Abstract
The molecular and cellular profile of liver cells during early human development is incomplete, complicating the isolation and study of hepatocytes, cholangiocytes, and hepatic stem cells from the complex amalgam of hepatic and hematopoietic cells, that is, the fetal liver. Epithelial cell adhesion molecule, CD326, has emerged as a marker of hepatic stem cells, and lipopolysaccharide receptor CD14 is known to be expressed on adult hepatocytes. Using flow cytometry, we studied the breadth of CD326 and CD14 expression in midgestation liver. Both CD45+ hematopoietic and CD45− nonhematopoietic cells expressed CD326. Moreover, diverse cell types expressing CD326 were revealed among CD45− cells by costaining for CD14. Fluorescence-activated cell sorting was used to isolate nonhematopoietic cells distinguished by expression of high levels of CD326 and low CD14 (CD326++CD14lo), which were characterized for gene expression associated with liver development. CD326++CD14lo cells expressed the genes albumin, α-fetoprotein, hepatic nuclear factor 3α, prospero-related homeobox 1, cytochrome P450 3A7, α1-antitrypsin, and transferrin. Proteins expressed included cell-surface CD24, CD26, CD29, CD34, CD49f, CD243, and CD324 and, in the cytoplasm, cytokeratins-7/8 (CAM 5.2 antigen) and some cytokeratin-19. Cultured CD326++CD14lo cells yielded albumin+ hepatocytes, cytokeratin-19+ cholangiocytes, and hepatoblasts expressing both markers. Using epifluorescence microscopy we observed CD326 and CD14 expression on fetal hepatocytes comprising the liver parenchyma, as well as on cells associated with ductal plates and surrounding large vessels. These findings indicate that expression of CD14 and CD326 can be used to identify functionally distinct subsets of fetal liver cells, including CD326++CD14lo cells, representing a mixture of parenchymal cells, cholangiocytes, and hepatoblasts.
Introduction
Study of the liver during fetal development offers a unique insight into the biology of hepatic stem cells and progenitors during a period of growth unparalleled in the rest of ontogeny. The human liver develops in the embryo with an endodermal outgrowth of primitive foregut in the 3rd to 4th weeks of gestation [1]. Ductal plates are formed, which are double-layered cylinders of cells that migrate into surrounding mesenchyme to form intrahepatic bile ducts. These ductal plate cells contain hepatoblasts, bipotent cells that give rise to both hepatocytes and biliary cells [2]. The degree to which hepatoblasts represent a hierarchy of precursor cells, including stem cells with extensive self-renewal capacity and progenitors committed to differentiation into hepatocytes and cholangiocytes, is not fully understood. Molecular identification of hepatic precursors, especially through characterization of cell-surface protein expression, is required to isolate and study the cells that contribute to the development of the liver.
CD326 is a promising marker for the identification and isolation of hepatic precursors. CD326 is a glycoprotein cell-adhesion molecule that does not belong to any of the 4 major cell adhesion molecule families, for example, cadherins, selectins, integrins, or the immunoglobulin cell adhesion molecules [3]. CD326 is known by many names, including the commonly used epithelial cell adhesion molecule and human epithelial antigen-125. CD326 is expressed by various cancers and is associated with cell proliferation and a poor survival prognosis as its expression is associated with the most aggressive proliferating tumors [4]. Under normal circumstances, CD326 is detected in fetal epithelial tissues and in some adult epithelia. In the adult liver, CD326 is expressed on biliary cells but not on hepatocytes [4,5]. During fetal development CD326 is expressed by hepatic precursors. Dan et al. mechanically isolated progenitor cells, giving rise to hepatic colonies in vitro that expressed CD326 [6]. Schmelzer et al. isolated CD326+ cells using immunomagnetic beads and differentiated them into albumin (ALB)+ hepatic cells, suggesting that CD326 is a marker for hepatic stem cells [7]. In the rat, one study suggests that fetal liver (FL) cells expressing CD326 represent the cell fraction committed to the biliary lineage [8]. However, more recent results demonstrated that CD326+ cells isolated from adult rat liver are bipotential hepatic progenitors that can repopulate injured liver [9].
The lipopolysaccharide receptor CD14 is expressed by a number of cell populations in the liver. CD14 is a well-recognized marker of monocyte differentiation. Monocytes and hematopoietic progenitors of all types are found in the FL throughout much of human gestation [10,11]. Monocytes are the precursors of tissue macrophages, including Browicz-Kupffer cells found in the liver, which also express CD14 [12]. CD14 was further shown to be expressed on nonhematopoietic cell populations in the adult liver such as on sinusoidal endothelial cells [13] and hepatocytes. CD14 expression was observed on hepatocytes in adult rats and is upregulated by LPS treatment in vitro and in vivo [14,15]. Soluble CD14 was further shown to be produced by rat hepatocytes [14] and is an acute phase protein present in human serum [16]. Moreover, CD14 expression was observed on human hepatocytes transplanted into immunodeficient mice [17–19]. Soluble human CD14 was detected in these animals, providing proof that adult hepatocytes are a source of CD14 in acute phase serum [19].
In this study we examined if CD14 is expressed by hepatocytes and their precursors present in the human midgestation liver. CD14 expression was used to differentiate a population of hepatic precursors among cells expressing CD326 that had a protein and gene expression pattern consistent with a hepatocyte and cholangiocyte origin. This study demonstrates a hitherto unappreciated diversity among CD326-expressing cells in FL and distinguishes a unique population of fetal hepatic cells in early human ontogeny.
Materials and Methods
Fetal human tissues and cell isolation
FLs were obtained from elective abortions with the approval of the Committee for Human Research at the University of California San Francisco and consent of the women undergoing the abortion. The gestational age of the specimens was estimated based on foot-length. Midgestation specimens up to 24 weeks of age were obtained for this study. Tissues were harvested shortly after termination of the pregnancy and transported to the laboratory on ice in Hank's balanced salt solution (HBSS) supplemented with 100 μg/mL gentamicin and 2.5 μg/mL amphotericin B (Invitrogen).
FL cells were isolated by enzymatic digestion of liver fragments generated by manual dicing using scissors or a scalpel. Fragments were digested in a volume of 20–30 mL HBSS with 1.0 mg/mL collagenase enzyme blend (Liberase CI) and 0.005% DNase (Roche Diagnostic Corporation) for 30 min at 37°C with gentle agitation. Digestion was inhibited with cold fetal bovine serum (FBS) and the cell suspension gently passed through a wire mesh to disrupt any remaining large tissue clumps. Erythrocytes were depleted with CD235a monoclonal antibody (mAb) and immunomagnetic beads as previously described [11,20]. FL cells were collected, washed with HBSS, and resuspended in buffer for antibody staining. The hematopoietic fraction of FL cells was studied for CD326 expression by isolation of CD235a− and CD235a+ light-density cells as previously reported [20].
FL cells were suspended in blocking buffer consisting of phosphate-buffered saline (PBS) supplemented with 0.01% NaN3 (Sigma Chemical Co.) and 5% normal mouse serum (Gemini-Bio-Products, Inc.) for mAb labeling and isolation by fluorescence-activated cell sorting (FACS). Antibodies used in this study are listed in Supplementary Table S1 (Supplementary Data are available online at www.liebertonline.com/scd). Cells were stained with CD45-fluorescein isothiocyanate, CD14-phycoerythrin (PE), and CD326-allophycocyanin for ≥30 min on ice, washed, and suspended in PBS with 0.3% bovine serum albumin (BSA; Roche Diagnostic Corporation) and 2 μg/mL propidium iodide (PI; Invitrogen). Live cells (PI−) were sorted on a FACSAria flow cytometer (BD Biosciences) using a large nozzle and low to moderate flow rates.
Phenotypic analyses
Freshly isolated cells or cells harvested after culture were analyzed for antigen expression using an LSR II flow cytometer (BD Biosciences). Cell-surface staining was performed as previously described [20]. Cytoplasmic staining was performed using Fix and Perm reagents (Invitrogen). Cells were first labeled with a fixable violet-fluorescent reactive dye (LIVE/DEAD Fixable Dead Cell Stain Kits; Invitrogen), and then cell surface staining was performed before cell fixation and permeabilization for cytoplasmic staining. Analyses of flow cytometric data were performed only on live cells using an electronic gate to identify PI− or violet dye− events using FlowJo software (Tree Star, Inc.).
Quantitative real-time reverse-transcriptase polymerase chain reaction analysis
Total RNA was isolated by RNeasy mini kit (QIAGEN). The iScripttm cDNA Synthesis kit (Bio-Rad Laboratories Inc.) was used for synthesis of cDNA. Quantitative polymerase chain reaction (qPCR) was performed by the ABI Prism 7500 sequence detection system (Applied Biosystems) using SYBR Green I DNA-binding dye. Human 18S mRNA gene was used as endogenous control. Primers are listed in Supplementary Table S2. Samples were run in triplicate 15 μL reaction volumes. The size of product was verified by gel electrophoresis. Results were obtained as threshold cycle (CT) values. Expression levels were calculated using the ΔCT method. The values were calculated as the mean values of 3 independent measurements, and the expression levels of mRNA in all samples were defined as a ratio to 18S mRNA gene expression. The significance of differences in the levels of gene expression measured by qPCR was determined using the nonparametric 2-tailed Mann–Whitney U-test.
Cell culture
Cells were cultured in William's E medium supplemented as described [21] with 5% FBS (Gemini-Bio-Products) and 20 ng/mL epidermal growth factor (R&D Systems, Inc.) in 24-well collagen-coated plates (Biocoat® BD Labware) for 3 weeks. The fresh medium was added weekly. Cells were collected for RNA isolation or fixed for immunofluorescence staining after 3 weeks.
Immunofluorescence staining for epifluorescence microscopy
Small pieces of FL were fixed in 10% formalin overnight, washed in PBS, passed through sucrose gradient, and embedded into optimal cutting temperature (OCT) medium. About 5 μm cryostat sections were generated for staining. Cultured cells were fixed with 4% formaldehyde, washed in PBS with 0.1% BSA, and permealized in 0.3% Triton in PBS with 10% FBS and 0.1% BSA.
Sample slides were incubated with primary antibodies for 1 h at room temperature, washed with 0.1% BSA in PBS thrice, and then incubated with the appropriate secondary antibodies. Samples were covered with ProLong® Gold antifade reagent with 4′,6-diamidino-2-phenylindole (Invitrogen). Images were analyzed with a Leica CTR6500 (Leica Microsystems). Colocalization analysis was performed using software In Vivo (Media Cybernetics, Inc.)
Results
Expression of CD326 by hematopoietic cells in the FL
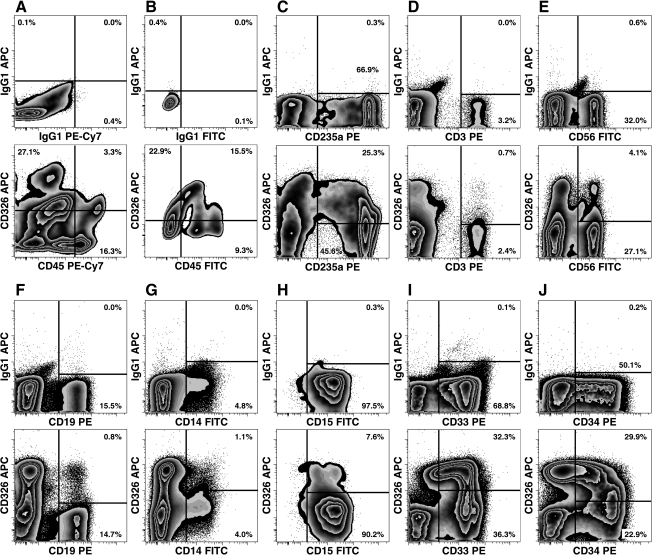
Analysis of midgestation liver indicated CD326 expression on CD45+ hematopoietic cells, although no CD326 expression was observed on adult peripheral blood cells (data not shown). Two different isolation procedures were used to prepare FLs for flow cytometric analyses. First, enzymatic digestion and erythrocyte depletion was used to isolate CD235a− FL cells of which 17% of CD45+ cells expressed CD326 (Fig. 1A). Second, light-density CD45+ FL cells had an even higher level of CD326 expression (Fig. 1B). Analyses of different hematopoietic lineages, found among the light-density FL cells, indicated that CD326-expressing cells included subsets of CD235a+ immature erythrocytes, CD3+ T-cells, CD56+ natural killer cells, CD19+ B-cells, CD14+ monocyte/macrophages, CD15+ granulocytes, CD33+ myeloid and progenitor cells, and CD34+ hematopoietic precursors (Fig. 1C–J, respectively), thus indicating widespread expression of CD326 by the hematopoietic fraction of the FL.
FIG. 1.
Expression of CD326 by hematopoietic cells. CD326 is expressed on CD45+ cells isolated from a 23 weeks' gestation FL by enzymatic digestion and erythrocyte (CD235a) depletion (A). CD326 expression is observed on hematopoietic cells enriched by density centrifugation of 20 weeks' gestation FL cells to remove hepatic parenchymal cells (B). Analyses of CD326 expression on different hematopoietic lineages isolated from a 21 weeks' gestation FL are shown in (C–J). Light-density erythroid precursors are shown in (C). Erythroblasts were depleted using immunomagnetic bead depletion to enrich leukocytes in for analysis (D–J). Leukocytes and erythroid precursors were further enriched in the analyses by electronic gating based on their characteristic forward and side light-scatter as well as their characteristic level of CD45 PE-Cy7 expression (not shown). Note, CD15 expression is shown only on high side scatter cells (granulocytes), whereas CD14 and CD33 expression levels are shown for both low and high scatter cells. In all analyses, dead cells were excluded based on positive PI staining. FL, fetal liver; PE, phycoerythrin; PI, propidium iodide.
CD14 and CD326 expression by nonhematopoietic cells in the FL
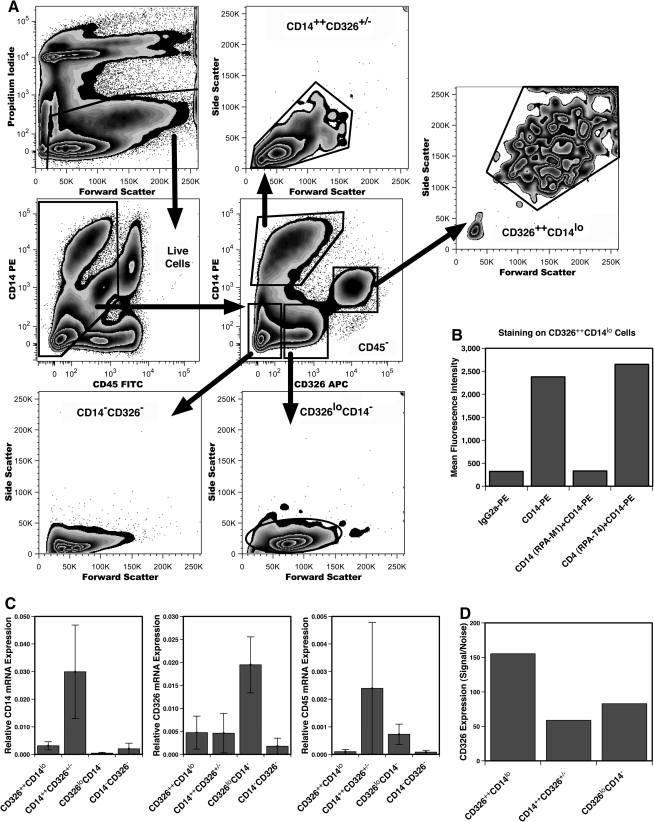
Expression of CD14 and CD326 was studied in a 4-color staining of CD235a− FL cells (Fig. 2A). Distinct populations of cells distinguished by their levels of CD14 and CD326 expression were evident among CD45− FL cells (Fig. 2A). Four subpopulations were defined that differed in size and cellular complexity as indicated by their forward and side light-scatter profiles. Cells expressing the highest levels of CD326, referred to as CD326++CD14lo cells, are among the largest cells in the FL. A second population of cells was defined by higher levels of CD14 with some of these cells expressing CD326 (CD14++CD326+/− cells). Low levels of CD326 expression were also observed on a subset that did not express CD14 (CD326loCD14− cells). Lastly, a fraction of cells remained that did not express any detectable CD14 or CD326, referred to as CD14−CD326− cells.
FIG. 2.
Expression of CD14 and CD326 on nonhematopoietic FL cells. Antigen expression was analyzed on 23 weeks' gestation CD235a− FL cells (A). Viable (PI−) nonhematopoietic cells (CD45−) were analyzed for CD14 and CD326 expression by sequential electronic gating, using gates 1 and 2 as shown. The flow of electronic gating of the data is shown using arrows. Differences in the size and complexity of the 4 cell populations defined by CD14 and CD326 expression (center plot) are indicated by their respective light-scatter profiles. Expression of CD14 on CD326++ cells was confirmed by specific blocking of CD14-PE binding by unconjugated anti-CD14 mAb (B). Relative levels of CD14, CD45, and CD326 mRNA expression determined by quantitative polymerase chain reaction analyses of samples collected from n = 2–10 (CD14) and n = 3–11 (CD326 and CD45) sorted tissues. Data are shown as the mean ± standard error (C). Signal-to-noise ratios of CD326 expression are compared among the 3 cell populations (D). Mean fluorescence intensity for CD326-allophycocyanin (signal) and IgG1-allophycocyanin (noise) staining was determined on PI−CD45− cells also gated based on the light-scatter characteristics of each cell population as indicated by gates 3, 4, and 5 shown in the light-scatter plots in (A). mAb, monoclonal antibody.
Expression of CD14 by CD326++CD14lo cells was evident when compared to CD45−CD326++ cells stained with IgG2a-PE (Fig. 2B). However, CD14 expression was not readily observed by these cells using a less-sensitive staining with fluorescein isothiocyanate-labeled mAb (data not shown). Therefore, CD14 expression was confirmed by specifically blocking staining using a different clone of unlabeled-mAb recognizing CD14, clone RPA-M1. Blocking with unlabeled anti-CD4 mAb (clone RPA-T4) had no effect on CD14-PE staining. CD14 gene expression was also measured by qPCR (Fig. 2C). Not surprisingly, the highest levels of CD14 gene expression were detected in CD14++CD326+/− cells and the least expression level was observed in CD326loCD14− cells. CD14 gene expression was observed in CD326++CD14lo cells at a 10th the levels of CD14++CD326+/− cells (P = 0.053) but at 6.8-fold higher than observed from CD326loCD14− cells (P = 0.007). These findings indicate a low level CD14 gene and protein expression by CD326++CD14lo cells.
Analysis of CD326 gene expression (Fig. 2C) unexpectedly indicated the highest level of expression by CD326loCD14− cells rather than CD326++CD14lo cells (P = 0.008). This suggested the possibility that the high levels of CD326 protein expression measured by flow cytometry on CD326++CD14lo cells was in part due to the high autofluorescence displayed by these large cells. However, when background levels of fluorescence were used to calculate a signal:noise ratio, CD326++CD14lo cells displayed an ∼2-fold higher level of CD326 expression than CD326loCD14− cells (Fig. 2D). Thus, the levels of gene expression did not correlate with cell-surface CD326 expression between these 2 sets of FL cells, a phenomenon that although not typical is also not unusual [22].
CD326++CD14lo cells express the highest levels of hepatocyte-associated genes and proteins
We analyzed gene expression among 4 cell populations isolated by FACS, based on their levels of CD14 and CD326 expression, to determine which population contains hepatocytes, cholangiocytes, and their precursors (Fig. 3). Preliminary studies indicated no hepatocyte-associated gene expression among CD14−CD326− cells (data not shown); thus, we did not further isolate these cells for the analyses shown in Fig. 3. Instead, we chose to differentially sort CD14++ cells based on CD326 expression.
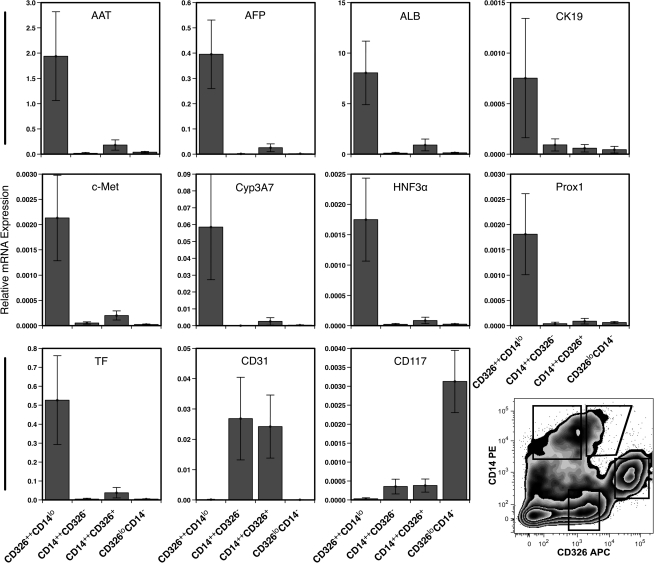
FIG. 3.
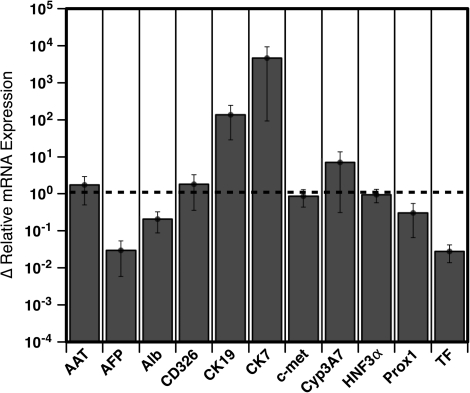
Gene expression profile of 4 sorted cell populations. Relative levels of gene expression were measured by quantitative polymerase chain reaction analyses. Cells were isolated by fluorescence-activated cell sorting based on their expression of CD14 and CD326 as shown in the zebra plot from livers ranging in gestational age from 19 to 24 weeks. Data represent the mean ± standard error from 7 experiments.
CD326++CD14lo cells expressed the highest levels of genes associated with hepatocytes (Fig. 3): α1-antitrypsin (AAT), α-fetoprotein (AFP), ALB, c-Met, cytochrome P450 3A7 (Cyp3A7) [23], hepatic nuclear factor 3α (HNF3α) [24], prospero-related homeobox 1 (Prox1) [25], and transferrin (TF). These cells also expressed the greatest amount of gene transcript for the biliary and hepatoblast marker cytokeratin (CK)-19.
CD14++ cells, whether expressing CD326 or not, expressed high levels of the gene coding for CD31 (platelet endothelial cell adhesion molecule-1). We believe that these cells are primarily comprised of sinusoidal endothelial cells and have found no conclusive evidence that they contain hepatocytes, biliary cells, or their precursors (work in progress). CD326loCD14− cells also did not express any significant amounts of hepatocyte-associated genes. Ongoing work in our laboratory suggests that CD326loCD14− cells are a heterogeneous population of cells containing immature erythrocytes and mesenchymal/fibroblast-like cells. In summary, these gene expression data indicate that fetal hepatocytes, cholangiocytes, and their precursors are likely found among CD326++CD14lo cells.
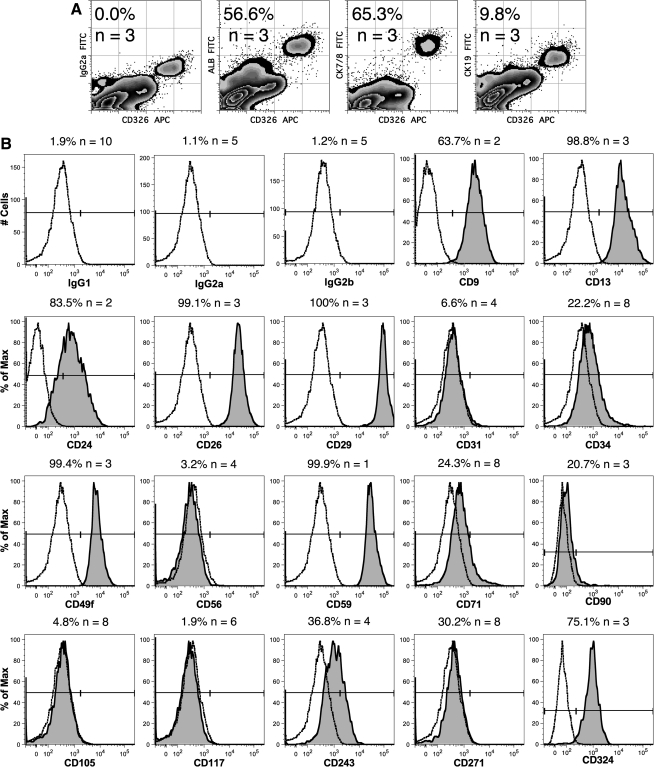
Flow cytometric analyses were performed to confirm expression of hepatocyte-associated markers and examine the diversity of cell types represented by CD326++CD14lo cells. Cytoplasmic staining yielded low but detectable levels of ALB among CD326++CD14lo cells (Fig. 4A). The mAb CAM5.2, which binds CK7 and CK8, also bound the majority of these cells but few other CD45− cells. Low expression of the cholangiocyte and hepatoblast marker CK19 was detected on freshly isolated cells, consistent with low expression of CK19 mRNA by freshly isolated cells measured by qPCR analyses (Fig. 3). Staining of FL sections revealed that CK19+ cells were concentrated in ductal plates and around blood vessels on cells that expressed high levels of CD326 (Fig. 5N). CK19 gene expression notably increased after culture (Figs. 6 and 7). Similar results were obtained for another cholangiocyte marker, CK7.
FIG. 4.
Flow cytometric analysis of surface and cytoplasmic antigens on CD326++CD14lo cells. Cytoplasmic antigen expression on viable CD45− FL cells is shown in (A). Fluorescein isothiocyanate-labeled cytoplasmic staining is shown together with cell-surface CD326 expression. Representative cell-surface expression of antigens stained with PE-labeled mAb on PI−CD45−CD326++CD14lo FL cells is shown in (B). Expression of the indicated antigens is shown using filled histograms, whereas staining with the corresponding isotype control antibody is shown using an unfilled histogram. The mean percentage expression level of the indicated antigen and number of experiments performed are shown for gated CD326++CD14lo cells.
FIG. 5.
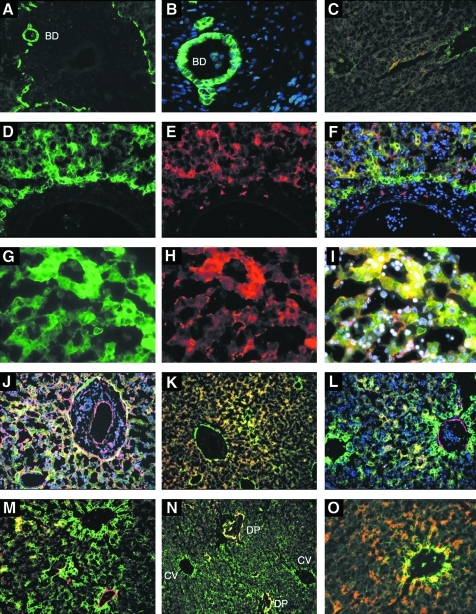
Immunolocalization of CD326++/+ cell subsets in FL. FL fixed and frozen sections of 21 weeks' gestation age were stained with mAbs against CD326 (green, all panels but E and H) and CD14 (C, E, F, H, and I), CD29 (J), CD49f (K), CD34 (L), CD90 (M), CK19 (N), and CK7/8 (O) shown in red. 4′,6-Diamidino-2-phenylindole staining of nuclei is shown in blue (B, F, I, J, and L). Note strong expression of CD326 around a ductal plate (DP) seen at 100 × magnification (A), including in bile duct (BD) cells seen in (A) and in (B) at 630 × magnification. CD326 and CD14 coexpression (yellow) in the parenchyma and around blood vessels is seen at 100 × magnification (C). Higher magnification of CD326 and CD14 expression is seen in (D–F) (200 × ) and (G–I) (400 × ). In these 6 panels, CD326 expression is shown by itself in the right column and CD14 expression is shown in the center column. Merged photomicrographs with 4′,6-diamidino-2-phenylindole staining are shown in (F) and (I). CD29 and CD326 are coexpressed by most cells in parenchyma (J, 200 × ) though some cells around blood vessels express only one marker. CD49f is coexpressed on CD326 positive cells in the parenchyma and with low-intensity on CD326bright cells (K, 100 × ). CD34 strongly marks endothelial cells and some CD326 positive parenchymal cells (L, 200 × ). CD90 is found mostly on vascular cells (M, 200 × ). CK19 is expressed around DPs but not around central vein (CV) (N, 100 × ). CK7/8 is coexpressed with CD326 around blood vessels and in the parenchyma (O, 200 × ).
FIG. 6.
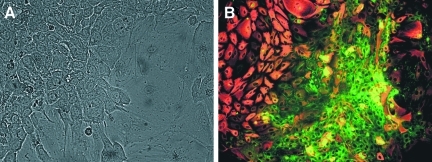
Morphology and marker expression on cultured CD326++CD14lo cells. Cultured CD326++CD14lo cells form a layer of polygonal cells (A) and express high levels of albumin (green) or/and CK19 (red) (B).
FIG. 7.
Changes in mRNA expression by CD326++CD14lo cells after 3 weeks of culture. The dashed line at 10° represents no change in expression; higher values reflect increased expression compared to mRNA levels in freshly isolated cells. Results are shown as the mean ± standard error of 4 independent experiments.
Analysis of cell surface antigen expression revealed a pattern of antigen expression by CD326++CD14lo cells that is mostly consistent with a parenchymal cell phenotype (Fig. 4B). CD326++CD14lo cells are the major nonhematopoietic cell population in the FL expressing heat-stable antigen (CD24), α6 integrin (CD49f), P-glycoprotein (CD243), and E-cadherin (CD324). Additionally, these cells expressed CD9, CD13, dipeptidyl peptidase IV (CD26), and β1 integrin (CD29), although CD26 and CD29 were widely expressed by different populations of FL cells and were not specific for CD326++CD14lo cells (data not shown). Variable levels of CD34, TF receptor (CD71), and nerve-growth factor receptor (CD271) expression suggest some degree of diversity among CD326++CD14lo cells.
Localization by immunofluorescence of CD326++CD14lo cells in the FL
CD326 staining of FL sections indicated widespread but weak expression by parenchymal cells (Fig. 5). At least 3 types of cells expressing CD326 could be distinguished by their location within the FL. Strong expression was observed on cholangiocytes forming bile ducts (Fig. 5A, B) and on cells surrounding blood vessels and ductal plates (Fig. 5A, J–O). We refer to these cells as CD326bright cells to distinguish them from the CD326++ cell population defined by flow cytometry. Parenchymal hepatocytes also expressed CD326 (Fig. 5C, D, G), but at a lower intensity.
Our goal was to compare the phenotypic profile of cells expressing CD326 detected by immunofluorescence microscopy and by flow cytometry to understand their relationship and location within the FL. Accordingly, CD14 expression was analyzed on FL sections and was weakly expressed by most parenchymal cells (Fig. 5C, E, H). The CD326bright cells in ductal plates and in the perivascular areas also expressed CD14. Thus, it is likely that these CD326bright cells are contained within the CD326++CD14lo cell population defined by flow cytometry, but that CD326++CD14lo cells also comprise the FL parenchyma. Fluorescence detected by flow cytometry is represented on a logarithmic scale and that the levels of CD326 expression can vary about 10-fold among cells in the CD326++CD14lo population.
The link between CD326++CD14lo cells and CD326-expressing cells in FL sections was further examined using antigens identified as markers of CD326++CD14lo cells. CD29 was expressed by multiple cell types, including CD326bright cells and parenchymal cells (Fig. 5J). CD49f and CD326 coexpression was also observed in parenchyma (Fig. 5K). CD34 coexpression with CD326 was seen in liver sections (Fig. 5L), revealing that some parenchymal cells were positive for both these markers, but CD326bright cells expressed mostly low or undetectable levels of CD34. These results are in agreement with the variable levels of CD34 expression observed on CD326++CD14lo cells by flow cytometry. CD90 stained only a few parenchymal cells, but appeared to stain endothelial cells, including cells adjacent to CD326bright cells (Fig. 5M). CK19 expression was not detected in the parenchyma but was expressed in some CD326bright cells in ductal plates and around some small vessels but not central veins (Fig. 5N). This suggests that CD326bright cells around central veins and in ductal plates might have different developmental potentials. CK7/8, the CAM5.2 antigen, was coexpressed with CD326 in parenchymal cells as well as around perivascular CD326bright cells (Fig. 5O). Together, these data indicate that CD326++CD14lo cells not only comprise the FL parenchyma but also contain different subsets of perivascular and biliary CD326bright cells.
Heterogeneity of cell types observed in cultures of CD326++CD14lo cells
Two types of cells were observed in cultures of CD326++CD14lo cells: large polygonal cells with a complex cytoplasm and small cells (Fig. 6A). Cultured cells expressed ALB and CK19, revealing cells positive for either or both markers (Fig. 6B). The small cells tended to express comparatively more ALB, whereas the larger cells exhibited more CK19 staining. CD326++CD14lo cells continued to express approximately the same mRNA levels of AAT, CD326, c-Met, Cyp3A7, and HNF3α after 3 weeks of culture as before culture (Fig. 7). AFP, ALB, Prox1, and TF expression decreased with culture. CK19 and CK7 expression was notably increased with culture.
Discussion
CD326 is of interest as a marker of liver progenitors and stem cells and has been used to positively select fetal hepatic precursor [7]. We sought to better understand the diversity of CD326 expression on freshly isolated cells from midgestation human FL. Among the cells that expressed CD326 were CD45+ hematopoietic cells representing multiple lineages. Moreover, 3 CD45− cell populations expressed CD326 as distinguished by CD14 expression. We demonstrate for the first time that this protein, CD14, is expressed by human fetal hepatocytes extending the findings that CD14 is expressed by adult hepatocytes in rodents and humans [14–19]. CD326++CD14lo cells expressed both genes and cell surface antigens associated with mature hepatocytes.
Specifically, CD326++CD14lo cells expressed a number of genes associated with functional hepatocytes: ALB, AFP, c-Met, HNF3α, Prox1, Cyp3A7, AAT, and TF. Cultured CD326++CD14lo cells also expressed these genes although the levels for some declined in culture. Antigen expression revealed similarities with adult hepatocytes, including expression of CD9 [26], CD13 [27], and CD26 [28]. Most CD326++CD14lo cells also expressed CD24, which is expressed by human hepatocellular carcinomas [29] and by rat oval cells [9]. The integrins CD29 and CD49f, which as heterodimers can bind laminin and are found on murine hepatic progenitors [30,31], were both highly expressed by CD326++CD14lo cells. Additionally, CD243, CD324, and cytoplasmic CK7/8 were primarily expressed by CD326++CD14lo cells, and further helped to define these cells as a distinct cell population in the FL. The ATP-dependent efflux pump CD243 is expressed by proliferating hepatocytes and hepatic neoplasms [32] and it was previously observed to be expressed on the canalicular membrane of FL hepatocytes and on the apical membrane of cholangiocytes [33]. The adhesion molecule CD324 has been identified as a marker of fetal hepatocytes in mice [34]. Taken together, these findings establish CD326++CD14lo cells as the population of FL cells containing the bulk of fetal parenchymal hepatocytes.
Despite the evidence that CD326++CD14lo cells are a largely homogeneous population of cells, we did uncover indications for a diversity of cell types and gene expression within this cell population. At least 3 cell populations exhibited clear CD326 expression in liver sections that appear all to be contained within the CD326++CD14lo cell population. Two types of CD326bright cells were identified by epifluorescence microscopy, cholangiocytes lining bile ducts and perivascular CD326bright cells. The latter population can be further divided into cells in close contact with endothelial cells and those separate from the endothelial cells by surrounding perivascular mesenchyme. The third population of cells represented by CD326++CD14lo cells comprises the parenchyma. The evidence for all these populations being represented by CD326++CD14lo cells is coexpression of CD14, CD34, CD49f, and CK7/8 detected by both flow cytometry and epifluorescence microscopy on CD326-expressing cells. Additionally, the published findings of parenchymal and cholangiocyte expression of CD243 further support this conclusion [33].
Variable levels of CD34, CD271, and CK19 expression suggest heterogeneity among CD326++CD14lo cells. CD34 coexpression with CD326 appeared highest on parenchymal cells, although not all parenchymal cells appeared to express CD34, which matches the results using flow cytometry that found only about a fifth of CD326++CD14lo cells expressed CD34. CK19 was clearly expressed on only a subpopulation of CD326bright cells associated with ductal plates. These CK19+CD326bright cells are likely to represent previously described ductal plate hepatoblasts [35]. CK19 expression was notably increased upon culture, suggesting caution in using this marker as an indicator of hepatoblast potential on cultured cells.
Schmelzer et al. isolated cells expressing CD326 using immunomagnetic beads [7]. Although they did not observe any CD14 expression, it is, nonetheless, likely that CD326++CD14lo cells were among the cells isolated by these investigators and shown to contain hepatic progenitors and likely stem cells. Our cultured CD326++CD14lo cells produced different cells expressing CK19 and ALB or both markers, which likely resulted from the growth of cholangiocytes, hepatocytes, and their progenitors, all residing among CD326++CD14lo cells.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge the contributions and helpful discussions of Drs. Tzong-Hae Lee and Holger Willenbring. We also thank Dale Hirschkorn for assistance with cell sorting, Cierra Sullivan for help with histology staining of liver sections, and the administrative staff at our institute, particularly JoAnn Yates, Jerry Michaelson, and Barbara Johnson.
This work was supported by Blood Systems Inc., grants from Novartis Pharmaceuticals Corporation, the National Institutes of Health (R21 DK068441 and R21 HD055328), and a pilot and feasibility grant awarded by the University of California San Francisco Liver Center (P30 DK026743).
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Crosby HA. Nijjar SS. de V de Goyet J. Kelly DA. Strain AJ. Progenitor cells of the biliary epithelial cell lineage. Semin Cell Dev Biol. 2002;13:397–403. doi: 10.1016/s108495210200126x. [DOI] [PubMed] [Google Scholar]
- 2.Shiojiri N. Development and differentiation of bile ducts in the mammalian liver. Microsc Res Tech. 1997;39:328–335. doi: 10.1002/(SICI)1097-0029(19971115)39:4<328::AID-JEMT3>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 3.Trzpis M. McLaughlin PMJ. de Leij LMFH. Harmsen MC. Epithelial cell adhesion molecule. More than a carcinoma marker and adhesion molecule. Am J Pathol. 2007;171:386–395. doi: 10.2353/ajpath.2007.070152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Momburg F. Moldenhauer G. Hämmerling GJ. Möller P. Immunohistochemical study of the expression of a Mr 34,000 human epithelium-specific surface glycoprotein in normal and malignant tissues. Cancer Res. 1987;47:2883–2891. [PubMed] [Google Scholar]
- 5.Joplin R. Strain AJ. Neuberger JM. Immuno-isolation and culture of biliary epithelial cells from normal human liver. In Vitro Cell Dev Biol. 1989;25:1189–1192. doi: 10.1007/BF02621273. [DOI] [PubMed] [Google Scholar]
- 6.Dan YY. Riehle KJ. Lazaro C. Teoh N. Haque J. Campbell JS. Fausto N. Isolation of multipotent progenitor cells from human fetal liver capable of differentiating into liver and mesenchymal lineages. Proc Natl Acad Sci USA. 2006;103:9912–9917. doi: 10.1073/pnas.0603824103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schmelzer E. Zhang L. Bruce A. Wauthier E. Ludlow J. Yao HL. Moss N. Melhem A. McClelland R. Turner W. Kulik M. Sherwood S. Tallheden T. Cheng N. Furth ME. Reid LM. Human hepatic stem cells from fetal and postnatal donors. J Exp Med. 2007;204:1973–1987. doi: 10.1084/jem.20061603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yovchev MI. Grozdanov PN. Joseph B. Gupta S. Dabeva MD. Novel hepatic progenitor cell surface markers in the adult rat liver. Hepatology. 2007;45:139–149. doi: 10.1002/hep.21448. [DOI] [PubMed] [Google Scholar]
- 9.Yovchev MI. Grozdanov PN. Zhou H. Racherla H. Guha C. Dabeva MD. Identification of adult hepatic progenitor cells capable of repopulating injured rat liver. Hepatology. 2008;47:636–647. doi: 10.1002/hep.22047. [DOI] [PubMed] [Google Scholar]
- 10.Muench MO. Cupp J. Polakoff J. Roncarolo MG. Expression of CD33, CD38, and HLA-DR on CD34+ human fetal liver progenitors with a high proliferative potential. Blood. 1994;83:3170–3181. [PubMed] [Google Scholar]
- 11.Suskind DL. Muench MO. Searching for common stem cells of the hepatic and hematopoietic systems in the human fetal liver: CD34+ cytokeratin 7/8+ cells express markers for stellate cells. J Hepatol. 2004;40:261–268. doi: 10.1016/j.jhep.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 12.Su GL. Goyert SM. Fan MH. Aminlari A. Gong KQ. Klein RD. Myc A. Alarcon WH. Steinstraesser L. Remick DG. Wang SC. Activation of human and mouse Kupffer cells by lipopolysaccharide is mediated by CD14. Am J Physiol Gastrointest Liver Physiol. 2002;283:G640–G645. doi: 10.1152/ajpgi.00253.2001. [DOI] [PubMed] [Google Scholar]
- 13.Couvelard A. Scoazec JY. Dauge MC. Bringuier AF. Potet F. Feldmann G. Structural and functional differentiation of sinusoidal endothelial cells during liver organogenesis in humans. Blood. 1996;87:4568–4580. [PubMed] [Google Scholar]
- 14.Liu S. Khemlani LS. Shapiro RA. Johnson ML. Liu K. Geller DA. Watkins SC. Goyert SM. Billiar TR. Expression of CD14 by hepatocytes: upregulation by cytokines during endotoxemia. Infect Immun. 1998;66:5089–5098. doi: 10.1128/iai.66.11.5089-5098.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li SW. Gong JP. Wu CX. Shi YJ. Liu CA. Lipopolysaccharide induced synthesis of CD14 proteins and its gene expression in hepatocytes during endotoxemia. World J Gastroenterol. 2002;8:124–127. doi: 10.3748/wjg.v8.i1.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bas S. Gauthier BR. Spenato U. Stingelin S. Gabay C. CD14 is an acute-phase protein. J Immunol. 2004;172:4470–4479. doi: 10.4049/jimmunol.172.7.4470. [DOI] [PubMed] [Google Scholar]
- 17.Su GL. Dorko K. Strom SC. Nüssler AK. Wang SC. CD14 expression and production by human hepatocytes. J Hepatol. 1999;31:435–442. doi: 10.1016/s0168-8278(99)80034-8. [DOI] [PubMed] [Google Scholar]
- 18.Pan Z. Zhou L. Hetherington CJ. Zhang DE. Hepatocytes contribute to soluble CD14 production, and CD14 expression is differentially regulated in hepatocytes and monocytes. J Biol Chem. 2000;275:36430–36435. doi: 10.1074/jbc.M003192200. [DOI] [PubMed] [Google Scholar]
- 19.Meuleman P. Steyaert S. Libbrecht L. Couvent S. Van Houtte F. Clinckspoor F. de Hemptinne B. Roskams T. Vanlandschoot P. Leroux-Roels G. Human hepatocytes secrete soluble CD14, a process not directly influenced by HBV and HCV infection. Clin Chim Acta. 2006;366:156–162. doi: 10.1016/j.cca.2005.09.022. [DOI] [PubMed] [Google Scholar]
- 20.Muench MO. Suskind DL. Bárcena A. Isolation, growth and identification of colony-forming cells with erythroid, myeloid, dendritic cell and NK-cell potential from human fetal liver. Biol Proced Online. 2002;4:10–23. doi: 10.1251/bpo29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lázaro CA. Croager EJ. Mitchell C. Campbell JS. Yu C. Foraker J. Rhim JA. Yeoh GC. Fausto N. Establishment, characterization, and long-term maintenance of cultures of human fetal hepatocytes. Hepatology. 2003;38:1095–1106. doi: 10.1053/jhep.2003.50448. [DOI] [PubMed] [Google Scholar]
- 22.Gry M. Rimini R. Strömberg S. Asplund A. Pontén F. Uhlén M. Nilsson P. Correlations between RNA and protein expression profiles in 23 human cell lines. BMC Genomics. 2009;10:365. doi: 10.1186/1471-2164-10-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Komori M. Nishio K. Fujitani T. Ohi H. Kitada M. Mima S. Itahashi K. Kamataki T. Isolation of a new human fetal liver cytochrome P450 cDNA clone: evidence for expression of a limited number of forms of cytochrome P450 in human fetal livers. Arch Biochem Biophys. 1989;272:219–225. doi: 10.1016/0003-9861(89)90213-0. [DOI] [PubMed] [Google Scholar]
- 24.Costa RH. Grayson DR. Darnell JE. Multiple hepatocyte-enriched nuclear factors function in the regulation of transthyretin and alpha 1-antitrypsin genes. Mol Cell Biol. 1989;9:1415–1425. doi: 10.1128/mcb.9.4.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Song KH. Li T. Chiang JY. A Prospero-related homeodomain protein is a novel co-regulator of hepatocyte nuclear factor 4alpha that regulates the cholesterol 7alpha-hydroxylase gene. J Biol Chem. 2006;281:10081–10088. doi: 10.1074/jbc.M513420200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Charrin S. Le Naour F. Labas V. Billard M. Le Caer JP. Emile JF. Petit MA. Boucheix C. Rubinstein E. EWI-2 is a new component of the tetraspanin web in hepatocytes and lymphoid cells. Biochem J. 2003;373:409–421. doi: 10.1042/BJ20030343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Röcken C. Licht J. Roessner A. Carl-McGrath S. Canalicular immunostaining of aminopeptidase N (CD13) as a diagnostic marker for hepatocellular carcinoma. J Clin Pathol. 2005;58:1069–1075. doi: 10.1136/jcp.2005.026328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feller AC. Radzun HJ. Heymann E. Haas H. Scholz W. Parwaresch MR. A monoclonal antibody detecting dipeptidylpeptidase IV in human tissue. Virchows Arch A Pathol Anat Histopathol. 1986;409:263–273. doi: 10.1007/BF00708333. [DOI] [PubMed] [Google Scholar]
- 29.Huang LR. Hsu HC. Cloning and expression of CD24 gene in human hepatocellular carcinoma: a potential early tumor marker gene correlates with p53 mutation and tumor differentiation. Cancer Res. 1995;55:4717–4721. [PubMed] [Google Scholar]
- 30.Kamiya A. Gonzalez FJ. Nakauchi H. Identification and differentiation of hepatic stem cells during liver development. Front Biosci. 2006;11:1302–1310. doi: 10.2741/1884. [DOI] [PubMed] [Google Scholar]
- 31.Suzuki A. Zheng Y. Kondo R. Kusakabe M. Takada Y. Fukao K. Nakauchi H. Taniguchi H. Flow-cytometric separation and enrichment of hepatic progenitor cells in the developing mouse liver. Hepatology. 2000;32:1230–1239. doi: 10.1053/jhep.2000.20349. [DOI] [PubMed] [Google Scholar]
- 32.Thorgeirsson SS. Huber BE. Sorrell S. Fojo A. Pastan I. Gottesman MM. Expression of the multidrug-resistant gene in hepatocarcinogenesis and regenerating rat liver. Science. 1987;236:1120–1122. doi: 10.1126/science.3576227. [DOI] [PubMed] [Google Scholar]
- 33.Cízková D. Morký J. Micuda S. Osterreicher J. Martínková J. Expression of MRP2 and MDR1 transporters and other hepatic markers in rat and human liver and in WRL 68 cell line. Physiol Res. 2005;54:419–428. [PubMed] [Google Scholar]
- 34.Ogou SI. Yoshida-Noro C. Takeichi M. Calcium-dependent cell-cell adhesion molecules common to hepatocytes and teratocarcinoma stem cells. J Cell Biol. 1983;97:944–948. doi: 10.1083/jcb.97.3.944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roskams T. Desmet V. Embryology of extra- and intrahepatic bile ducts, the ductal plate. Anat Rec (Hoboken) 2008;291:628–635. doi: 10.1002/ar.20710. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.