSS for 24 h, and the association of c-Jun with promoter regions of miR-21 was analyzed by ChIP assay. *P < 0.05 vs. static controls.
We next explored whether AP-1 is involved in PPARα-mediated miR-21 biosynthesis. Reporter constructs with the full-length wild-type, truncation, or site-specific mutants of the miPPR-21 promoter were transfected into HeLa cells in the presence or absence of Ad-PPARα, followed by TPA treatment to activate AP-1. The strongest induction of promoter activity was observed in cells with the full-length and wild-type miPPR-21 (Fig. 5C, a). Deletion between nt −246 and −150 (Fig. 5C, c) or mutation of the AP-1 binding sites between −195 and −185 (Fig. 5C, d) led to a decrease of TPA-induced luciferase activities (Fig. 5C, c and d). Further mutation of the two binding sites (−195 to −185 and −95 to −74) caused an additional decrease of the TPA-induced luciferase activities (Fig. 5C, e). These results indicate that these three AP-1 binding sites in miPPR21 play important roles for its TPA inducibility. It is noted that PPARα overexpression suppressed the TPA activation of promoter in all of the plasmids studied (Fig. 5C, a–d) except for the one with all three AP-1 binding sites mutated (Fig. 5C, e). These results indicate that PPARα negatively regulates miR-21 transcription, which is mediated by AP-1.
OSS Induces miR-21 Biosynthesis via c-Jun Signaling.
OSS induced pri-miR-21 expression in ECs as early as 30 min of shearing (Fig. 5D). This OSS induction of pri-miR-21 reached a maximal level at 1 h and was sustained for 24 h after shearing, suggesting that OSS induction of miR-21 occurs at the transcriptional level. The maximal expression of pre-miR-21 occurred at 3 h of shearing (Fig. 5D). In concert with these findings, OSS induction of mature miR-21 reached a peak at 6 h of shearing (Fig. 1A).
We sought whether the PPARα/AP-1 signaling cascade can modulate OSS induction of miR-21, in addition to being the signaling event responsible for the OSS/miR induction of inflammatory gene expression. This hypothesis was verified by ChIP assay, which showed that OSS stimulated a 2.18 ± 0.43 fold enrichment of c-Jun binding to the endogenous miPPR-21 in ECs after 24 h of shearing (Fig. 5E). These data provide evidence that OSS regulates miR-21 biosynthesis via an increase in AP-1 activation. Together with the results that AP-1 mediates miR-21–induced EC inflammatory responses, our findings suggest a role of PPARα/AP-1 in a positive feedback loop in regulating EC gene expression and functions.
Discussion
In the present study, we have identified a direct link between miR-21 and nuclear receptor PPARα and demonstrated that OSS induction of miR-21 represses PPARα translation to promote AP-1 activation and, hence, the proinflammatory molecule VCAM-1 and MCP-1 expressions, as well as the adhesion of monocytes to ECs (as summarized in Fig. 6). Thus, miR-21 acts as an epigenetic mediator of proinflammatory phenotype of ECs in response to OSS. We also found that AP-1 can induce pri-miR-21 and its mature form, thus providing an autoregulatory feedback loop that may contribute to the sustained induction of inflammatory responses to OSS.
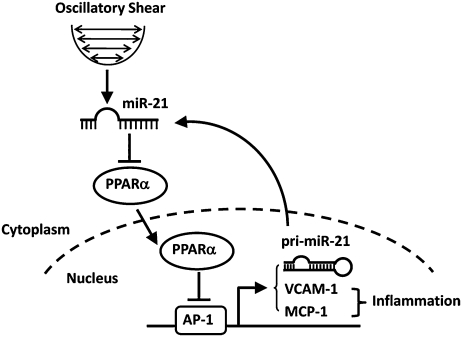
Fig. 6.
Schematic representation of miR-21–mediated positive feedback loop that regulates OSS-induced endothelial inflammation. OSS induction of miR-21 represses PPARα through direct targeting at its 3′-UTRs. Decreased expression of PPARα reduces the inhibitory effects of PPARα on AP-1 activation and, hence, promotes the expression of adhesion molecules VCAM-1 and MCP-1 and EC inflammation, as well as miR-21 transcription. The increase in miR transcripts would further repress PPARα to constitute a positive feedback circuit.
We have reported the differential expression profiles of miRs in ECs in response to OSS vs. PSS identified by microarray (27). The expression of miR-21 was found to be significantly up-regulated by OSS relative to PSS (P = 0.00001). Our quantitative RT-PCR assay in the current study has confirmed these microarray results and further demonstrated that OSS induces a sustained expression of miR-21 (Fig. 1A). The time course of miR-21 expression in ECs in response to PSS was opposite to that of OSS, with a transient down-regulation (Fig. S1A). Specific mechanosensors and signal pathways may contribute to the diversity of miR-21 regulation by OSS vs. PSS. These potential pathways include integrins, mitogen-activated protein kinases, AMP-activated protein kinase, krüppel-like factor 2, nuclear factor κ B (NF-κB), bone morphogenic protein 4, as well as other signaling pathways whose activations have been shown differentially correlated to different types of shear stress (1). Weber et al. (28), using a cone-and-plate system, recently reported that miR-21 is up-regulated by steady laminar shear stress (LSS; 15 dynes/cm2) in ECs. The difference reported by Weber et al. vs. our study on miR-21 expression may be attributable to the difference in the shear stress patterns generated (steady laminar vs. pulsatile), as well as the cell culture and shearing systems used. Our in vitro results on the OSS induction and PSS inhibition of miR-21 expression are consistent with the findings in a recent in vivo study by Fang et al. (19), who reported that miR-21 expression is elevated in ECs in the inner curvature of the aortic arch in swine, where the blood flow is oscillatory, compared with the cells in athero-protected regions of straight segments, where the local flow is pulsatile. These in vitro and in vivo results suggest the importance of miR-21 in modulating EC responses to hemodynamic forces.
MiR-21 was predicted by in silico analyses to target an antiinflammatory factor, PPARα, in the vascular system. PPARα negatively regulates the expression of many proinflammatory genes by inhibiting the NFκB and AP-1 pathways (13). Our results on miR expression patterns and in silico predictions led us to propose a regulatory mechanism in which miR-21 represses PPARα to modulate OSS-induced proinflammatory responses. This hypothesis is supported by our finding that miR-21 targets 3′-UTRs of PPARα mRNA to result in its posttranscriptional repression. Manipulation of miR-21 altered the enrichment of PPARα mRNA in miRISCs without changing its total mRNA levels (Fig. 2D), suggesting a functional role of miR-21 via its inhibition of translation of PPARα mRNA, but not via cleavage of the mRNA strand.
An important transcription factor that has been shown to contribute to EC proinflammatory phenotype is AP-1, whose transcriptional activity is known to be down-regulated by PPARα through its direct association with AP-1 component c-Jun in COS cells (13). The PPARα agonist, fenofibrate, inhibits de novo synthesis of c-Jun, but not c-Fos, in cardiac fibroblasts (29). Recent studies by Dong et al. (25) demonstrated that miR-21 promotes AP-1 activity in cultured cardiac myocytes through inhibition of its downstream target programmed cell death 4. Here, we demonstrated that miR-21 induces AP-1 activation as well as c-Jun expression by repressing PPARα translation in ECs, indicating a unique mechanism by which a miR acts on a specific target within its multiple target pool to modulate specific responses.
An important finding of the present study is that OSS may regulate miR-21 expression through an autoregulatory feedback loop involving miR-21, PPARα, and AP-1. Regulation of miR expression has been documented at the transcriptional level (30, 31), in which the transcription of the miR gene could be coordinated by transcription factors binding to the promoter regions of the miR gene. We demonstrated that OSS enhances association of c-Jun with miPPR-21 (Fig. 5E); induction of miR-21 by AP-1 activation is suppressed by overexpression of PPARα (Fig. 5C). These results on AP-1 regulation of miR-21 are in agreement with the previous study by Fujita et al. (32), who showed that TPA stimulation of HL60 cells induces miR-21 promoter activity. Here, we reveal that hemodynamic forces act through a feedback circuit to modulate miR-21 expression at the transcriptional level via the PPARα/AP-1 signaling cascade. MiR-21 is positively regulated by AP-1 and is also required for AP-1 activation; this reverberating relationship ensures the signal transduction of the upstream triggering events, leading to the sustained induction of AP-1, as well as the responses of the downstream proinflammatory molecules VCAM-1 and MCP-1. Whether OSS exerts additional effects on posttranscriptional regulation of the processing and maturation of miR-21 remains an important issue that requires further investigations. Additionally, enhanced expressions of VCAM-1 and MCP-1 have been observed at the inner curvature of the aortic arch (2, 4), where the blood flow is low and oscillatory. MiR-21 is highly expressed compared with that at the atheroprotective regions (19), implicating a correlation of in vivo VCAM-1 and MCP-1 expressions and miR-21 activation. The OSS induction of miR-21/PPARα/AP-1/VCAM-1 and MCP-1 cascades presented here is consistent with these in vivo studies.
Our results suggest that miR regulation is not a simple linear pathway, but is instead a complex regulatory network containing check and balance. This study of miR expression profiles in response to different flow patterns and the miR-mediated EC inflammatory responses has revealed essential roles of miRs in regulating atherosclerotic EC phenotype. Our finding suggests a therapeutic potential for targeting miR-21 as the treatment of vascular disorders related to hemodynamic force-induced EC inflammation or dysfunction, such as atherosclerosis.
Materials and Methods
Antibodies and Reagents.
Rabbit polyclonal antibody (pAb) against AGO2 was purchased from Cell Signaling Technology. Goat pAbs against VCAM-1 and MCP-1, rabbit pAbs against c-Jun and c-Fos, and mouse monoclonal antibody (mAb) against β-actin were obtained from Santa Cruz Biotechnology. Mouse mAb against PPARα was from R&D Systems. AMR21, PreR21, and the respective negative control inhibitor and mimic were purchased from Ambion. The PPARα-specific and control siRNA were purchased from Santa Cruz Biotechnology.
Cell Culture.
Human umbilical vein ECs were cultured in medium 199 (Gibco) supplemented with 10% FBS (Gibco) and 10% endothelial growth medium (Cell Applications). HeLa cells were cultured in DMEM (Gibco) supplemented with 10% FBS. Human monocytic cells, THP-1, were maintained in culture medium RPMI 1640 (Gibco) supplemented with 10% FBS.
Statistical Analysis.
Data are expressed as mean ± SEM from three independent experiments. Statistical analysis was performed by Student's t test for two groups of data and by one-way ANOVA for multiple comparisons. Statistical significance among multiple groups was determined by post hoc analysis (Tukey Honestly Significant Difference test). P < 0.05 was considered statistically significant.
The detailed methods are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
This work was supported in part by National Institutes of Health Research Grants HL064382, HL080518, HL085159, and R21HL104402 (to S.C.), and HL106579 (to S.C., S.S., and J.Y.-J.S.), and National Science Council (Taiwan) Grant 99-2321-B-400-002 /NRPB-100CV013 and NSC-99-2911-I-009-101 (to J.-J.C.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1107052108/-/DCSupplemental.
References
- 1.Chiu JJ, Chien S. Effects of disturbed flow on vascular endothelium: Pathophysiological basis and clinical perspectives. Physiol Rev. 2011;91:327–387. doi: 10.1152/physrev.00047.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huo Y, Wischgoll T, Kassab GS. Flow patterns in three-dimensional porcine epicardial coronary arterial tree. Am J Physiol Heart Circ Physiol. 2007;293:H2959–H2970. doi: 10.1152/ajpheart.00586.2007. [DOI] [PubMed] [Google Scholar]
- 3.Xu Q. Disturbed flow-enhanced endothelial turnover in atherosclerosis. Trends Cardiovasc Med. 2009;19:191–195. doi: 10.1016/j.tcm.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 4.Shyy YJ, Hsieh HJ, Usami S, Chien S. Fluid shear stress induces a biphasic response of human monocyte chemotactic protein 1 gene expression in vascular endothelium. Proc Natl Acad Sci USA. 1994;91:4678–4682. doi: 10.1073/pnas.91.11.4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chappell DC, Varner SE, Nerem RM, Medford RM, Alexander RW. Oscillatory shear stress stimulates adhesion molecule expression in cultured human endothelium. Circ Res. 1998;82:532–539. doi: 10.1161/01.res.82.5.532. [DOI] [PubMed] [Google Scholar]
- 6.Hsiai TK, et al. Monocyte recruitment to endothelial cells in response to oscillatory shear stress. FASEB J. 2003;17:1648–1657. doi: 10.1096/fj.02-1064com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Orr AW, Hahn C, Blackman BR, Schwartz MA. p21-activated kinase signaling regulates oxidant-dependent NF-kappa B activation by flow. Circ Res. 2008;103:671–679. doi: 10.1161/CIRCRESAHA.108.182097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ahmad M, Theofanidis P, Medford RM. Role of activating protein-1 in the regulation of the vascular cell adhesion molecule-1 gene expression by tumor necrosis factor-alpha. J Biol Chem. 1998;273:4616–4621. doi: 10.1074/jbc.273.8.4616. [DOI] [PubMed] [Google Scholar]
- 9.Wang N, et al. Adenovirus-mediated overexpression of c-Jun and c-Fos induces intercellular adhesion molecule-1 and monocyte chemoattractant protein-1 in human endothelial cells. Arterioscler Thromb Vasc Biol. 1999;19:2078–2084. doi: 10.1161/01.atv.19.9.2078. [DOI] [PubMed] [Google Scholar]
- 10.Nagel T, Resnick N, Dewey CF, Jr, Gimbrone MA., Jr Vascular endothelial cells respond to spatial gradients in fluid shear stress by enhanced activation of transcription factors. Arterioscler Thromb Vasc Biol. 1999;19:1825–1834. doi: 10.1161/01.atv.19.8.1825. [DOI] [PubMed] [Google Scholar]
- 11.Marx N, Sukhova GK, Collins T, Libby P, Plutzky J. PPARalpha activators inhibit cytokine-induced vascular cell adhesion molecule-1 expression in human endothelial cells. Circulation. 1999;99:3125–3131. doi: 10.1161/01.cir.99.24.3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tedgui A, Mallat Z. Anti-inflammatory mechanisms in the vascular wall. Circ Res. 2001;88:877–887. doi: 10.1161/hh0901.090440. [DOI] [PubMed] [Google Scholar]
- 13.Delerive P, et al. Peroxisome proliferator-activated receptor alpha negatively regulates the vascular inflammatory gene response by negative cross-talk with transcription factors NF-kappaB and AP-1. J Biol Chem. 1999;274:32048–32054. doi: 10.1074/jbc.274.45.32048. [DOI] [PubMed] [Google Scholar]
- 14.Fernandez AZ. Peroxisome proliferator-activated receptors in the modulation of the immune/inflammatory response in atherosclerosis. PPAR Res. 2008;2008:285842. doi: 10.1155/2008/285842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 16.Urbich C, Kuehbacher A, Dimmeler S. Role of microRNAs in vascular diseases, inflammation, and angiogenesis. Cardiovasc Res. 2008;79:581–588. doi: 10.1093/cvr/cvn156. [DOI] [PubMed] [Google Scholar]
- 17.Harris TA, Yamakuchi M, Ferlito M, Mendell JT, Lowenstein CJ. MicroRNA-126 regulates endothelial expression of vascular cell adhesion molecule 1. Proc Natl Acad Sci USA. 2008;105:1516–1521. doi: 10.1073/pnas.0707493105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suárez Y, Wang C, Manes TD, Pober JS. Cutting edge: TNF-induced microRNAs regulate TNF-induced expression of E-selectin and intercellular adhesion molecule-1 on human endothelial cells: Feedback control of inflammation. J Immunol. 2010;184:21–25. doi: 10.4049/jimmunol.0902369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fang Y, Shi C, Manduchi E, Civelek M, Davies PF. MicroRNA-10a regulation of proinflammatory phenotype in athero-susceptible endothelium in vivo and in vitro. Proc Natl Acad Sci USA. 2010;107:13450–13455. doi: 10.1073/pnas.1002120107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ni CW, Qiu H, Jo H. MicroRNA-663 upregulated by oscillatory shear stress plays a role in inflammatory response of endothelial cells. Am J Physiol Heart Circ Physiol. 2011;300:H1762–H1769. doi: 10.1152/ajpheart.00829.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng Y, Zhang C. MicroRNA-21 in cardiovascular disease. J Cardiovasc Transl Res. 2010;3:251–255. doi: 10.1007/s12265-010-9169-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jazbutyte V, Thum T. MicroRNA-21: From cancer to cardiovascular disease. Curr Drug Targets. 2010;11:926–935. doi: 10.2174/138945010791591403. [DOI] [PubMed] [Google Scholar]
- 23.Suárez Y, Fernández-Hernando C, Pober JS, Sessa WC. Dicer dependent microRNAs regulate gene expression and functions in human endothelial cells. Circ Res. 2007;100:1164–1173. doi: 10.1161/01.RES.0000265065.26744.17. [DOI] [PubMed] [Google Scholar]
- 24.Ji R, et al. MicroRNA expression signature and antisense-mediated depletion reveal an essential role of MicroRNA in vascular neointimal lesion formation. Circ Res. 2007;100:1579–1588. doi: 10.1161/CIRCRESAHA.106.141986. [DOI] [PubMed] [Google Scholar]
- 25.Dong S, et al. MicroRNA expression signature and the role of microRNA-21 in the early phase of acute myocardial infarction. J Biol Chem. 2009;284:29514–29525. doi: 10.1074/jbc.M109.027896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tugwood JD, et al. The mouse peroxisome proliferator activated receptor recognizes a response element in the 5′ flanking sequence of the rat acyl CoA oxidase gene. EMBO J. 1992;11:433–439. doi: 10.1002/j.1460-2075.1992.tb05072.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang KC, et al. Role of microRNA-23b in flow-regulation of Rb phosphorylation and endothelial cell growth. Proc Natl Acad Sci USA. 2010;107:3234–3239. doi: 10.1073/pnas.0914825107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weber M, Baker MB, Moore JP, Searles CD. MiR-21 is induced in endothelial cells by shear stress and modulates apoptosis and eNOS activity. Biochem Biophys Res Commun. 2010;393:643–648. doi: 10.1016/j.bbrc.2010.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ogata T, et al. The peroxisome proliferator-activated receptor alpha activator fenofibrate inhibits endothelin-1-induced cardiac fibroblast proliferation. J Cardiovasc Pharmacol. 2004;44(Suppl 1):S279–S282. doi: 10.1097/01.fjc.0000166274.24797.0e. [DOI] [PubMed] [Google Scholar]
- 30.Kim VN, Nam JW. Genomics of microRNA. Trends Genet. 2006;22:165–173. doi: 10.1016/j.tig.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 31.Löffler D, et al. Interleukin-6 dependent survival of multiple myeloma cells involves the Stat3-mediated induction of microRNA-21 through a highly conserved enhancer. Blood. 2007;110:1330–1333. doi: 10.1182/blood-2007-03-081133. [DOI] [PubMed] [Google Scholar]
- 32.Fujita S, et al. miR-21 Gene expression triggered by AP-1 is sustained through a double-negative feedback mechanism. J Mol Biol. 2008;378:492–504. doi: 10.1016/j.jmb.2008.03.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.