Abstract
Understanding the heterogeneous genetic mechanisms of tumor initiation in lymphoid leukemias (LL) will lead to improvements in prognostic classification and treatment regimens. In previous studies of mouse leukemias, we showed that retroviral insertion at the Evi32 locus leads to increased expression of Prdm14, a pluripotency gene implicated in the self-renewal capacity of embryonic stem cells and the early stages of breast cancer. Here we show that PRDM14 is also overexpressed in ~25% of human lymphoid neoplasms, with increased frequencies in T-cell acute LL (T-ALL) and hyperdiploid precursor B-cell acute LL (pre-B ALL). To test if Prdm14 overexpression could initiate leukemia, mice were transduced with bone marrow (BM) cells transfected with a Prdm14 expression vector. Lymphoid leukemias developed in 96% of female mice and 42% of male mice. Prior to the onset of leukemia, differentiation of transduced cells was biased up to 1000-fold towards cells with features of common lymphoid progenitors (CLP), and lymphoid differentiation showed a relative block at the pro-B stage. Microarray gene expression analysis of expanded CLP-like cells prior to the onset of leukemia demonstrated upregulation of genes involved in pluripotency, tumor initiation, early B-lineage commitment, Wnt/Ras signaling, and the epithelial-to-mesenchymal transition. Among the dysregulated genes were imprinted genes and non-coding RNAs including Dlk1 and Meg3, which are also key pluripotency mediators. Heightened expression of the estrogen-dependent oncogene, Myb, in tumors suggests a basis for the increased frequency of cancer in female mice. These data provide the first direct evidence for the association of Prdm14 with cancer initiation in an in vivo mouse model and in human lymphoid malignancies, while suggesting mechanisms for Prdm14’s mode of action.
Keywords: leukemia, Prdm14, leukemia-initiating cell, pluripotency, common lymphoid progenitor, acute lymphoblastic leukemia
Introduction
Acute lymphoblastic leukemias (ALLs), neoplasms of lymphoid precursors of the T and B cell lineages, are variably curable based on age of presentation, disease burden at diagnosis, immunophenotype, and cytogenetic abnormalities (Pizzo and Poplack 2006). Genomic aberrations detected by standard karyotypic analyses involve large chromosomal rearrangements (such as translocations), aneuploidy, and the gain or loss of large chromosomal regions. The advent of high throughput genomic technologies changed our view of leukemias, however, by showing that large numbers of more subtle genetic abnormalities are present in tumor cells. Key genes such as PAX5 and IKZF1, important for normal lymphoid lineage commitment, are altered in ALL, often by copy number variations (CNV) such as deletion or amplification rather than translocation (Downing and Mullighan 2006, Mullighan et al 2005, Mullighan et al 2007, Mullighan et al 2008, Mullighan et al 2009). Nevertheless, genetic aberrations that trigger the initiation of lymphoid leukemias and lymphomas other than the balanced translocations involving immunoglobulin and T cell receptor genes remain largely unknown.
In mice, retroviral insertion is a powerful strategy to identify genes implicated in the initiation of leukemia (Akagi et al 2004, Hansen et al 2000, Jonkers and Berns 1996, Suzuki et al 2002). In certain inbred strains, the somatic insertion of retroviruses may activate expression of nearby proto-oncogenes or disrupt tumor suppressor genes. In these mouse tumors, common proviral insertion sites (CIS) identify loci involved in the onset and progression of hematologic malignancies. We previously showed that mouse lymphoid malignancies containing retroviral insertions at the CIS designated ecotropic viral insertion site 32 (Evi32) had very high expression of the PRDI-BF1 RIZ homology domain (PR) domain gene Prdm14, implicating this gene in the pathogenesis of mouse lymphoblastic leukemia (LL) for the first time (Dettman and Justice 2008, Weiser et al 2007).
Prdm14 (human ortholog PRDM14) is a member of the PR domain-containing family of transcription factors, which are crucial for normal hematopoiesis and can behave as tumor suppressors or as proto-oncogenes (Fumasoni et al 2007, Jiang and Huang 2000, Kim and Huang 2003, Kinameri et al 2008). While the molecular function of PRDM14 is largely unknown, sequence homology analyses suggest it may have histone methyltransferase (HMT) activity, similar to other PRDM family members (Baudat et al 2010, Hayashi et al 2005, Kim et al 2003, Myers et al 2010, Parvanov et al 2010). PRDM14 contains an N-terminal PR domain followed by six DNA binding C2H2 zinc fingers (ZF) (Fumasoni et al 2007). The PR domain is similar to the Su(var)3–9, enhancer of zeste [E(z)], and trithorax (SET) domain, which has global HMT activity.
Expression of Prdm14 is normally restricted to pluripotent cells. In embryonic stem (ES) cells, PRDM14 suppresses the expression of molecular markers associated with differentiation (Tsuneyoshi et al 2008). PRDM14 mediates pluripotency in human ES cells by directly binding the proximal enhancer of the key pluripotency gene POU5F1 (OCT3/4) (Chia et al 2010). Prdm14 is also expressed in primordial germ cells (PGCs) of mouse embryos from embryonic day (E) 6.5 until E 14.5, when it is required to establish the pluripotency of nascent PGCs by epigenetic reprogramming. As a result, mice with a null mutation of Prdm14 fail to develop germ cells and are infertile (Kurimoto et al 2008, Yamaji et al 2008).
In addition to its normal role in resetting pluripotency, PRDM14 has oncogenic activity with overexpression correlating with early stages of breast cancer. Importantly, expression of PRDM14 reduced the chemosensitivity of cultured cancer cells and enhanced their proliferation, while siRNA-mediated knockdown of gene expression augmented their chemosensitivity (Nishikawa et al 2007). Furthermore, the 8q13.3 region containing PRDM14 is amplified in 34–75% of human breast cancers (Moelans et al 2010, Nishikawa et al 2007).
Given the oncogenic potential of this pluripotency gene in mouse and human malignancy, we investigated the possible contribution of Prdm14 as an oncogene in mouse bone marrow (BM)-derived hematopoietic cells. Prior to leukemic transformation, overexpression of Prdm14 caused a dramatic developmental shift characterized by expansion of pre-leukemic cells that share an immunophenotype with common lymphoid progenitors (CLP). Preleukemic and leukemic cells with dysregulated Prdm14 overexpressed markers of stem cells and activators of oncogenic pathways. Thus, Prdm14 may initiate leukemia in a manner consistent with its role in the epigenetic regulation of pluripotency.
Results
Expression of Prdm14 in mouse hematopoietic cells
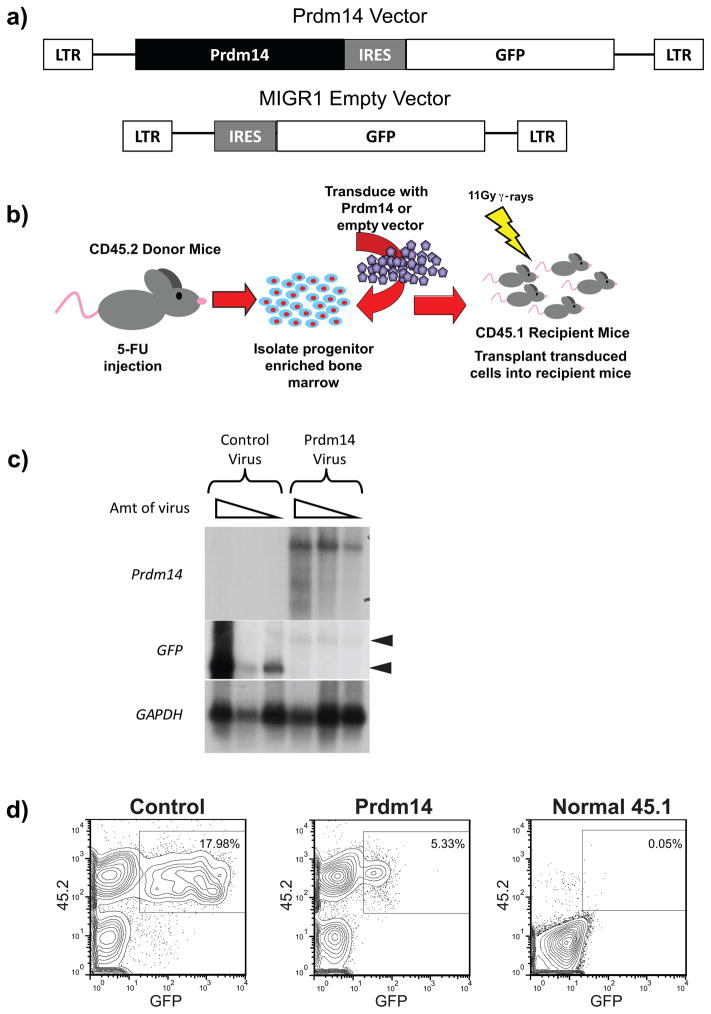
The coding sequence of Prdm14 was subcloned into the Murine Stem Cell Virus Internal Ribosomal Entry Site Green Fluorescent Protein R1 (MIGR1) retroviral vector (Figure 1a) followed by an internal ribosome entry site (IRES) for co-translation of green fluorescent protein (GFP) to permit tracking of transduced cells. The same MIGR1 empty vector (EV) without Prdm14 was used as a control. BM cells from 5-fluorouracil (5-FU)-treated CD45.2 mice were infected with each retrovirus (Figure 1b). Lethally irradiated CD45.1 mice were reconstituted with the transduced cells, allowing for the identification of donor cells with antibodies specific for CD45.2 and transduced cells by GFP fluorescence. Northern blot analyses identified GFP and Prdm14 transcripts of expected sizes, although GFP transcripts expressed from the Prdm14-MIGR1 vector were present at lower levels than those from the control vector in cells transduced with equivalent multiplicities of infection (MOI) (Figure 1c). Successful engraftment of CD45.2+ donor cells was observed at four weeks after reconstitution (Figure 1d). However, the frequency and fluorescence intensity of GFP+ cells in the peripheral blood (PB) of Prdm14-MIGR1 recipients were lower than for EV-MIGR1 recipients (Figure 1d).
Figure 1. Overexpression of Prdm14 in vivo.
1a) Murine stem cell virus vectors were used to overexpress Prdm14 and serve as an empty vector control. Both vectors contain an IRES sequence and express GFP as a marker of transduced cells. 1b) An overview shows the experimental method used to express Prdm14 in vivo. Mice expressing the CD45.2 cell surface antigen are treated with 5-fluorouracil (5-FU) to induce cycling and enrichment of BM hematopoietic stem cells (HSCs). These CD45.2 donor cells are transduced with retroviral particles containing either Prdm14-MIGR1 or the EV-MIGR1 control and then injected intravenously into lethally irradiated recipient mice that express the CD45.1 allele. Engraftment of transduced cells allows for long-term overexpression of Prdm14. 1c) Northern blot analysis of transcripts produced from NIH3T3 cells transduced with Prdm14-MIGR1 or the control vector. A GFP probe detects both constructs and shows a higher level of expression in cells transduced with the control vector than with the Prdm14-MIGR1 vector. A second probe detects the Prdm14 transcript only. Both vectors produce transcripts of the expected sizes, shown as arrowheads. 1d) Four weeks after transplantation, fewer GFP+ cells are detected in PB of mice transduced with Prdm14-MIGR1 than with the control. A normal CD45.1 mouse has no GFP fluorescence.
Overexpression of Prdm14 causes skewing of lymphoid progenitor cell populations
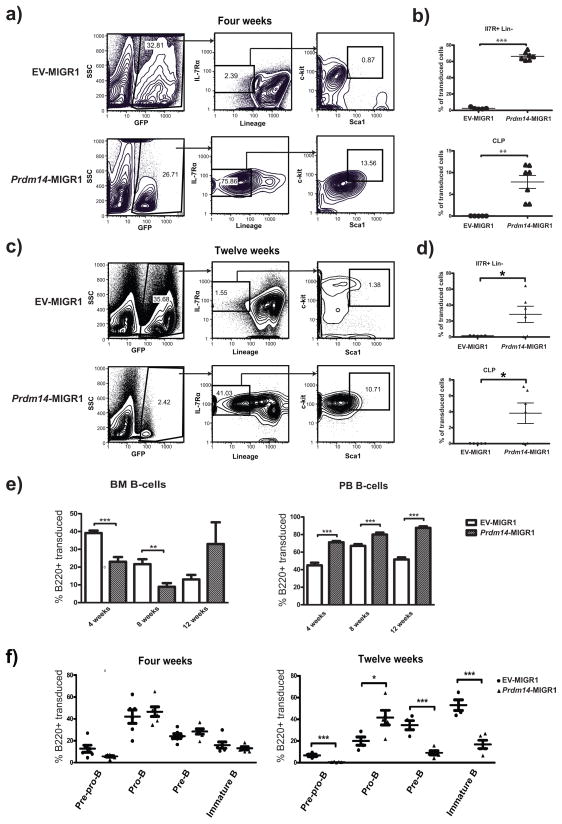
Recipients of Prdm14-MIGR1 or EV-MIGR1-transduced BM cells were examined at four, eight, and twelve weeks after transfer to evaluate effects on hematopoiesis. Studies of recipient BM cells obtained at four weeks after reconstitution showed that although the frequencies of GFP+ cells were generally comparable for recipients of either transfected population (Figure 2a, left panels), they differed drastically in immunophenotype. The BM of normal mice contains a small population of cells (~0.02% of total BM cells) termed common lymphoid progenitors (CLP) (Kondo et al 1997) that do not express lineage-specific markers of mature hematopoietic cells (lin−) but express the IL-7 receptor alpha chain (IL-7Rα), the Kit receptor tyrosine kinase (c-Kit, CD117), and Sca1 (Ly6a). The BM of Prdm14 recipients had a population of early lymphoid progenitor (IL-7Rα+lin−) cells that was over 30-fold greater than in EV recipients (Figure 2a). IL-7Rα+lin− cells expressing both Kit and Sca1 at high levels were also 15-fold more frequent in Prdm14 recipients (Figure 2a), with the great majority of cells being clearly Kit+ while expressing Sca1 at intermediate levels. Data from multiple mice indicate that the BM of Prdm14 recipients had a dramatically expanded population of these early lymphoid and CLP-like cells four weeks after reconstitution, ranging from 3 – 12% of the transduced BM population, a frequency 250–1000-fold greater than in EV recipients (Figure 2b). This expansion was specific to the GFP+ Prdm14-expressing subset of cells in these mice; both the GFP− population and BM from EV recipients contained only rare early lymphocyte progenitors and CLPs (Supplementary Figure 1).
Figure 2. Aberrant expression of Prdm14 expands an early lymphoid cell population.
2a) Flow cytometry of BM at four weeks post transduction shows skewing of Prdm14-expressing cells towards an early lymphoid population of IL-7Rα+lin− cells, many of which express intermediate levels of Kit and Sca1, similar to normal CLP. Serial gating of cells was performed as designated by the arrows. 2b) Relative frequencies of GFP+ cells that are IL7Rα+Lin− and CLP (IL7R+Lin−Kit+Sca1+) are profoundly expanded in Prdm14 recipients at 4 weeks. 2c,d) Recapitulation of flow cytometry of BM at twelve weeks post-transplantation demonstrates smaller fraction of early lymphoid progenitors and CLPs in Prdm14 recipients than at four weeks, but still markedly increased compared to EV recipients. 2e) BM and PB cells underwent flow cytometry for B220+ elements of the B cell lineage at 4, 8, and 12 weeks. GFP+ PB cells from Prdm14 recipients were skewed towards the B cell lineage at all time points compared to cells from EV recipients. GFP+ BM cells were not committed to the B cell lineage at four weeks compared to EV BM cells but later appear to skew towards the B cell lineage (but not significantly) at twelve weeks after transplantation. 2f) Flow cytometry was carried out on BM cells from transduced mice at successive time points after transplantation using early B cell lineage markers (B220, CD43, CD19, and IgM). At twelve weeks after transplantation, cells accumulate at the pro-B stage, but differentiation past this point is markedly limited (Pre-Pro-B B220+CD43+CD19−IgM−, Pro-B B220+CD43+CD19+IgM−, Pre-B B220+CD43−CD19+IgM−, Immature B B220+CD43−CD19+IgM+). (* indicates p<0.05, ** indicates p<0.01, and *** indicates p<0.001).
The percentages of IL-7Rα+lin− and KithiSca1hi cells among GFP+ cells in Prdm14 recipient BM was lower in mice examined at twelve weeks after reconstitution than those evaluated at four weeks (Figure 2c), although the magnitude of difference for the IL-7Rα+lin− population compared to that for EV recipients was still quite high at 26.5-fold. At this time point, 25% of GFP+ cells in PRDM14 recipient BM were lin+, in keeping with significant increases in both the BM and PB of cells expressing B220, a marker expressed by B lineage cells from the pre-pro-B to mature B cell stages of differentiation. The proportions of B220+ cells were significantly lower in Prdm14 recipients at four and eight weeks after reconstitution, but were higher in BM at twelve weeks (Figure 2e). The proportions of B220+ cells in PB were significantly higher for Prdm14 recipients at all time points examined (Figure 2e).
We then determined if the altered B220+ BM populations were comparable for all stages of B cell differentiation in the BM. FACS analyses of B cell subsets at four weeks after reconstitution with Prdm14- or EV-transfected cells revealed no significant differences in the cell populations at any stage of differentiation (Figure 2f). At 12 weeks, however, the populations of pre-pro-B, pre-B and immature B cells were significantly reduced, and the proportions of pro-B cells were significantly increased in BM of Prdm14 recipients compared to EV recipients (Figure 2f). These findings indicated that the dominant CLP-like population, present at four weeks in the BM of Prdm14 recipients, had undergone differentiation within the B cell lineage by 12 weeks after reconstitution, but that there was a relative block in maturation at the pro-B to pre-B transition. Consistent with the findings of a biased maturation within the B cell lineage, transduced cells from Prdm14 recipients exhibited reduced myelo-erythroid developmental potential (Supplementary Figure 2a, b, c). Furthermore, Prdm14-transduced cells were less likely to contribute to thymic and peripheral T-cells (Supplementary Figure 2d, e).
Mice transduced with Prdm14 develop LL
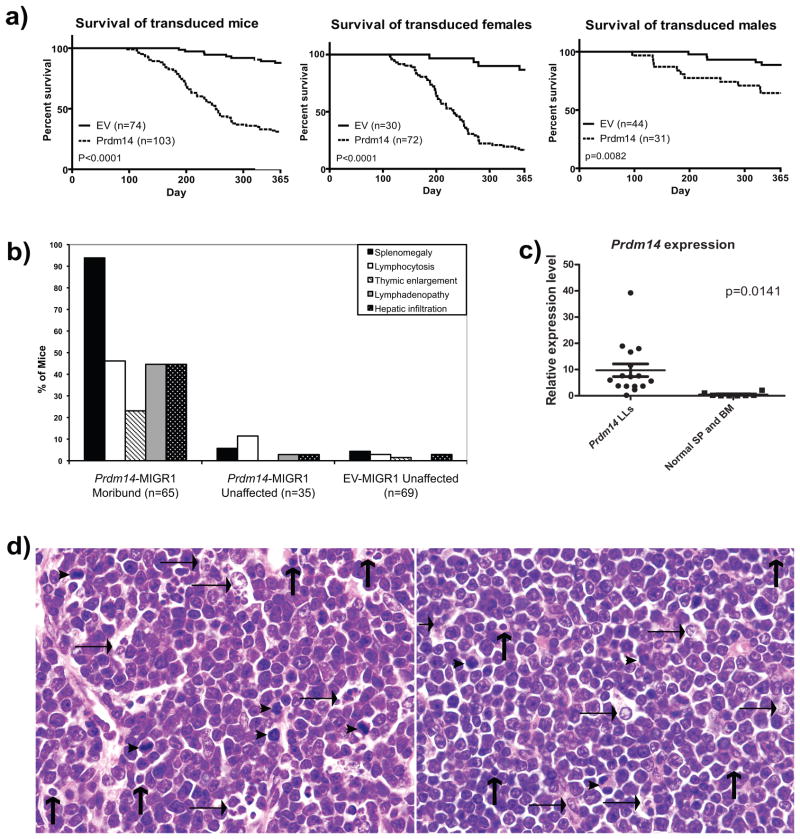
One hundred three Prdm14 recipient mice and 74 EV recipient mice were followed for up to one year after reconstitution. Beginning as early as 100 days after BM transplantation, Prdm14 recipients developed labored breathing, lethargy, cachexia, hind limb paralysis, and kyphosis, ultimately progressing to death. By 52 weeks, 80% of the Prdm14 recipients had been euthanized when moribund or had died compared to only 12% of EV recipients. Notably, 96% (69/72) of female recipients had died by one year after reconstitution compared to only 42% (13/31) of male recipients (Figure 3a). At necropsy, almost all moribund Prdm14 recipients had splenomegaly, with lower frequencies exhibiting lymphadenopathy, hepatomegaly, enlarged thymi, and PB lymphoblasts (Figure 3b).
Figure 3. Overexpression of Prdm14 in vivo causes LL.
3a) The survival of Prdm14 recipients is significantly reduced compared to EV recipient mice (p<0.00001). Moribund mice were first observed at approximately 100 days post-transplantation and necropsied as they became ill during the one year aging study. Female mice succumbed to disease more readily than male mice. 3b) All mice diagnosed as leukemic at least two or more of the following characteristics: splenomegaly (92%), lymphocytosis (46%), thymic enlargement (22%), lymphadenopathy (44%) or hepatic infiltration (44%). Prdm14 recipients (“Prdm14-MIGR unaffected”) that did not develop overt leukemia did not develop these symptoms at a significant rate. 3c) Prdm14 was overexpressed in most of the tumors as assessed by quantitative real time PCR. 3d) Histologic features of LL in Prdm14 recipients. Left panel: Spleen section with lymphoblasts containing nuclei with central prominent nucleoli interspersed among tingible body macrophages (horizontal arrows), apoptotic cells (vertical arrows), and histiocytes. There are several mitotic figures (arrowheads). Right panel:Section of a thymic mass showing a uniform population of lymphoblasts. There are several large tingible body macrophages with ingested apoptotic debris.
Analysis of slides stained with H&E prepared from tissues obtained at necropsy showed that all moribund Prdm14 recipients died with LL involving the spleen and lymph nodes and often associated with involvement of the thymus and BM (Figure 3d). Diagnoses of overt leukemia were based on the presence of lymphoblasts in hepatic blood vessels as well as blasts seen in PB. Hyperactivity of splenic red pulp elements was evidenced by expanded populations of megakaryocytes in about 17% of cases (data not shown) but without established features of megakaryocytic leukemia (Hao et al 2006). Erythroblastosis was found in a small number of other cases. In contrast, moribund EV recipients had no evidence of LL or proliferative expansions of other lineages.
Clonality of the LL was confirmed by the detection of clonal proviral insertions and immunoglobulin (Ig) and T-cell receptor (TCR) rearrangements by Southern analysis. In addition, qPCR analyses showed that the tumors overexpressed Prdm14 (Figure 3c). Furthermore, syngeneic mice injected intraperitoneally with tumor cells from Prdm14 recipients rapidly succumbed to leukemias that were clonally identical to the parental tumors (Supplementary Figure 3a).
The histopathologic diagnosis of LL is consistent with the origins of tumors from thymic T cells, BM-derived precursor B cells, or more mature peripheral B cells (Morse et al 2002). To discriminate among these possibilities, DNA prepared from the tumors was examined by Southern blotting for the organization of Ig heavy chain (IgH) and Ig kappa light chain (IgK) gene segments. The results of these studies showed that 65% of evaluated malignancies had clonal rearrangements at the IgH locus, but none had IgK rearrangements (Supplementary Figure 3b and data not shown). This indicated that the tumors with clonal IgH rearrangements were arrested at the pro-B cell stage of differentiation, which is characterized by V(D)J rearrangements at IgH with the IgK locus in germline configuration. Studies of the same DNAs revealed that 12% of the cases had clonal rearrangements at the TCR Jβ1 or Jβ2 loci, indicating a T-cell origin (Supplementary Figure 3c and data not shown). Co-existing clonal rearrangements of IgH and TCR loci were identified in 19% of cases, indicating the presence of composite tumors comprised of distinct neoplasms of T and B cell origin, B lineage tumors that have undergone non-productive rearrangements at TCR loci, or T lineage tumors with DJ only rearrangements in IgH. Of note, one tumor had no Ig or TCR rearrangement but was classified as LL by histopathology.
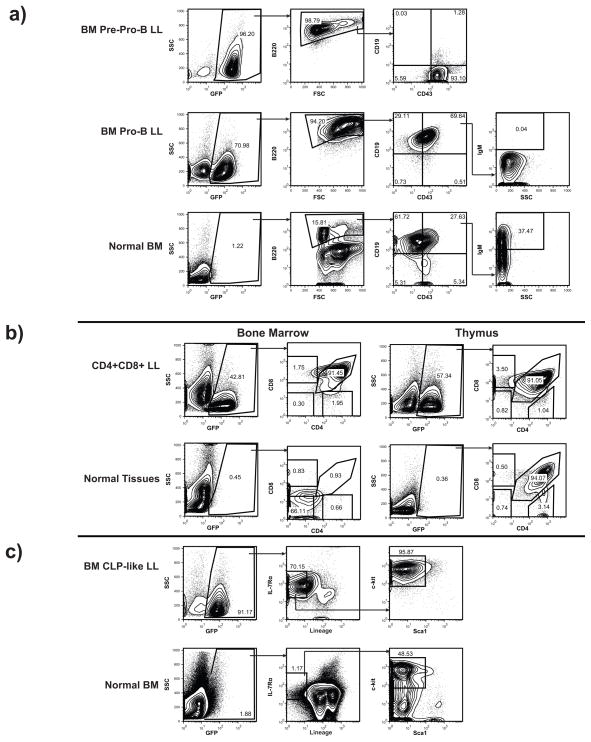
Flow cytometric studies showed that the LL expressed cell surface markers characteristic of lymphoid progenitors, B-cells, or T-cells, consistent with lineage assignments inferred from clonal rearrangements of Ig and TCR loci. The percentages of GFP+ tumor cells in BM, thymus, blood and peripheral lymphoid tissues ranged from ~40% to >95% of all WBCs (Figure 4a,b,c and data not shown). Most tumors analyzed were B220+, indicative of origins from B lineage cells. More detailed analyses indicated that the B220+ tumors were arrested at different stages of early B cell development including CD19− CD43+IgM− pre-pro-B cells as well as CD19+CD43+IgM− pro-B cells (Figure 4a). BM and thymi from mice that developed T-lineage tumors were populated by CD4+CD8+ cells similar to the “double-positive” population found in normal thymus but not in spleen (Figure 4b). Notably, all LL with thymic involvement carried clonal TCR rearrangements, and were classified as either pre-T-cell or mixed tumors by molecular rearrangements. One LL examined by flow cytometry was lin− but expressed IL-7Rα and Kit at high levels and Sca1 at intermediate levels (Figure 4c), a phenotype like that of normal CLPs (Kondo et al 1997).
Figure 4. Prdm14 overexpression causes a range of lymphoid leukemia cell types.
4a) B220+ B-cell tumors were isolated from Prdm14 transplanted mice. Here, transduced cells are overtaking the BM, with 96% and 71% GFP+ cells in the BM in the examples. Tumors were found at the pre-pro-B stage (B220+CD43+CD19−) and the pro-B cell stage (B220+CD43+CD19+IgM−). 4b) T-cell leukemias also occurred in Prdm14 recipients. BM and thymus show a high percentage of GFP+ cells (57% in thymus and 43% in the bone marrow). These cells are CD4+CD8+ Double Positive (DP) T-cells, which are normally very rare in the BM of mice. 4c) Prdm14 transduction also caused CLP-like tumors. Again, GFP+ cells are overtaking the bone marrow, with 91% being GFP+. The cells were lin−, IL-7Rα+, Kit+, with intermediate levels of Sca1, similar to the immunophenotype of normal CLPs.
Expanded CLP-like cells and tumors in Prdm14-transduced mice express genes associated with pluripotency, early B-cell development, and oncogenesis
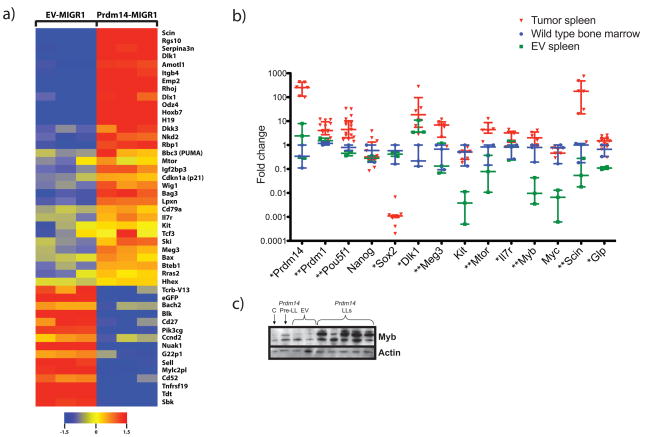
In order to determine downstream effects of Prdm14 expression, populations of CLP-like cells (IL-7Rα+lin−Kit+Sca1+) were isolated from Prdm14-transduced and EV-transduced mice at four weeks post transplantation, and examined for transcript profiles using microarrays. 1450 genes were differentially expressed (p ≤ 0.05) between the two groups. Of genes with fold change differences of 1.5 and higher, 284 were upregulated and 125 were downregulated in CLP-like cells from Prdm14-transduced mice (Supplementary Table 1). Correlation of differential expression distinguished between Prdm14-transduced and EV-transduced IL-7Rα+lin−Kit+Sca1+ cells (Supplementary Figure 4). Gene ontology and pathway analyses of differentially expressed genes revealed genes involved in early B-cell lineage commitment; regulators and mediators of Wnt, AKT, and Ras pathways; epithelial-to-mesenchymal transition; and previously well-characterized oncogenes (Figure 5a). Strikingly, a number of the transcripts distinguishing the two cell populations examined were for genes or non-coding RNAs that are regulated by imprinting. Both maternally and paternally imprinted genes were overexpressed, including genes expressed from the same locus such as Dlk1 and Meg3 (Gtl2). Downregulated genes included signal transduction kinases and B-cell activation genes (Figure 5a).
Figure 5. Prdm14 alters expression of pluripotency genes and oncogenes in pre-leukemic and leukemic cells.
5a) Heatmap of gene expression in flow-sorted CLP-like cells obtained at four weeks post transplantation from Prdm14-transduced and EV-transduced mice. Each column represents one sample. Genes depicted are relevant in oncogenesis, tumor suppression, pluripotency, signal transduction, or B cell development. 5b) Expression of Pou5f1, Prdm1, Dlk1, Meg3, Mtor, Il7r, Myb, Scin, and Glp is increased in tumors from Prdm14-transduced mice. Sox2 is downregulated. Levels in tumors are compared to both wild-type whole bone marrow and spleen from EV recipients using Mann-Whitney analysis. Error bars represent median with interquartile range (* indicates p<0.05 and ** indicates p<0.01). 5c) Western blot analysis shows increased MYB protein in Prdm14-induced leukemias, which is seen neither in EV controls nor in pre-leukemic cells. C = 8 week old spleen control; Prdm14 Pre-LL = spleen from a Prdm14 recipient that had not developed a tumor.
Data from gene expression profiles of selected genes in individual splenic tumors from Prdm14-transduced mice showed sustained upregulation of imprinted genes such as Meg3 and Dlk1. Strikingly, additional genes were upregulated in tumors, including the pluripotency gene Pou5f1 and Prdm14’s PGC partner Prdm1 (Figure 5b). Myb was highly expressed in Prdm14-induced LLs, but not in pre-leukemic cells (Figure 5b). Myb protein was also upregulated in tumors compared to pre-leukemic cells overexpressing Prdm14 (Figure 5c). Sox2 was downregulated, but other pluripotency genes (Nanog) and early B-cell genes (Pax5, Ebf1, Pu.1) were expressed at similar levels in tumors compared to controls (Figure 5b, Supplementary Figure 5).
PRDM14 is overexpressed in a subset of human ALLs
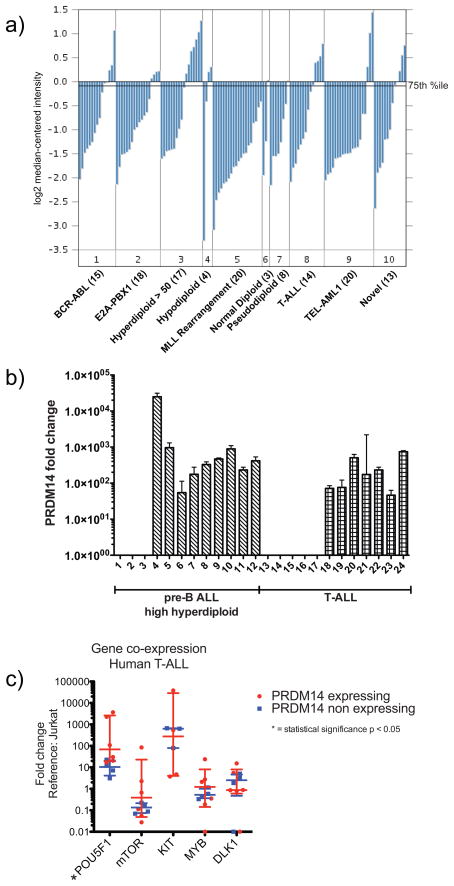
We next determined the degree and frequency and levels of PRDM14 expression in human ALL. Outlier analyses of microarray gene expression data from multiple previous studies of pediatric ALL showed that the levels of PRDM14 expression in a subset of 10–25% of patients placed PRDM14 among the top 1–2% of most overexpressed of all genes in these cohorts (Bhojwani et al 2006, Maser et al 2007, Ross et al 2003, Sorich et al 2008). In one study of 132 patients, in which cytogenetic abnormalities and histology were available, overexpression of PRDM14 was associated most frequently with high hyperdiploid (chromosome number greater than 50) tumors as well as T-ALL (Figure 6a) (Ross et al 2003).
Figure 6. PRDM14 is overexpressed in a subset of pediatric ALL samples.
6a) PRDM14 relative expression was evaluated in 132 pediatric ALL samples (Ross et al 2003) using Affymetrix Human Genome U133A arrays, normalized to median-centered fluorescence intensity. Samples were sorted by cytogenetics and histologic subtype. Line marked “75th %ile” denotes the 75th percentile of fluorescence intensity within this cohort; samples with expression intensity above this line were compared to samples below for outlier analysis of relative degree of overexpression. 6b) PRDM14 relative expression determined by qPCR using TaqMan gene expression assays. Expression normalized to tata-box binding protein (TBP); fold change compared to Jurkat cell line. Error bars represent 95% confidence interval of replicates. 6c) In T-ALL, POU5F1 expression is increased in samples expressing Prdm14. Other genes evaluated showed no significant difference, although mTOR expression trended higher in the PRDM14-expressing group. Error bars represent median with interquartile range (* indicates p<0.05).
In light of these findings, we examined PRDM14 expression levels in primary tumor samples from patients with high hyperdiploid ALL and T-ALL. Preliminary analysis of 24 samples confirmed PRDM14 expression in 9/12 (75%) of high hyperdiploid pre-B ALLs and 7/12 (58%) of T-ALL samples (Figure 6b). Although the levels of PRDM14 expression were low relative to expression levels of reference genes (data not shown), the transcript levels in PRDM14-expressing samples were at least 50-fold greater than the lowest levels detectable by our assay (Figure 6b). Comparisons of PRDM14 transcript levels with those of other pluripotency genes demonstrated statistically significant overexpression of POU5F1 (OCT4) in PRDM14-expressing T-ALL samples (Figure 6c; Supplementary Figure 6).
Discussion
Here we show for the first time that Prdm14 acts as a proto-oncogene in vivo. Overexpression of Prdm14 via retroviral transduction of mouse BM cells used to reconstitute lethally irradiated mice resulted in the development of LLs. Interestingly, the incidence of disease in female mice (96%) was more than twice that in male mice (42%). The onset of LL was preceded by a marked expansion of cells with the immunophenotype of CLPs. Both pre-leukemic cells and LLs from Prdm14 recipients expressed a series of pluripotency markers and oncogenes at high levels. Additionally, we detected overexpression of PRDM14 in the majority of high hyperdiploid and T-ALL samples, both in analyses of previously published array data as well as in our own analysis of primary patient samples. Significantly, little or no PRDM14 expression has been previously detected in normal mouse or human tissues other than in pluripotent cells (Dettman and Justice 2008, Nishikawa et al 2007, Surani et al 2007, Tsuneyoshi et al 2008, Yamaji et al 2008). Combined with our leukemogenic mouse model, our discovery of PRDM14 expression in human leukemias suggests a novel mechanism of cancer initiation and possibly tumor progression in human hematopoietic malignancies.
Importantly, we have shown that a PR domain-containing protein can initiate cancer. Usually, isoforms of genes that contain a PR domain act as tumor suppressors in vivo. Even though Prdm1 functions in a signaling cascade with Prdm14 in establishing pluripotency in primordial germ cell (PGC) development (Ohinata et al 2009), PRDM1 functions as a tumor suppressor in DLBCLs (Pasqualucci et al 2006). Perhaps the differences between Prdm14 and Prdm1 in cancer are related to their normal functions in PGC development: Prdm1 represses somatic cell gene transcription in PGCs, while Prdm14 is essential for epigenetic reprogramming events required to establish the pluripotency of nascent PGCs (Yamaji et al 2008). The high ratio of upregulated to downregulated genes in our gene expression array analyses also suggests that Prdm14 functions in transcriptional activation, unlike other PR-domain containing proteins, which primarily function as transcriptional repressors. Our expression data are consistent with expression profiles of human ES cells depleted of PRDM14 (Chia et al 2010).
Prdm14 acts as a proto-oncogene by interfering with normal hematopoietic development. Despite levels of engraftment similar to those of EV-transduced marrow cells at four weeks post reconstitution, Prdm14-expressing marrow cells contributed a smaller fraction of PB cells. By twelve weeks post reconstitution, engraftment of Prdm14-transduced cells had declined, and the contribution of Prdm14-expressing cells to T-cell, myeloid cell, and erythrocytic lineages was negligible. While these findings could suggest innately impaired engraftment potential due to Prdm14 expression, it is more likely that they simply reflect the skewed cellular differentiation that occurs in Prdm14-transduced marrow. Most significantly, marrow populations of IL7R+lin− early lymphoid progenitors, especially CLP-like cells, were dramatically increased at four weeks over baseline. By twelve weeks, the frequency of these IL-7Rα+lin−Kit+Sca1+ cells started to decline. However, a concomitant expansion of B-lineage cells and developmental arrest at the pro-B cell stage both suggest that a fraction of these cells attempted but failed to complete B-cell differentiation. Gene expression analysis of pre-leukemic cells was consistent with this block, with upregulation of genes associated with commitment to the B-lineage (Tcf3, Il7r) and downregulation of genes associated with B-cell maturation and activation (Cd27, Lat2, Blnk). Despite waning overall engraftment of transduced cells, 80% of Prdm14 recipients eventually developed a spectrum of clonal, GFP-expressing LLs arrested at varying stages of lymphoid development including pre-pro-B, pro-B, precursor T, and even CLP. This diversity of tumors suggests that the expansion of a hardy immature lymphoid population derived from Prdm14-transduced cells led directly to the development of leukemias in these mice.
Epigenetic events that would normally lead to a pluripotent state in germ cells seem to be perturbed in cells aberrantly expressing Prdm14, resulting in expanded populations of stem-like progenitor cells. Although global epigenetic changes were not observed in Prdm14-expressing tumors (data not shown), the ZFs in PR domain proteins target the protein to specific DNA sequences (Chia et al 2010, Myers et al 2010). Prdm14-expressing pre-leukemic cells overexpressed genes at imprinted loci, including H19, Odz4, Dlk1, and Meg3. Dlk1 and Meg3 are encoded within a complex locus, Dlk1-Dio3, which regulates the pluripotency of mouse ES cells and iPS cells (Liu et al 2010, Stadtfeld et al 2010). Differentially methylated regions (DMR) throughout the locus regulate reciprocal imprinting of Dlk1 and adjacent non-coding RNAs including Meg3. In human disease, abnormal methylation at DLK1 correlates with biallelic overexpression in 61% of acute myelogenous leukemias (Khoury et al 2010). Further, DLK1 overexpression is associated with myelodysplastic syndrome as well as megakaryocytic and erythroleukemias (Sakajiri et al 2005). Given that Prdm14 resets the epigenetic programming of PGCs, the overexpression of imprinted genes in our tumors indicates that genes or long non-coding RNAs within loci such as Dlk1-Dio3 may be targets of Prdm14 in resetting pluripotency. Surprisingly, Prdm14 overexpression induced upregulation of the early embryonic histone methyltransferase Glp. Glp is also upregulated in Prdm14−/− mice, leading the authors to conclude that Prdm14 directly inhibits Glp (Yamaji et al 2008). However, our finding may reflect a more complex regulatory scheme for expression of Glp.
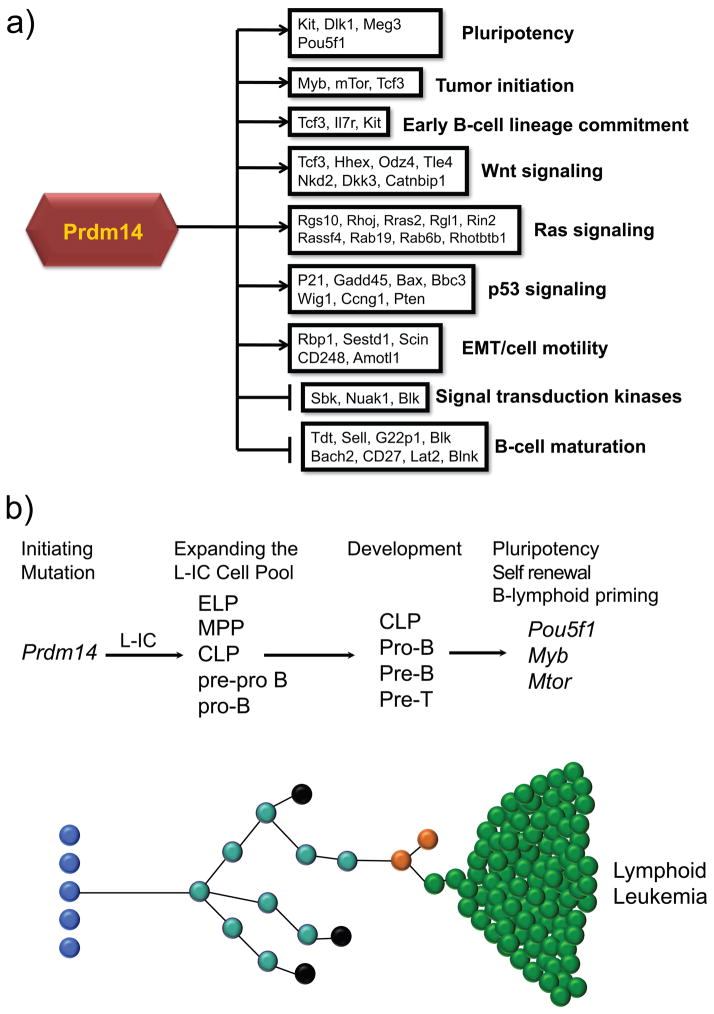
Based on our gene expression profiling of the pre-leukemic population, Prdm14 triggered a cascade of altered gene expression induced via multiple pathways involved in oncogenesis (Figure 7a). Upregulation of genes involved in the EMT suggests a phenotype of cancer stem cell activation, tumor invasiveness, increased metastasis, and increased drug resistance (Hennessy et al 2009, Singh and Settleman 2010). Self-renewal pathways (Wnt, Ras, AKT) exhibited altered expression of pathway initiators (i.e. Hhex, Tcf3) and mediators (i.e. Mtor) suggestive of partial constitutive pathway activation. Notably, stem cell markers such as Kit and Myc were overexpressed in tumors compared to normal spleen, but had the same expression levels as in bone marrow, reflective of stem cell-like characteristics in mature tumors. These genes are associated with both PGC development and a stem cell phenotype in human leukemias and breast cancers (Besmer et al 1993, Ko et al 2003, Palmu et al 2002, Tsuda et al 2005).
Figure 7. A model for the development of lymphoblastic leukemia.
7a) Schematic of dyregulated pathways induced by Prdm14 overexpression. 7b) Overexpression of PRDM14 initiates leukemia by expanding a progenitor cell pool after its misexpression in hematopoietic stem cells. This pool of leukemia-initiating cells (L-ICs) has an immunophenotype and developmental capacity that is similar to the CLP. Other cells may also be expanded, including multipotential progenitors (MPPs), lymphoid primed multipotential progenitors (LMPPs), early lymphoid progenitors (ELPs), pre-pro B or pro-B cells. Development of these pools occurs as clones are expanded. Most clones likely undergo apoptosis (black), but a rare subclone, expressing GFP, acquires transformed properties with the expression of Pou5f1, Mtor, and Myb.
Although pre-leukemic cell populations expanded soon after transplantation, Prdm14-transduced mice exhibited an extended latent period prior to developing leukemia. Activation of the p53 pathway and the pro-apoptotic Bcl-family genes Bax and Bbc3 (PUMA) may prevent the rapid development of tumors (Figure 7a). Unlike the pre-leukemic cells, however, mature tumors exhibited further gene expression changes that likely overwhelm normal anti-proliferative controls. Eventual overexpression of Pou5f1 and Prdm1 in mature tumors, in combination with previously expressed genes from the Dlk1-Dio3 locus, point to a powerful trend toward pluripotent drive independent of Sox2 and Nanog expression. POU5F1 overexpression in both mouse LLs and human T-ALL with PRDM14 overexpression suggests activation of pluripotency as a means of self-renewal. Prdm14-induced LLs also expressed Myb at high levels, both as transcript and protein. Myb induces increased self-renewal of hematopoietic cells and is required for the up-regulation of many lymphoid-associated genes, including Il7r, as well as for the earliest stages of lymphoid commitment in lymphoid primed progenitors (Greig et al 2010, Ramsay and Gonda 2008). Given the essential role of Myb in the development of the CLP population (Greig et al 2010), eventual expansion of and tumor evolution from our abnormal CLP-like cells may well be triggered once Myb expression increases. MYB is also highly expressed in estrogen receptor-positive breast cancer cells, and its transcription is activated when estradiol binds estrogen receptor alpha (Drabsch et al 2007, Guerin et al 1990). The linkage between estrogen and levels of Myb expression may help explain the higher frequency of Prdm14-induced LLs in female than in male mice observed in this study.
Identification of cells with cancer-initiating properties has become a driving force in tumor biology. It has been hypothesized that stem cells with the ability to self-renew, a process by which cells maintain the ability to undergo extensive proliferation while preserving the undifferentiated state, lie at the origins of all cancers with tumors developing from maturational arrest of tissue-specific stem cells. The results of the current study outline a model of an ancestral, pre-leukemic clone in the mouse from a population of IL-7Rα+lin−Kit+Sca1+ lymphoid progenitor cells with impaired ability to complete maturation. Evolution from an apparent ancestral clone has been demonstrated in childhood ALL in multiple analyses of paired diagnosis and relapse samples, in which tumors from a subset of patients lost some chromosomal abnormalities at relapse, while other abnormalities were retained (Davi et al 1996, Davidsson et al 2010, Ford et al 2001, Mullighan et al 2008). We therefore propose a model for the development of PRDM14-expressing lymphoid leukemias: an initiating mutation in a susceptible progenitor cell perturbs its self-renewal or differentiation to expand a pool of leukemia-initiating cells (L-ICs), which initially proliferate slowly (Figure 7b). The L-ICs attempt but fail to complete the developmental program, which then goes awry (Castor et al 2005, Jordan 2006, Tremblay et al 2010). In human disease, these persistent and abnormal L-IC clones could resist therapy simply due to their indolent state, with their propensity to self-renew and acquire new mutations leading to subsequent malignant retransformation and disease relapse.
Although LLs arise from an expanded progenitor pool in our mouse model of Prdm14-induced leukemogenesis, PRDM14 may be a cancer stem cell oncogene that is dependent on the cellular milieu in which it is expressed to trigger cancer. For instance, PRDM14 is overexpressed in a large proportion of human breast cancers as a result of gene amplification and in association with aberrant CpG methylation of the second intron (Hu et al 2005, Nishikawa et al 2007). If PRDM14 acts similarly in breast tissue and in hematopoietic cells, it may activate a pluripotency cascade in differentiated cells that leads to the development of self-renewing, de-differentiated, pre-cancerous clones, if not a “cancer stem cell” (Villadsen et al 2007). We conclude that in light of its broad relevance to cancer initiation, as well as its limited expression in normal tissues, PRDM14 and its regulatory targets are likely to provide prime candidates for therapeutic interventions in cancer.
Materials and Methods
Animal care
All mouse experiments were carried out under the approval of the Institutional Animal Care and Use Committee at Baylor College of Medicine (BCM). Mice were housed in the barrier facility at BCM, under the care of the Center for Comparative Medicine, which is accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International.
Human samples
Patient samples were obtained between 1996 and 2004 through an institutional review board-approved protocol (Molecular Characterization of Normal and Abnormal Hematopoiesis, H3342) at Texas Children’s Hospital (Houston, TX) after informed consent was obtained. Samples from PB via venipuncture or leukopheresis, or from BM via aspiration, were lysed in Ultraspec RNA reagent (Biotecx, Houston, TX) for long-term storage.
Molecular cloning of Prdm14
The coding sequence of Prdm14 was amplified using Platinum Pfx (Invitrogen, Carlsbad, CA) using cDNA isolated from the Evi32 insertion-containing tumor, 27-001, and subcloned into the MIGR1 vector, a gift from Robert Hawley (Hawley et al 1992) using XhoI restriction enzyme sites designed in the primer sequences. The murine stem cell virus-based vector was chosen due to its known efficiency in infecting hematopoietic stem cells (HSCs) (Persons et al 1997, Sauvageau et al 1995, Yan et al 1996). The recombinant virus was sequenced to verify integrity and orientation.
Retroviral transductions
Retroviral supernatants were produced via transient transfection of 293T cells with the Prdm14-MIGR1 vector or a MIGR1-EV along with the PCL-Eco packaging construct using Lipofectamine 2000 (Invitrogen) (Naviaux et al 1996). Retrovirus-containing supernatants were collected 48 hours post-transfection and were titered by infection of NIH3T3 fibroblasts followed by flow cytometric analyses to determine percentages of GFP-positive cells. Donor C57BL/6J-CD45.2 mice were injected intraperitoneally with 150 mg/kg of 5-FU. The mice were killed six days later, and total BM cells were isolated. Donor BM cells were incubated in Stempro 34 media (Invitrogen) containing 6 ng/ml IL-3, 10 ng/ml IL-6, and 100 ng/ml stem cell factor (SCF) (Peprotech, Rocky Hill, NJ). Polybrene was added to the media at a final concentration of 4 μg/mL, and retroviral supernatants were added to the cells at a MOI of one. Cells were spin-infected at 1266xg for two hours at room temperature. Sex-matched recipient C57BL/6J-CD45.1 mice given a split dose of 11 gray of γ-irradiation were injected via the retro-orbital sinus with 105 BM cells. Control mice were subjected to irradiation to ensure lethal levels. Irradiated mice were monitored daily for signs of morbidity.
Flow cytometry
Flow cytometry was carried out on PB and on cell suspensions prepared from tumors, BM, spleen, and thymus. PB was collected from the retro-orbital sinus followed by treatment with ammonium-chloride lysing buffer to remove red blood cells. Cells were stained with cocktails of fluorescently-labeled antibodies for lineage (CD45.2, B220, CD4, CD8, Mac1, and Gr1), for B-cell development (CD19, CD43, IgM, and B220), for late T-cell development (CD3, CD4, and CD8), for early T-cell development (CD25, CD44, CD4, and CD8), for CLP (IL7-Rα, Kit, Sca-1, CD4, CD8, CD3, B220, Ter119, Mac1, and Gr1), or for erythroid and natural killer (NK) cells (Ter119 and NK1.1) (BD Biosciences, San Jose, CA; eBiosciences, San Diego, CA). Nucleated cells were gated based on forward scatter (FSC) and side scatter (SSC), with propidium iodide (PI) staining used to exclude dead cells. Cells were analyzed using a four laser BD Special Order LSR-II (BD Biosciences). FlowJo Software was used for data analyses (Tree Star Inc., Ashland, OR).
Gene expression analysis
RNA was extracted, and cDNA generated, according to standard protocols (Supplementary Methods). Quantitative reverse transcription polymerase chain reaction (qPCR) was carried out in triplicate using the ABI Prism 7000 Sequence Detection System and ABI 7900HT Fast Real Time PCR system (Applied Biosystems [ABI], Carlsbad, CA). Gene expression studies were conducted with SybrGreen Master Mix (ABI) and custom primers (Supplementary Table 2), designed using Primer Express 3.0 (ABI) and RealTime PCR design tool (Integrated DNA Technologies, Coralville, IA). Human PRDM14 expression was determined using TaqMan gene expression assay Hs01119055_m1 with Gene Expression Master Mix (ABI). PCR and fold change analysis were performed as described previously (Dettman and Justice 2008). GAPDH and 18S rRNA served as reference genes in mouse tissue; TBP (ABI assay 4326322E) was used as the reference in human tissue (Lossos et al 2003).
Gene expression microarray
30,000 IL-7Ra+lin−Kit+Sca1+ cells were sorted from whole bone marrow obtained from Prdm14-transduced and EV-transduced mice four weeks post transplantation using BD FACSAira II (BD Biosciences), with >92% post-sort cell purity. Total RNA was obtained using RNAqueous Micro (Ambion). RNA quality was assessed with the Experion bioanalyzer using the Stdsens Chip (BioRad, Hercules, CA). RNA samples were amplified with in vitro transcription using the Illumina TotalPrep RNA Amplification kit (Ambion). RNA was then labeled and hybridized to MouseWG-6 v2.0 Expression BeadChip arrays (Illumina, San Diego, CA). Data was normalized using variance-stabilizing transformation, robust spine normalization. Quality control (Supplementary Figure 4), hybridization, and data generation were performed by the Laboratory for Translational Genomics at BCM. Data were analyzed by R (www.r-project.org) and visualized in Genespring (Agilent, Santa Clara, CA). Gene ontology and pathway analysis was performed with the Database for Annotation, Visualization, and Integrated Discovery at http://david.abcc.ncifcrf.gov (Dennis et al 2003, Huang da et al 2009).
Histology/Pathology
Tumors were fixed using 10% aqueous buffered zinc formalin overnight at room temperature. Samples were dehydrated in ethanol and embedded in paraffin. Tumors were stained with hematoxylin and eosin (Sigma-Aldrich, St. Louis, MO) and analyzed according to established parameters in mouse tumor identification (Fredrickson et al 1995, Morse et al 2002).
Western analysis
Western blots were performed using protein extracts from homogenized tumor cells and sorted cells, as previously described (Boles et al 2009). Antibodies directed against actin and Myb were obtained commercially (Santa Cruz Biotechnology, Santa Cruz, CA).
Statistical analysis
Publicly available expression data from four pediatric ALL studies (Bhojwani et al 2006, Maser et al 2007, Ross et al 2003, Sorich et al 2008) were evaluated for PRDM14 expression levels. Outlier analysis was conducted at the 75th, 90th, and 95th percentiles of expression levels to evaluate for presence of a subset of tumors overexpressing PRDM14. Oncomine™ (Compendia Bioscience, Ann Arbor, MI) was used for analysis and visualization.
Comparative analysis of gene expression fold change was performed with DataAssist v2.0 (ABI) and Prism v5.0 (GraphPad, La Jolla, CA). Mann-Whitney tests were used to compare relative gene expression levels; error bars represent the median with interquartile range. Logrank analysis of overall survival was performed using Prism v5.0.
Supplementary Material
Acknowledgments
The work by MJJ was supported by NIH R01 CA63229, R01 CA15530 and a Baylor College of Medicine Interim funding award. SJS was supported by an NIH National Research Service Award Institutional Training Grant 3 T32 CA115303-04, “Pediatric Oncology Research Training Program.” HCM was supported by the Intramural Research Program of the NIH, National Institute of Allergy and Infectious Diseases. We thank John Belmont and Molly Bray at the Laboratory for Translational Genomics at BCM for microarray data production; Jill Crowe for excellent technical assistance; and Frank Probst for the critical reading of the manuscript.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Supplementary information is available at Oncogene’s website (http://www.nature.com/onc).
References
- Akagi K, Suzuki T, Stephens RM, Jenkins NA, Copeland NG. RTCGD: retroviral tagged cancer gene database. Nucleic Acids Res. 2004;32:D523–527. doi: 10.1093/nar/gkh013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudat F, Buard J, Grey C, Fledel-Alon A, Ober C, Przeworski M, et al. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science. 2010;327:836–840. doi: 10.1126/science.1183439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besmer P, Manova K, Duttlinger R, Huang EJ, Packer A, Gyssler C, et al. The kit-ligand (steel factor) and its receptor c-kit/W: pleiotropic roles in gametogenesis and melanogenesis. Dev Suppl. 1993:125–137. [PubMed] [Google Scholar]
- Bhojwani D, Kang H, Moskowitz NP, Min DJ, Lee H, Potter JW, et al. Biologic pathways associated with relapse in childhood acute lymphoblastic leukemia: a Children’s Oncology Group study. Blood. 2006;108:711–717. doi: 10.1182/blood-2006-02-002824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boles MK, Wilkinson BM, Wilming LG, Liu B, Probst FJ, Harrow J, et al. Discovery of candidate disease genes in ENU-induced mouse mutants by large-scale sequencing, including a splice-site mutation in nucleoredoxin. PLoS Genet. 2009;5:e1000759. doi: 10.1371/journal.pgen.1000759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castor A, Nilsson L, Astrand-Grundstrom I, Buitenhuis M, Ramirez C, Anderson K, et al. Distinct patterns of hematopoietic stem cell involvement in acute lymphoblastic leukemia. Nat Med. 2005;11:630–637. doi: 10.1038/nm1253. [DOI] [PubMed] [Google Scholar]
- Chia NY, Chan YS, Feng B, Lu X, Orlov YL, Moreau D, et al. A genome-wide RNAi screen reveals determinants of human embryonic stem cell identity. Nature. 2010 doi: 10.1038/nature09531. [DOI] [PubMed] [Google Scholar]
- Davi F, Gocke C, Smith S, Sklar J. Lymphocytic progenitor cell origin and clonal evolution of human B-lineage acute lymphoblastic leukemia. Blood. 1996;88:609–621. [PubMed] [Google Scholar]
- Davidsson J, Paulsson K, Lindgren D, Lilljebjorn H, Chaplin T, Forestier E, et al. Relapsed childhood high hyperdiploid acute lymphoblastic leukemia: presence of preleukemic ancestral clones and the secondary nature of microdeletions and RTK-RAS mutations. Leukemia. 2010;24:924–931. doi: 10.1038/leu.2010.39. [DOI] [PubMed] [Google Scholar]
- Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- Dettman EJ, Justice MJ. The zinc finger SET domain gene Prdm14 is overexpressed in lymphoblastic lymphomas with retroviral insertions at Evi32. PLoS One. 2008;3:e3823. doi: 10.1371/journal.pone.0003823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downing JR, Mullighan CG. Tumor-specific genetic lesions and their influence on therapy in pediatric acute lymphoblastic leukemia. Hematology/the Education Program of the American Society of Hematology American Society of Hematology. 2006:118–122. 508. doi: 10.1182/asheducation-2006.1.118. [DOI] [PubMed] [Google Scholar]
- Drabsch Y, Hugo H, Zhang R, Dowhan DH, Miao YR, Gewirtz AM, et al. Mechanism of and requirement for estrogen-regulated MYB expression in estrogen-receptor-positive breast cancer cells. Proc Natl Acad Sci U S A. 2007;104:13762–13767. doi: 10.1073/pnas.0700104104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford AM, Fasching K, Panzer-Grumayer ER, Koenig M, Haas OA, Greaves MF. Origins of “late” relapse in childhood acute lymphoblastic leukemia with TEL-AML1 fusion genes. Blood. 2001;98:558–564. doi: 10.1182/blood.v98.3.558. [DOI] [PubMed] [Google Scholar]
- Fredrickson TN, Hartley JW, Morse HC, 3rd, Chattopadhyay SK, Lennert K. Classification of mouse lymphomas. Current topics in microbiology and immunology. 1995;194:109–116. doi: 10.1007/978-3-642-79275-5_14. [DOI] [PubMed] [Google Scholar]
- Fumasoni I, Meani N, Rambaldi D, Scafetta G, Alcalay M, Ciccarelli FD. Family expansion and gene rearrangements contributed to the functional specialization of PRDM genes in vertebrates. BMC evolutionary biology. 2007;7:187. doi: 10.1186/1471-2148-7-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greig KT, de Graaf CA, Murphy JM, Carpinelli MR, Pang SH, Frampton J, et al. Critical roles for c-Myb in lymphoid priming and early B-cell development. Blood. 2010;115:2796–2805. doi: 10.1182/blood-2009-08-239210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerin M, Sheng ZM, Andrieu N, Riou G. Strong association between c-myb and oestrogen-receptor expression in human breast cancer. Oncogene. 1990;5:131–135. [PubMed] [Google Scholar]
- Hansen GM, Skapura D, Justice MJ. Genetic profile of insertion mutations in mouse leukemias and lymphomas. Genome Res. 2000;10:237–243. doi: 10.1101/gr.10.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao X, Shin MS, Zhou JX, Lee CH, Qi CF, Naghashfar Z, et al. Histologic and molecular characterizations of megakaryocytic leukemia in mice. Leuk Res. 2006;30:397–406. doi: 10.1016/j.leukres.2005.08.021. [DOI] [PubMed] [Google Scholar]
- Hawley RG, Fong AZ, Burns BF, Hawley TS. Transplantable myeloproliferative disease induced in mice by an interleukin 6 retrovirus. The Journal of experimental medicine. 1992;176:1149–1163. doi: 10.1084/jem.176.4.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K, Yoshida K, Matsui Y. A histone H3 methyltransferase controls epigenetic events required for meiotic prophase. Nature. 2005;438:374–378. doi: 10.1038/nature04112. [DOI] [PubMed] [Google Scholar]
- Hennessy BT, Gonzalez-Angulo AM, Stemke-Hale K, Gilcrease MZ, Krishnamurthy S, Lee JS, et al. Characterization of a naturally occurring breast cancer subset enriched in epithelial-to-mesenchymal transition and stem cell characteristics. Cancer Res. 2009;69:4116–4124. doi: 10.1158/0008-5472.CAN-08-3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu M, Yao J, Cai L, Bachman KE, van den Brule F, Velculescu V, et al. Distinct epigenetic changes in the stromal cells of breast cancers. Nat Genet. 2005;37:899–905. doi: 10.1038/ng1596. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Jiang GL, Huang S. The yin-yang of PR-domain family genes in tumorigenesis. Histol Histopathol. 2000;15:109–117. doi: 10.14670/HH-15.109. [DOI] [PubMed] [Google Scholar]
- Jonkers J, Berns A. Retroviral insertional mutagenesis as a strategy to identify cancer genes. Biochimica et biophysica acta. 1996;1287:29–57. doi: 10.1016/0304-419x(95)00020-g. [DOI] [PubMed] [Google Scholar]
- Jordan CT. Searching for leukemia stem cells--not yet the end of the road? Cancer Cell. 2006;10:253–254. doi: 10.1016/j.ccr.2006.09.010. [DOI] [PubMed] [Google Scholar]
- Khoury H, Suarez-Saiz F, Wu S, Minden MD. An upstream insulator regulates DLK1 imprinting in AML. Blood. 2010;115:2260–2263. doi: 10.1182/blood-2009-03-212746. [DOI] [PubMed] [Google Scholar]
- Kim KC, Geng L, Huang S. Inactivation of a histone methyltransferase by mutations in human cancers. Cancer Res. 2003;63:7619–7623. [PubMed] [Google Scholar]
- Kim KC, Huang S. Histone methyltransferases in tumor suppression. Cancer Biol Ther. 2003;2:491–499. doi: 10.4161/cbt.2.5.629. [DOI] [PubMed] [Google Scholar]
- Kinameri E, Inoue T, Aruga J, Imayoshi I, Kageyama R, Shimogori T, et al. Prdm proto-oncogene transcription factor family expression and interaction with the Notch-Hes pathway in mouse neurogenesis. PLoS ONE. 2008;3:e3859. doi: 10.1371/journal.pone.0003859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko CD, Kim JS, Ko BG, Son BH, Kang HJ, Yoon HS, et al. The meaning of the c-kit proto-oncogene product in malignant transformation in human mammary epithelium. Clin Exp Metastasis. 2003;20:593–597. doi: 10.1023/a:1027323210736. [DOI] [PubMed] [Google Scholar]
- Kondo M, Weissman IL, Akashi K. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell. 1997;91:661–672. doi: 10.1016/s0092-8674(00)80453-5. [DOI] [PubMed] [Google Scholar]
- Kurimoto K, Yamaji M, Seki Y, Saitou M. Specification of the germ cell lineage in mice: a process orchestrated by the PR-domain proteins, Blimp1 and Prdm14. Cell Cycle. 2008;7:3514–3518. doi: 10.4161/cc.7.22.6979. [DOI] [PubMed] [Google Scholar]
- Liu L, Luo GZ, Yang W, Zhao X, Zheng Q, Lv Z, et al. Activation of the imprinted Dlk1-Dio3 region correlates with pluripotency levels of mouse stem cells. J Biol Chem. 2010 doi: 10.1074/jbc.M110.131995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lossos IS, Czerwinski DK, Wechser MA, Levy R. Optimization of quantitative real-time RT-PCR parameters for the study of lymphoid malignancies. Leukemia. 2003;17:789–795. doi: 10.1038/sj.leu.2402880. [DOI] [PubMed] [Google Scholar]
- Maser RS, Choudhury B, Campbell PJ, Feng B, Wong KK, Protopopov A, et al. Chromosomally unstable mouse tumours have genomic alterations similar to diverse human cancers. Nature. 2007;447:966–971. doi: 10.1038/nature05886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moelans CB, de Weger RA, Monsuur HN, Vijzelaar R, van Diest PJ. Molecular profiling of invasive breast cancer by multiplex ligation-dependent probe amplification-based copy number analysis of tumor suppressor and oncogenes. Mod Pathol. 2010 doi: 10.1038/modpathol.2010.84. [DOI] [PubMed] [Google Scholar]
- Morse HC, 3rd, Anver MR, Fredrickson TN, Haines DC, Harris AW, Harris NL, et al. Bethesda proposals for classification of lymphoid neoplasms in mice. Blood. 2002;100:246–258. doi: 10.1182/blood.v100.1.246. [DOI] [PubMed] [Google Scholar]
- Mullighan CG, Flotho C, Downing JR. Genomic assessment of pediatric acute leukemia. Cancer journal (Sudbury, Mass) 2005;11:268–282. doi: 10.1097/00130404-200507000-00003. [DOI] [PubMed] [Google Scholar]
- Mullighan CG, Goorha S, Radtke I, Miller CB, Coustan-Smith E, Dalton JD, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446:758–764. doi: 10.1038/nature05690. [DOI] [PubMed] [Google Scholar]
- Mullighan CG, Phillips LA, Su X, Ma J, Miller CB, Shurtleff SA, et al. Genomic analysis of the clonal origins of relapsed acute lymphoblastic leukemia. Science. 2008;322:1377–1380. doi: 10.1126/science.1164266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullighan CG, Su X, Zhang J, Radtke I, Phillips LA, Miller CB, et al. Deletion of IKZF1 and prognosis in acute lymphoblastic leukemia. The New England journal of medicine. 2009;360:470–480. doi: 10.1056/NEJMoa0808253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers S, Bowden R, Tumian A, Bontrop RE, Freeman C, MacFie TS, et al. Drive against hotspot motifs in primates implicates the PRDM9 gene in meiotic recombination. Science. 2010;327:876–879. doi: 10.1126/science.1182363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naviaux RK, Costanzi E, Haas M, Verma IM. The pCL vector system: rapid production of helper-free, high-titer, recombinant retroviruses. J Virol. 1996;70:5701–5705. doi: 10.1128/jvi.70.8.5701-5705.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa N, Toyota M, Suzuki H, Honma T, Fujikane T, Ohmura T, et al. Gene amplification and overexpression of PRDM14 in breast cancers. Cancer Res. 2007;67:9649–9657. doi: 10.1158/0008-5472.CAN-06-4111. [DOI] [PubMed] [Google Scholar]
- Ohinata Y, Ohta H, Shigeta M, Yamanaka K, Wakayama T, Saitou M. A signaling principle for the specification of the germ cell lineage in mice. Cell. 2009;137:571–584. doi: 10.1016/j.cell.2009.03.014. [DOI] [PubMed] [Google Scholar]
- Palmu S, Soderstrom KO, Quazi K, Isola J, Salminen E. Expression of C-KIT and HER-2 tyrosine kinase receptors in poor-prognosis breast cancer. Anticancer Res. 2002;22:411–414. [PubMed] [Google Scholar]
- Parvanov ED, Petkov PM, Paigen K. Prdm9 controls activation of mammalian recombination hotspots. Science. 2010;327:835. doi: 10.1126/science.1181495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasqualucci L, Compagno M, Houldsworth J, Monti S, Grunn A, Nandula SV, et al. Inactivation of the PRDM1/BLIMP1 gene in diffuse large B cell lymphoma. The Journal of experimental medicine. 2006;203:311–317. doi: 10.1084/jem.20052204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persons DA, Allay JA, Allay ER, Smeyne RJ, Ashmun RA, Sorrentino BP, et al. Retroviral-mediated transfer of the green fluorescent protein gene into murine hematopoietic cells facilitates scoring and selection of transduced progenitors in vitro and identification of genetically modified cells in vivo. Blood. 1997;90:1777–1786. [PubMed] [Google Scholar]
- Pizzo PA, Poplack DG. Principles and practice of pediatric oncology. 5. Lippincott Williams & Wilkins; Philadelphia: 2006. [Google Scholar]
- Ramsay RG, Gonda TJ. MYB function in normal and cancer cells. Nature reviews. 2008;8:523–534. doi: 10.1038/nrc2439. [DOI] [PubMed] [Google Scholar]
- Ross ME, Zhou X, Song G, Shurtleff SA, Girtman K, Williams WK, et al. Classification of pediatric acute lymphoblastic leukemia by gene expression profiling. Blood. 2003;102:2951–2959. doi: 10.1182/blood-2003-01-0338. [DOI] [PubMed] [Google Scholar]
- Sakajiri S, O’Kelly J, Yin D, Miller CW, Hofmann WK, Oshimi K, et al. Dlk1 in normal and abnormal hematopoiesis. Leukemia. 2005;19:1404–1410. doi: 10.1038/sj.leu.2403832. [DOI] [PubMed] [Google Scholar]
- Sauvageau G, Thorsteinsdottir U, Eaves CJ, Lawrence HJ, Largman C, Lansdorp PM, et al. Overexpression of HOXB4 in hematopoietic cells causes the selective expansion of more primitive populations in vitro and in vivo. Genes Dev. 1995;9:1753–1765. doi: 10.1101/gad.9.14.1753. [DOI] [PubMed] [Google Scholar]
- Singh A, Settleman J. EMT, cancer stem cells and drug resistance: an emerging axis of evil in the war on cancer. Oncogene. 2010;29:4741–4751. doi: 10.1038/onc.2010.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorich MJ, Pottier N, Pei D, Yang W, Kager L, Stocco G, et al. In vivo response to methotrexate forecasts outcome of acute lymphoblastic leukemia and has a distinct gene expression profile. PLoS Med. 2008;5:e83. doi: 10.1371/journal.pmed.0050083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Apostolou E, Akutsu H, Fukuda A, Follett P, Natesan S, et al. Aberrant silencing of imprinted genes on chromosome 12qF1 in mouse induced pluripotent stem cells. Nature. 2010;465:175–181. doi: 10.1038/nature09017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surani MA, Hayashi K, Hajkova P. Genetic and epigenetic regulators of pluripotency. Cell. 2007;128:747–762. doi: 10.1016/j.cell.2007.02.010. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Shen H, Akagi K, Morse HC, Malley JD, Naiman DQ, et al. New genes involved in cancer identified by retroviral tagging. Nat Genet. 2002;32:166–174. doi: 10.1038/ng949. [DOI] [PubMed] [Google Scholar]
- Tremblay M, Tremblay CS, Herblot S, Aplan PD, Hebert J, Perreault C, et al. Modeling T-cell acute lymphoblastic leukemia induced by the SCL and LMO1 oncogenes. Genes Dev. 2010;24:1093–1105. doi: 10.1101/gad.1897910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda H, Tani Y, Weisenberger J, Kitada S, Hasegawa T, Murata T, et al. Frequent KIT and epidermal growth factor receptor overexpressions in undifferentiated-type breast carcinomas with ‘stem-cell-like’ features. Cancer Sci. 2005;96:333–339. doi: 10.1111/j.1349-7006.2005.00060.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuneyoshi N, Sumi T, Onda H, Nojima H, Nakatsuji N, Suemori H. PRDM14 suppresses expression of differentiation marker genes in human embryonic stem cells. Biochem Biophys Res Commun. 2008;367:899–905. doi: 10.1016/j.bbrc.2007.12.189. [DOI] [PubMed] [Google Scholar]
- Villadsen R, Fridriksdottir AJ, Ronnov-Jessen L, Gudjonsson T, Rank F, LaBarge MA, et al. Evidence for a stem cell hierarchy in the adult human breast. The Journal of cell biology. 2007;177:87–101. doi: 10.1083/jcb.200611114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiser KC, Liu B, Hansen GM, Skapura D, Hentges KE, Yarlagadda S, et al. Retroviral insertions in the VISION database identify molecular pathways in mouse lymphoid leukemia and lymphoma. Mamm Genome. 2007;18:709–722. doi: 10.1007/s00335-007-9060-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaji M, Seki Y, Kurimoto K, Yabuta Y, Yuasa M, Shigeta M, et al. Critical function of Prdm14 for the establishment of the germ cell lineage in mice. Nat Genet. 2008;40:1016–1022. doi: 10.1038/ng.186. [DOI] [PubMed] [Google Scholar]
- Yan XQ, Lacey D, Hill D, Chen Y, Fletcher F, Hawley RG, et al. A model of myelofibrosis and osteosclerosis in mice induced by overexpressing thrombopoietin (mpl ligand): reversal of disease by bone marrow transplantation. Blood. 1996;88:402–409. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.