Abstract
Systemic lupus erythematosus is a chronic-relapsing autoimmune disease of incompletely understood etiology. Recent evidence strongly supports an epigenetic contribution to the pathogenesis of lupus. To understand the extent and nature of dysregulated DNA methylation in lupus T cells, we performed a genome-wide DNA methylation study in CD4+ T cells in lupus patients compared to normal healthy controls. Cytosine methylation was quantified in 27,578 CG sites located within the promoter regions of 14,495 genes. We identified 236 hypomethylated and 105 hypermethylated CG sites in lupus CD4+ T cells compared to normal controls, consistent with widespread DNA methylation changes in lupus T cells. Of interest, hypomethylated genes in lupus T cells include CD9, which is known to provide potent T-cell co-stimulation signals. Other genes with known involvement in autoimmunity such as MMP9 and PDGFRA were also hypomethylated. The BST2 gene, an interferon-inducible membrane-bound protein that helps restrict the release of retroviral particles was also hypomethylated in lupus patients. Genes involved in folate biosynthesis, which plays a role in DNA methylation, were overrepresented among hypermethylated genes. In addition, the transcription factor RUNX3 was hypermethylated in patients, suggesting an impact on T-cell maturation. Protein-protein interaction maps identified a transcription factor, HNF4a, as a regulatory hub affecting a number of differentially methylated genes. Apoptosis was also an overrepresented ontology in these interaction maps. Further, our data suggest that the methylation status of RAB22A, STX1B2, LGALS3BP, DNASE1L1 and PREX1 correlates with disease activity in lupus patients.
Key words: lupus, T cells, CD4+ T cells, methylation, methylome
Introduction
Systemic lupus erythematosus (SLE or lupus) is a chronic relapsing autoimmune disorder of complex etiology. A number of genetic polymorphisms have been associated with lupus;1 however, discordance in the development of lupus in twins demonstrated that the disease is not exclusively genetic in origin. Many non-genetic factors (like environmental) have now been linked to lupus,2 and in a recent report DNA methylation differences between twins discordant for lupus was demonstrated.
DNA methylation as a regulator of transcription generally occurs on cytosines in cytosine-guanosine (CG) dinucleotides located within the promoter, most often between 1,000 bp and 800 bp upstream of the transcription start site.4 Transcription of genes with methylated CG sites in this region is usually repressed. Although DNA methylation was initially thought to be a stable modification with only passive removal of methyl groups possible, accumulating evidence suggests that DNA demethylation occurs actively.5 In fact, DNA methylation appears to be key in a variety of cellular responses to environmental stimuli including hypoxia,6 hormonal signaling7 and viral latency and reactivation.8 Furthermore, aberrant hypo- and hypermethylation may occur in specific genes within the same cell, as is the case in some neoplastic cells.9
DNA methylation is emerging as an important contributing factor in lupus. Lupus patients exhibit global T-cell hypomethylation,10 notably including a number of autoimmune-related genes such as ITGAL (CD11a)11 and TNFSF7 (CD70).12 Interestingly, these changes correlate with increased disease activity.13 T-cell DNA hypomethylation has been implicated in the development of drug-induced lupus by hydralazine and procainamide14 and contributes to disease in the lupus-prone MRLlpr mouse model.15 Interruption of the ERK signaling pathway in T cells and subsequent suppression of the maintenance DNA methyltransferase DNMT1 induces anti-dsDNA antibody production and a lupus-like gene expression profile in mice.16
Another gene that is hypomethylated in lupus T cells is CD40LG, located on the X chromosome. CD40LG is hypomethylated in T cells from healthy men, while healthy women have one methylated and one hypomethylated allele.17 Treatment of CD4+ T cells with the DNA methylation inhibitor 5-azacytidine demethylated CD40LG and doubled its expression in normal healthy women but not men.17,18 When DNA methylation is inhibited by 5-azacytidine, the inactive CD40LG allele in women (located on the inactive, heavily methylated X chromosome) becomes available for transcription, as its promoter sequence demethylates. CD40LG is hypomethylated in CD4+ T cells from women with active lupus as compared to healthy controls, similar to T cells treated with 5-azacytidine.17 These findings suggest that hypomethylation of the normally inactivated and silenced X chromosome might be related to the higher prevalence of lupus in women. Indeed, data from Scofield et al. suggest that men with Klinefelter's syndrome (47, XXY) have a similar risk to develop lupus as normal women (46, XX) and ∼14 times higher risk to develop lupus compared to normal men (46, XY).19,20 These data reinforce the idea of a gene-dose effect on the X-chromosome for the risk to develop lupus. This gene-dose effect may be achieved by hypomethylation of the X chromosome, making more than one X chromosome available for transcription.
The extent of DNA methylation changes in lupus CD4+ T cells has never been examined on a genome-wide level. In this study, we provide the most detailed description of DNA methylation changes associated with lupus to date. This detailed analysis provides a platform for pathway associations, with potential significance to the pathogenesis of lupus.
Results
We studied genome-wide CD4+ T-cell DNA methylation in lupus patients and controls using a high-throughput method based on bead microarrays that allow simultaneous screening of 27,578 CG sites spanning the promoter region of 14,495 genes. Twelve biological replicates (12 lupus patients and 12 controls) were studied (Table 1). One lupus patient, Case 2, was excluded from subsequent analysis secondary to methotrexate use, as methotrexate may increase DNA methylation.29
Table 1.
Demographic characteristics of lupus patients and controls included in this study
| Case ID# | Case age | Case race | SLEDAI | Control ID# | Control age | Control race |
| 1 | 32 | African-American | 10 | 1 | 31 | African-American |
| 2 | 38 | European-American | 8 | 2 | 40 | European-American |
| 3 | 35 | African-American | 6 | 3 | 32 | African-American |
| 4 | 46 | African-American | 4 | 4 | 46 | African-American |
| 5 | 47 | European-American | 8 | 5 | 48 | European-American |
| 6 | 25 | Hispanic | 8 | 6 | 33 | Hispanic |
| 7 | 43 | European-American | 7 | 7 | 43 | Hispanic |
| 8 | 58 | European-American | 4 | 8 | 57 | African-American |
| 9 | 31 | African-American | 4 | 9 | 34 | African-American |
| 10 | 43 | African-American | 4 | 10 | 45 | African-American |
| 11 | 45 | Asian American | 2 | 11 | 44 | Asian American |
| 12 | 71 | European-American | 0 | 12 | 72 | European-American |
SLEDAI, Systemic Lupus Erythematosus Disease Activity Index.
Validation of our Illumina DNA methylation microarray data was performed using bisulfite sequencing and subsequent analysis of five representative genes including unmethylated, intermediately-methylated and heavily-methylated genes. Linear regression analysis revealed correlation between the microarray methylation data and bisulfite sequencing as expected (r2 = 0.83, n = 90) (Sup. Fig. 1).
To ensure that differential methylation patterns identified in our study were not influenced by potential differences in T-cell subset populations between lupus patients and controls, we examined the methylation status of a number of genes known to be demethylated in particular T-cell subsets, including IL4, IL5 and IL13 (Th2 cells), IFNG (Th1 cells), IL17 and IL17F (Th17 cells) and FOXP3 (regulatory T cells). This epigenetic immunophenotyping revealed no methylation differences in these loci, suggesting no difference in T-cell subsets between lupus patients and controls (Table 2). Epigenetic immunophenotyping has been recently shown to be as precise as FACS for T-cell quantification.30 Indeed, this is consistent with previous studies that showed no difference in the Th1:Th2 ratio between lupus patients and healthy controls in the peripheral blood.31
Table 2.
T-cell subset epigenetic immunophenotypes in lupus patients (cases) and controls
| Gene | Average methylation (%) | p value | |
| cases | controls | ||
| IL4 | 0.85 | 0.84 | 0.60 |
| IL5, site 1 | 0.80 | 0.78 | 0.16 |
| IL5, site 2 | 0.70 | 0.70 | 0.80 |
| IL13, site 1 | 0.79 | 0.76 | 0.25 |
| IL13, site 2 | 0.83 | 0.83 | 0.95 |
| IFNG | 0.52 | 0.50 | 0.43 |
| IL17 | 0.62 | 0.63 | 0.70 |
| IL17F, site 1 | 0.69 | 0.67 | 0.40 |
| IL17F, site 2 | 0.79 | 0.78 | 0.71 |
| FOXP3, site 1 | 0.77 | 0.75 | 0.22 |
| FOXP3, site 2 | 0.67 | 0.67 | 0.89 |
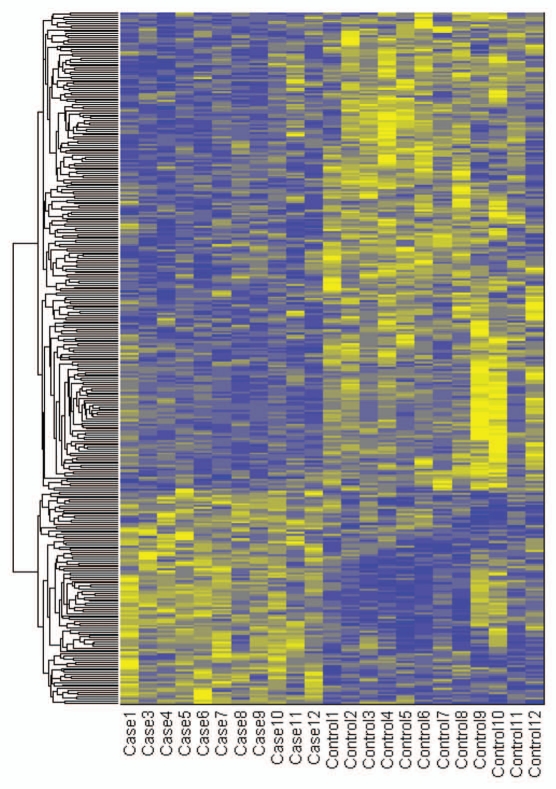
We identified 341 CG loci with differential methylation between cases and controls (with significance threshold of 1.2-fold difference and minimum methylation >6 standard deviations above technical noise). We observed over two times more hypomethylated loci compared to hypermethylated loci in lupus patients: 236 and 105, representing 232 and 104 unique genes, respectively (Fig. 1).
Figure 1.
Heatmap of differentially methylated genes between lupus patients and controls. Methylation values were standardized across rows and clustered with Euclidean distance metric and average linkage. Yellow/blue gradient represents standardized level of hypermethylation/hypomethylation in lupus patients compared to controls. The order of clustered genes is preserved in Supplemental Tables 1 and 2.
Hypomethylated genes in lupus patients.
Among the 232 hypomethylated genes in lupus patients (Sup. Table 1), 42 belong to the “arthritis” category (p = 7.80 × 10−4). Specifically, 11 genes were overrepresented in the “development of connective tissue” category (Table 3).
Table 3.
“Development of connective tissue” ontology overrepresented by hypomethylated genes in lupus patients
| ADAMTS1 | ADAM metallopeptidase with thrombospondin type 1 motif; 1 preproprotein |
| ALX4 | Aristaless-like homeobox 4 |
| CD9 | CD9 antigen |
| ESRRA | Estrogen-related receptor alpha |
| FGF8 | Fibroblast growth factor 8 isoform F precursor |
| HOXA13 | Homeobox protein A13 |
| HOXD11 | Homeobox D11 |
| MMP9 | Matrix metalloproteinase 9 preproprotein |
| MSX1 | Msh homeo box homolog 1 |
| PDGFRA | Platelet-derived growth factor receptor alpha precursor |
| SOX5 | SRY (sex determining region Y)-box 5 isoform c |
p = 1.43 × 10−5
Canonical pathway analysis identified CDK5 and PTEN signaling as the most significant. Metabolic signature was represented by “glycine, serine and threonine metabolism” and “insulin receptor signaling” pathways (Table 4).
Table 4.
Canonical pathways identified among hypomethylated genes in lupus CD4+ T cells
| Ingenuity canonical pathways | Molecules | p value |
| CDK5 signaling | NTRK2, ADCY5, ITGA2, PRKACA, HRAS | 4.68 × 10−3 |
| PTEN signaling | NTRK2, FOXO1, ITGA2, PDGFRA, HRAS | 1.12 × 10−2 |
| Glycine, serine and threonine metabolism | GAMT, CBS, SHMT1, ETNK2 | 1.38 × 10−2 |
| Insulin receptor signaling | FOXO1, PTPN1, PRKACA, HRAS, ACLY | 2.45 × 10−2 |
Hypermethylated genes in lupus patients.
Among the 104 hypermethylated genes in lupus CD4+ T cells (Sup. Table 2), seven were represented in the “response to nutrient” ontology (AACS, DDAH2, IGFBP7, LRP2, MEST, PTGS2, SKIV2L; p = 8.2 × 10−2). Metabolic signatures were also seen in overrepresented canonical pathways, where “inositol metabolism” (ALDOA, ALDOC; p = 6.38 × 10−4), “pentose phosphate pathway” (ALDOA, ALDOC, PGM5; p = 1.24 × 10−3) and “folate biosynthesis” (FOLH1, GGH; p = 6.65 × 10−3) were the most significant.
Besides metabolism, we observed genes overrepresented in cell cycle ontology. Specifically, GNB2L1, HOXB13 and IGFBP7 were overrepresented in “delay in G1 phase of cell lines” (p = 1.80 × 10−4). Two other cell cycle related genes, CCND2 and CDKN2C, were overrepresented in “disease of thymus gland” category (p = 2.32 × 10−3).
Interaction maps of differentially-methylated genes.
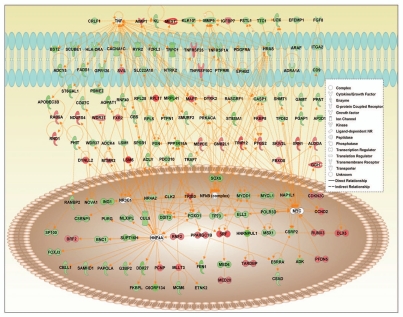
A common protein-protein interaction network for hypo- and hypermethylated genes in lupus (Fig. 2) reveals overrepresentation of functional annotations such as “growth of cells,” “developmental process of tissue,” “apoptosis” and “cell division process” (Table 5). Further, a surprising number of genes are regulated by hepatocyte nuclear factor 4a (HNF4a), a transcription factor previously undescribed with relation to lupus. Interestingly, TNF and NFκB signaling are also prominent in this network (Fig. 2).
Figure 2.
Interaction map between hypermethylated and hypomethylated genes in lupus CD4+ T cells and transcription factors affecting/affected by them. Green and red colors indicate hypomethylated and hypermethylated genes in lupus patients, respectively. White color indicates genes that are not user specified but incorporated into the network through relationships with other genes.
Table 5.
Main functional annotations overrepresented by protein-protein interaction network (shown in Fig. 2) of hypomethylated and hypermethylated genes in lupus CD4+ T cells
| Function annotation | Molecules | Number of molecules | p value |
| Growth of cells | ADRA1A, AIMP1, ARAF, BRF2, CASP1, CCND2, CDKN2C, CLK2, DDIT3, ESRRA, FADS1, FGF8, FHIT, FOXO1, GNB2L1, HNF4A, HRAS, IGFBP7, ING1, LOX, LSM1, MAPT, MEST, MLLT3, MMP9, MYC, MYCL1, MYOD1, NR3C1, NR4A2, NTRK2, PDCD10, PDGFRA, PFDN5, PPAT, PPP1R15A, PRKACA, PSME2, PTGS2, PTPN1, RUNX3, SHMT1, SLC22A18, SRF, ST8SIA1, TNF, TNFRSF25, TNFRSF1A, TP73, TPD52 | 50 | 3.07 × 10−13 |
| Developmental process of tissue | CCND2, CD9, DLX5, EPHB2, ESRRA, F2RL3, FGF8, FKBP8, FOXO1, HRAS, KL, MEST, MMP9, MSX1, MYOD1, NR3C1, NR4A2, NTRK2, PDGFRA, PRKACA, PTGS2, PXN, RNF2, SOX5 (includes EG:6660), SRF, ST8SIA1, SVIL, TNF, TNFRSF1A | 29 | 5.80 × 10−11 |
| Apoptosis | ADCY5, ADRA1A, AIMP1, ALDOA, CACNA1C, CASP1, CCND2, CD9, CDKN2C, CUL5, DDIT3, FEN1, FGF8, FHIT, FKBP8, FOXO1, FSTL1, GNB2L1, HRAS, IGFBP7, ING1, ITGA2, KL, MAPT, MLLT3, MMP9, MSX1, MYC, MYOD1, NOVA1, NR3C1, NR4A2, NTRK2, PDCD10, PDGFRA, PPP1R15A, PRKACA, PTGS2, PTPN1, PXN, RUNX3, RYR2, SOX5 (includes EG:6660), SRF, ST6GAL1, ST8SIA1, TNF, TNFRSF25, TNFRSF10C, TNFRSF1A, TP73, TRAF7, TRIB3, TRPC1 | 54 | 1.37 × 10−10 |
| Cell division process | ADRA1A, AIMP1, ARAF, CCND2, CDKN2C, CUL5, DDIT3, ESRRA, FGF8, FHIT, FOXO1, GNB2L1, HNF4A, HRAS, IGFBP7, ING1, ITGA2, LSM1, MMP9, MYC, MYOD1, NR3C1, NR4A2, NTRK2, PDGFRA, PPP1R15A, PRKACA, PTGS2, RNF2, RPL5, RUNX3, SRF, ST8SIA1, TARDBP, TNF, TNFRSF1A, TP73 | 37 | 4.59 × 10−10 |
Effect of medications on DNA methylation in lupus patients.
Five out of 11 patients included in our analysis were receiving hydroxychloroquine, two patients were receiving azathioprine, and one patient was receiving prednisone. None of the patients analyzed were being treated with other immunosuppressive agents. To ensure differential methylation between lupus patients and controls was not influenced by hydroxychloroquine treatment, we performed a subset analysis in hydroxychloroquine-treated and non-treated patients. Only eight out of 341 differentially methylated CG loci between lupus patients and controls were also differentially methylated when comparing hydroxychloroquine-treated versus non-treated patients.
Correlation between DNA methylation and disease activity.
To determine if variability in disease activity could be linked with variability in DNA methylation patterns, we first had to examine the degree of variability in methylation between cases and controls. The expectation would be that cases exhibit variable methylation as a group, whereas controls have a relatively stable pattern. This was indeed the case. Specifically, the methylation of 70 CG sites was highly variable in cases, compared to only 35 in controls (Sup. Fig. 2).
Genes with highly variable methylation status in cases and not in controls were tested for correlation with disease activity as measured by Systemic Lupus Erythematosus Disease Activity Index (SLEDAI) scores. The genes that correlate significantly with SLEDAI scores are listed in Table 6.
Table 6.
Genes in which methylation levels correlate with systemic lupus erythematosus disease activity index (SLEDAI) scores in lupus patients
| Target ID | Gene symbol | Gene name | r2 | p value |
| cg02635865 | RAB22A | RAB22A, member RAS oncogene family | 0.666 | 0.025 |
| cg26134665 | STX1B2 | Syntaxin 1B | 0.652 | 0.030 |
| cg14870271 | LGALS3BP | Lectin, galactoside-binding, soluble, 3 binding protein | 0.610 | 0.046 |
| cg09202373 | DNASE1L1 | Deoxyribonuclease I-like 1 | −0.615 | 0.044 |
| cg26206598 | PREX1 | Phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 | −0.686 | 0.020 |
Discussion
The majority of genes we found to be differentially methylated in lupus patients are hypomethylated, which is consistent with a global hypomethylation pattern previously described in lupus T cells.32 Further, a recent study of DNA methylation changes in total white blood cells also showed global hypomethylation in lupus patients compared to their healthy twins or siblings.3
Among the genes hypomethylated in lupus CD4+ T cells, the gene ontology category “development of connective tissue” was significantly overrepresented (Table 3). Matrix metalloproteinase 9 (MMP-9) is an important constituent of skin and connective tissue basement membranes, functioning to break down type IV and V collagen during tissue remodeling processes. MMP-9 has been previously linked to other autoimmune diseases such as rheumatoid arthritis and Sjögren's syndrome. MMP-9 levels are significantly higher in the synovial fluid of patients with rheumatoid arthritis compared to osteoarthritis, suggesting that MMP-9, along with other metalloproteinases, might contribute to cartilage destruction in rheumatoid arthritis.33 MMP-9 activity levels are increased in salivary glands from patients with Sjögren's syndrome compared to normal controls.34 The mRNA expression of MMP-9 is increased in the skin of the lupus prone MRLlpr mice during disease development, with highest levels of expression in skin with lupus-like involvement and dermal immune complex deposition.35
A second associated connective tissue-related gene found to be hypomethylated in lupus CD4+ T cells is platelet-derived growth factor receptor α (PDGFRα). Autoantibodies against PDGFRα are detected in 36% of patients with active lupus and 10% of patients with inactive disease.36 Interestingly, patients with anti-PDGFRα antibodies are far more likely to develop autoimmune hemolytic anemia (25 vs. 0% of anti-PDGFRα-negative patients),36 which is known to be correlated with both an earlier disease onset and a more aggressive disease in lupus patients.37 Imatinib, a specific PDGF receptor-targeting tyrosine kinase inhibitor initially developed to treat chronic myeloid leukemia, has been shown to significantly improve survival and delay the onset of proteinuria in lupus-prone mice.38
CD9 was hypomethylated in lupus CD4+ T cells in our study, consistent with a previous report that CD9 is hypomethylated in lupus total white blood cells.3 CD9 encodes for a cell surface glycoprotein that is a member of the transmembrane 4 superfamily (TM4SF) and is expressed on a variety of cells, including macrophages and lymphocytes.39 CD9 is expressed on T cells and a monoclonal antibody against CD9 results in T-cell activation. Indeed, anti-CD9 costimulation of T cells is independent of and as potent as anti-CD28 costimulation in the presence of anti-CD3 antibodies and the absence of antigen presentation.40 These data suggest that CD9 hypomethylation might play a role in T-cell autoreactivity described in lupus patients.
Another gene hypomethylated in lupus patients was BST2 (Tetherin), an interferon-inducible membrane-bound protein that restricts the release of nascent viral particles by tethering them to the cell surface.41 Analysis of gene expression profiles for CD4+ T cells from lupus patients compared to controls extracted from Gene Expression Omnibus (Lauwerys et al.; GEO accession: GSE4588) shows that BST2 is over two-fold overexpressed in patients, suggesting hypomethylation may be the cause of this upregulation. Several studies have proposed links between lupus and viruses including Epstein-Barr virus42 and endogenous retroviruses.43 The hypomethylation and overexpression of BST2 in CD4+ T cells from lupus patients is a novel observation and raises the possibility that the observed loss of methylation and overexpression of BST2 would explain the suggested low prevalence of HIV-1 infection in lupus patients.44
Among the hypermethylated genes identified in lupus CD4+ T cells, several genes are overrepresented in gene ontologies of metabolic pathways and response to micronutrients. This is potentially suggestive of a link between environmental factors and epigenetic modifications, specifically with relation to disease pathogenesis. Of particular interest is the “folate biosynthesis” pathway represented by the hypermethylation of FOLH1 (folate hydrolase 1 isoform 1) and GGH (gamma-glutamyl hydrolase precursor). DNA methylation requires S-adenosylmethionine (SAM) as a methyl donor, which is then converted to S-adenosylhomocysteine (SAH). SAM levels depend on dietary nutrients including folate and methionine, while SAH levels depend on homocysteine concentrations.45 The DNA methylation reaction is proportional to SAM levels and inversely proportional to SAH levels. Aging is associated with reduced DNMT1 expression levels similar to T cells from lupus patients.46 Indeed, stimulated T cells from older individuals (57–76 years of age) show DNA demethylation that is progressive with increased donor age when incubated in folate- or methionine-restricted media.45 DNMT1 knockdown in T cells from younger individuals reproduced similar demethylation effects and overexpression of methylation sensitive genes implicated in autoimmunity when incubated with folate- or methionine-restricted media.45 These data indicate that low levels of certain micronutrients involved in DNA methylation might work synergistically with low DNMT1 levels to increase the expression of genes implicated in autoimmunity in T cells.45
Aberrant expression of transcription factors (TFs) may have catastrophic consequences for the cell. We observed differential methylation in a number of transcription factors in lupus CD4+ T cells. Of particular interest is the hypermethylation of RUNX3. The runt domain-containing family of transcription factors (RUNX) function as cell proliferation repressors. Both RUNX1 and RUNX3 are required for T-cell maturation. RUNX3 is expressed in mouse mature dendritic cells, and knockout mice show a phenotype of accelerated dendritic cell maturation and increased efficiency of T-cell stimulation.47 Interestingly, RUNX3 is known to directly interact with the promoter of the aforementioned integrin gene ITGAL, whereby overexpression of RUNX3 is associated with upregulation of ITGAL expression.48 RUNX3 promoter hypermethylation has also been noted in patients with ANCA-associated vasculitis. Expression of RUNX3 is inversely related to the expression of the commonly seen ANCA antigens PR3 and MPO, mediated through binding of RUNX3 to intronic regions of both genes.49
Protein-protein interaction analysis among differentially methylated genes in lupus CD4+ cells highlights several important functional annotations including apoptosis (Table 5). Abnormal apoptosis is tightly linked to the pathogenesis of autoimmunity. It is postulated that apoptotic blebs help to expose autoantigens to the immune system, enhancing immune complex-mediated immune response. This has been demonstrated in keratinocytes after UV light exposure.50 Abnormal lymphocyte apoptosis is known to cause autoimmunity in MRLlpr lupus-prone mice, which exhibit a defect in Fas, a cell surface mediator of apoptosis.51 Mice overexpressing human BCL-2, a protein that blocks apoptosis, also develop autoimmunity.52 On the contrary, in human lupus, enhanced lymphocyte apoptosis and reduced clearance of apoptotic cells has been reported.53,54 More recently, it has been shown that T-cell apoptosis is increased in lupus patients compared to controls, and the proportion of apoptotic T cells is higher in patients with active compared to inactive disease. Our results suggest that abnormal methylation of genes involved in apoptosis might play a role in dysregulation of T-cell apoptosis observed in lupus patients.
Finally, our data suggest that DNA methylation changes in specific genetic loci correlate with disease activity in lupus patients (Table 6). If validated, DNA methylation changes might provide novel biomarkers for disease activity in lupus patients.
In summary, we performed a DNA methylation study in CD4+ T cells and demonstrated genome-wide DNA methylation changes in lupus patients compared to normal controls. Our data indicate the presence of hypomethylated and hypermethylated loci in lupus T cells, albeit, the former being twice as common. This work provides novel insights that could help to better understand the pathogenesis of lupus and identifies differentially methylated loci in key genes and pathways that will require further investigation in autoimmunity. Further, our findings provide a foundation to begin developing novel epigenetic biomarkers for disease activity in lupus.
Methods
Patients and controls.
Twelve female lupus patients and 12 female controls were studied (Table 1). The mean age was 42.83 ± 3.61 in patients and 43.75 ± 3.40 in controls (mean (years) ± SEM), p = 0.86. All patients fulfilled the American College of Rheumatology classification criteria for SLE.21 All protocols were approved by the institutional review board at our institution. Clinical data were collected at the time of sample collection.
Cell selection, DNA isolation.
Peripheral blood was collected from patient samples and controls. CD4+ T lymphocytes were obtained via Ficoll/Paque PBMC enrichment (GE Healthcare Life Sciences, Piscataway, NJ) followed by CD4+ T-cell isolation using a MACS magnetic bead CD4+ T-cell isolation kit (Miltenyi Biosystems, Auburn, CA). Whole genomic DNA was then prepared using a Qiagen DNEasy kit (Qiagen, Germantown, MD) and bisulfite-treated using a Zymo EZ DNA Methylation Kit (Zymo, Orange, CA).
Illumina Infinium Human Methylation27.
Bisulfite-converted patient and control DNA samples were prepared and quantified using a NanoDrop scanning spectrophotometer (Thermo, Wilmington, DE). For each sample, 500 ng of whole-genome bisulfite-converted DNA was denatured, fragmented and amplified using Illumina-supplied reagents according to manufacturer instructions. Following precipitation, DNA was hybridized to Illumina Infinium HumanMethylation27 arrays, containing 50-mer oligonucleotides designed to hybridize either methylated (N-CG-N following bisulfite conversion) or unmethylated (N-TG-N following bisulfite conversion) cytosine on each CG pair interrogated and coupled to beads mounted on glass slides. Microarrays were washed under high stringency, labeled with biotin (C and G nucleotides) or dinitrophenyl (A and T nucleotides) and scanned with an Illumina iScan. Raw data were exported for further analysis using Illumina BeadStudio software and the Methylation Module add-in (Illumina, San Diego, CA).
Illumina Infinium Methylation controls.
The Infinium Methylation system includes a variety of both sample-independent and sample-dependent controls, which were evaluated in each chip. Sample-independent controls included staining standards consisting of high- and background-intensity dinitrophenyl (DNP) and biotin, hybridization standards of low-, medium- and high-levels, target removal evaluation, and extension controls of both DNP (A/T) and biotin (C/G) fluorescence channels. Sample-dependent controls included both high- and background-level bisulfite conversion controls for each of the four basic oligomer types, specificity controls for mismatch and perfectly matched sequences in DNP and biotin channels, and 16 fully negative controls per chip.
CG methylation validation.
Representative CG loci were chosen from the Illumina assay repertoire for bisulfite sequencing. These sites were located within promoter regions of traditionally hypomethylated HOXA7 and HOXA9, intermediately-methylated HOXA3 and hypermethylated HOXA5. Primers were designed using Sequenom's EpiDesigner online tool (Sequenom, San Diego, CA) (primer sequences are available upon request). PCR was performed on a Bio-Rad MyCycler (Bio-Rad, Hercules, CA) using ZymoTaq one-step master mix (Zymo, Orange, CA). The cycling conditions were as follows: 95°C for 10 min followed by 40 cycles of 95°C for 30 seconds, then 52°C for 40 seconds, then 72°C for 1 min, followed by 72°C for 7 min. Each PCR product was confirmed with a small aliquot via agarose gel electrophoresis; the remainder was then purified using Zymo ZR-96 DNA Clean & Concentrate kits (Zymo, Orange, CA). Purified DNA was sequenced using an ABI 3730 sequencer and trace files were then used to calculate cytosine methylation percentages using the ESME software package (Epigenomics AG, Berlin). These values were correlated with those obtained by the Illumina assay for each of the above-mentioned genes for array validation.
Statistical analysis.
Methylation data pre-processing and identification of differentially methylated loci. The relative level of methylation for each CG site was calculated as the ratio of methylated-probe signal to total locus signal intensity and defined within 0 to 1 range, exported from the Illumina BeadStudio software package. The data were then normalized as described previously,22,23 using the variability of areas with low methylation level as a reference point. In order to find genes with methylation levels above the level of technical noise, a frequency histogram of raw methylation signal was determined for each array. The histogram yielded a right-skewed unimodal distribution curve with a mode of approximately 0.013. A normal distribution curve representing the variability of the data around zero was then fitted around the mode, mirroring the Gaussian profile of the left part of the histogram. Its parameters were then defined (mean, SD) and the data were normalized to the standard deviation of the noise after subtraction of the mean. The data were then log10-transformed and adjusted to each other by linear regression under the assumption that the methylation of most CG sites does not change. The data were then filtered to remove CG sites with a methylation level value less than 3.0, equivalent to setting a threshold at three standard deviations above the noise level. CG sites with methylation differential below the noise level under all experimental conditions (about 1,600) were excluded from consideration, as their methylation cannot be reliably assessed. Differentially methylated data were deposited in the Gene Expression Omnibus (GEO accession: GSE27895). All methods and analyses were performed in Matlab (Mathworks, Natick, MA), unless otherwise specified.
To identify differentially methylated CG sites between lupus patients and controls, we used the associative analysis as described by Dozmorov et al.23 CG sites with methylation signal greater than 6 SD above noise level and have at least 1.2-fold difference in methylation between cases and controls were identified. Our study had a 94% power to detect a methylation difference of 1.2-fold or greater between lupus patients and controls (α = 0.05).
Identification of methylation changes correlating with disease activity in lupus patients. Normalized data were converted back to the original scale of 0–1 corresponding to the percentage of loci methylation. Absolute standard deviation for each CG site included on the arrays was calculated separately for cases and controls and the absolute standard deviation ratios of cases versus controls was calculated. CG sites with standard deviation ratios above 4 were considered for subsequent analysis. Pearson's correlation coefficients and p values were calculated for correlation between methylation levels and SLEDAI scores in lupus patients. Genes associated with these CG sites were identified.
Functional enrichment and network analysis. To identify if differentially methylated genes shared common functional properties, we used the Database for Annotation, Visualization and Integrated Discovery (DAVID).24 The Gene Functional Classification tool in DAVID builds clusters of genes with significantly similar ontologies as tested against a complete list of genes in the database. Medium stringency was used to yield a broader set of ontological groups and to group genes with similar functions.
Lists of hypermethylated and hypomethylated genes were also submitted to the Ingenuity Pathway Analysis system (IPA; Ingenuity® Systems, Redwood City, CA, www.ingenuity.com). By default, p < 0.05 was used in all calculations. Gene lists from each group were analyzed for overrepresented general functions, canonical pathways and the networks they are associated with. Finally, we also used a large-scale literature analysis package called IRIDESCENT,25 which has been a useful method to identify published commonalities within a set of genes.26,27 Genes without published literature were analyzed by using a microarray meta-analytic approach to predict function.28 Each tool was used to help confirm and clarify the results from the other tools.
Acknowledgements
This work was made possible by NIH Grant Number R03AI076729 from the National Institute of Allergy and Infectious Diseases and NIH Grants Number P20RR020143 and P30AR053483 and the Lupus Research Institute. We are thankful to the genotyping core at OMRF and in particular to Adam Adler and Kenneth Kaufman, Ph.D., for their help with this study.
Supplementary Material
References
- 1.Delgado-Vega A, Sanchez E, Lofgren S, Castillejo-Lopez C, Alarcon-Riquelme ME. Recent findings on genetics of systemic autoimmune diseases. Curr Opin Immunol. 2010;22:698–705. doi: 10.1016/j.coi.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Poole BD, Templeton AK, Guthridge JM, Brown EJ, Harley JB, James JA. Aberrant Epstein-Barr viral infection in systemic lupus erythematosus. Autoimmun Rev. 2009;8:337–342. doi: 10.1016/j.autrev.2008.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Javierre BM, Fernandez AF, Richter J, Al-Shahrour F, Martin-Subero JI, Rodriguez-Ubreva J, et al. Changes in the pattern of DNA methylation associate with twin discordance in systemic lupus erythematosus. Genome Research. 2009;20:170–179. doi: 10.1101/gr.100289.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hughes T, Webb R, Fei Y, Wren JD, Sawalha AH. DNA methylome in human CD4+ T cells identifies transcriptionally repressive and non-repressive methylation peaks. Genes Immun. 2010;11:554–560. doi: 10.1038/gene.2010.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bruniquel D, Schwartz RH. Selective, stable demethylation of the interleukin-2 gene enhances transcription by an active process. Nat Immunol. 2003;4:235–240. doi: 10.1038/ni887. [DOI] [PubMed] [Google Scholar]
- 6.Watson JA, Watson CJ, McCrohan AM, Woodfine K, Tosetto M, McDaid J, et al. Generation of an epigenetic signature by chronic hypoxia in prostate cells. Hum Mol Genet. 2009;18:3594–3604. doi: 10.1093/hmg/ddp307. [DOI] [PubMed] [Google Scholar]
- 7.Chen P, Jiang T, Ouyang J, Cui Y, Chen Y. Epigenetic programming of diverse glucocorticoid response and inflammatory/immune-mediated disease. Med Hypotheses. 2009;73:657–658. doi: 10.1016/j.mehy.2009.08.013. [DOI] [PubMed] [Google Scholar]
- 8.Blazkova J, Trejbalova K, Gondois-Rey F, Halfon P, Philibert P, Guiguen A, et al. CpG methylation controls reactivation of HIV from latency. PLoS Pathogens. 2009;5:1000554. doi: 10.1371/journal.ppat.1000554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baylin SB, Herman JG, Graff JR, Vertino PM, Issa JP. Alterations in DNA methylation: a fundamental aspect of neoplasia. Adv Cancer Res. 1998;72:141–196. [PubMed] [Google Scholar]
- 10.Richardson B, Scheinbart L, Strahler J, Gross L, Hanash S, Johnson M. Evidence for impaired T cell DNA methylation in systemic lupus erythematosus and rheumatoid arthritis. Arthritis Rheum. 1990;33:1665–1673. doi: 10.1002/art.1780331109. [DOI] [PubMed] [Google Scholar]
- 11.Lu Q, Kaplan M, Ray D, Zacharek S, Gutsch D, et al. Demethylation of ITGAL (CD11a) regulatory sequences in systemic lupus erythematosus. Arthritis Rheum. 2002;46:1282–1291. doi: 10.1002/art.10234. [DOI] [PubMed] [Google Scholar]
- 12.Oelke K, Lu Q, Richardson D, Wu A, Deng C, Hanash S, et al. Overexpression of CD70 and overstimulation of IgG synthesis by lupus T cells and T cells treated with DNA methylation inhibitors. Arthritis Rheum. 2004;50:1850–1860. doi: 10.1002/art.20255. [DOI] [PubMed] [Google Scholar]
- 13.Deng C, Kaplan MJ, Yang J, Ray D, Zhang Z, McCune WJ, et al. Decreased Ras-mitogen-activated protein kinase signaling may cause DNA hypomethylation in T lymphocytes from lupus patients. Arthritis Rheum. 2001;44:397–407. doi: 10.1002/1529-0131(200102)44:2<397::AID-ANR59>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 14.Yung R, Chang S, Hemati N, Johnson K, Richardson B. Mechanisms of drug-induced lupus. IV. Comparison of procainamide and hydralazine with analogs in vitro and in vivo. Arthritis Rheum. 1997;40:1436–1443. doi: 10.1002/art.1780400811. [DOI] [PubMed] [Google Scholar]
- 15.Sawalha AH, Jeffries M. Defective DNA methylation and CD70 overexpression in CD4+ T cells in MRL/lpr lupus-prone mice. Eur J Immunol. 2007;37:1407–1413. doi: 10.1002/eji.200636872. [DOI] [PubMed] [Google Scholar]
- 16.Sawalha AH, Jeffries M, Webb R, Lu Q, Gorelik G, Ray D, et al. Defective T-cell ERK signaling induces interferon-regulated gene expression and overexpression of methylation-sensitive genes similar to lupus patients. Genes Immun. 2008;9:368–378. doi: 10.1038/gene.2008.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu Q, Wu A, Tesmer L, Ray D, Yousif N, Richardson B. Demethylation of CD40LG on the inactive X in T cells from women with lupus. J Immunol. 2007;179:6352–6358. doi: 10.4049/jimmunol.179.9.6352. [DOI] [PubMed] [Google Scholar]
- 18.Zhou Y, Yuan J, Pan Y, Fei Y, Qiu X, Hu N, et al. T cell CD40LG gene expression and the production of IgG by autologous B cells in systemic lupus erythematosus. Clin Immunol. 2009;132:362–370. doi: 10.1016/j.clim.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scofield RH, Bruner GR, Namjou B, Kimberly RP, Ramsey-Goldman R, Petri M, et al. Klinefelter's syndrome (47,XXY) in male systemic lupus erythematosus patients: support for the notion of a gene-dose effect from the X chromosome. Arthritis Rheum. 2008;58:2511–2517. doi: 10.1002/art.23701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sawalha AH, Harley JB, Scofield RH. Autoimmunity and Klinefelter's syndrome: when men have two X chromosomes. J Autoimmun. 2009;33:31–34. doi: 10.1016/j.jaut.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hochberg MC. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1997;40:1725. doi: 10.1002/art.1780400928. [DOI] [PubMed] [Google Scholar]
- 22.Dozmorov I, Knowlton N, Tang Y, Shields A, Pathipvanich P, Jarvis JN, et al. Hypervariable genes—experimental error or hidden dynamics. Nucleic Acids Res. 2004;32:147. doi: 10.1093/nar/gnh146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dozmorov I, Lefkovits I. Internal standard-based analysis of microarray data. Part 1: analysis of differential gene expressions. Nucleic Acids Res. 2009;37:6323–6339. doi: 10.1093/nar/gkp706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dennis G, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, et al. DAVID: Database for Annotation, Visualization and Integrated Discovery. Genome Biol. 2003;4:3. [PubMed] [Google Scholar]
- 25.Wren JD, Garner HR. Shared relationship analysis: ranking set cohesion and commonalities within a literature-derived relationship network. Bioinformatics. 2004;20:191–198. doi: 10.1093/bioinformatics/btg390. [DOI] [PubMed] [Google Scholar]
- 26.Xu Z, Patterson TA, Wren JD, Han T, Shi L, Duhart H, et al. A microarray study of MPP+-treated PC12 Cells: Mechanisms of toxicity (MOT) analysis using bioinformatics tools. BMC Bioinformatics. 2005;6:8. doi: 10.1186/1471-2105-6-S2-S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mizumoto N, Hui F, Edelbaum D, Weil MR, Wren JD, Shalhevet D, et al. Differential activation profiles of multiple transcription factors during dendritic cell maturation. J Invest Dermatol. 2005;124:718–724. doi: 10.1111/j.0022-202X.2005.23616.x. [DOI] [PubMed] [Google Scholar]
- 28.Wren JD. A global meta-analysis of microarray expression data to predict unknown gene functions and estimate the literature-data divide. Bioinformatics. 2009;25:1694–1701. doi: 10.1093/bioinformatics/btp290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim YI, Logan JW, Mason JB, Roubenoff R. DNA hypomethylation in inflammatory arthritis: reversal with methotrexate. J Lab Clin Med. 1996;128:165–172. doi: 10.1016/s0022-2143(96)90008-6. [DOI] [PubMed] [Google Scholar]
- 30.Sehouli J, Loddenkemper C, Cornu T, Schwachula T, Hoffmuller U, Grutzkau A, et al. Epigenetic quantification of tumor-infiltrating T-lymphocytes. Epigenetics. 2011;6:236–246. doi: 10.4161/epi.6.2.13755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Akahoshi M, Nakashima H, Tanaka Y, Kohsaka T, Nagano S, Ohgami E, et al. Th1/Th2 balance of peripheral T helper cells in systemic lupus erythematosus. Arthritis Rheum. 1999;42:1644–1648. doi: 10.1002/1529-0131(199908)42:8<1644::AID-ANR12>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 32.Richardson B, Scheinbart L, Strahler J, Gross L, Hanash S, Johnson M. Evidence for impaired T cell DNA methylation in systemic lupus erythematosus and rheumatoid arthritis. Arthritis Rheum. 1990;33:1665–1673. doi: 10.1002/art.1780331109. [DOI] [PubMed] [Google Scholar]
- 33.Yoshihara Y, Nakamura H, Obata K, Yamada H, Hayakawa T, Fujikawa K, et al. Matrix metalloproteinases and tissue inhibitors of metalloproteinases in synovial fluids from patients with rheumatoid arthritis or osteoarthritis. Ann Rheum Dis. 2000;59:455–461. doi: 10.1136/ard.59.6.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perez P, Goicovich E, Alliende C, Aguilera S, Leyton C, Molina C, et al. Differential expression of matrix metalloproteinases in labial salivary glands of patients with primary Sjogren's syndrome. Arthritis Rheum. 2000;43:2807–2817. doi: 10.1002/1529-0131(200012)43:12<2807::AID-ANR22>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 35.Hedberg A, Fismen S, Fenton KA, Mortensen ES, Rekvig OP. Deposition of chromatin-IgG complexes in skin of nephritic MRL-lpr/lpr mice is associated with increased local matrix metalloprotease activities. Exp Dermatol. 2010;19:265–274. doi: 10.1111/j.1600-0625.2010.01064.x. [DOI] [PubMed] [Google Scholar]
- 36.Kurasawa K, Arai S, Owada T, Maezawa R, Kumano K, Fukuda T. Autoantibodies against platelet-derived growth factor receptor alpha in patients with systemic lupus erythematosus. Mod Rheumatol. 2010;20:458–465. doi: 10.1007/s10165-010-0310-x. [DOI] [PubMed] [Google Scholar]
- 37.Jeffries M, Hamadeh F, Aberle T, Glenn S, Kamen DL, Kelly JA, et al. Haemolytic anaemia in a multi-ethnic cohort of lupus patients: a clinical and serological perspective. Lupus. 2008;17:739–743. doi: 10.1177/0961203308090990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zoja C, Corna D, Rottoli D, Zanchi C, Abbate M, Remuzzi G. Imatinib ameliorates renal disease and survival in murine lupus autoimmune disease. Kidney Int. 2006;70:97–103. doi: 10.1038/sj.ki.5001528. [DOI] [PubMed] [Google Scholar]
- 39.Kaji K, Takeshita S, Miyake K, Takai T, Kudo A. Functional association of CD9 with the Fc gamma receptors in macrophages. J Immunol. 2001;166:3256–3265. doi: 10.4049/jimmunol.166.5.3256. [DOI] [PubMed] [Google Scholar]
- 40.Tai XG, Yashiro Y, Abe R, Toyooka K, Wood CR, Morris J, et al. A role for CD9 molecules in T cell activation. J Exp Med. 1996;184:753–758. doi: 10.1084/jem.184.2.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kuhl BD, Sloan RD, Donahue DA, Bar-Magen T, Liang C, Wainberg MA. Tetherin restricts direct cell-to-cell infection of HIV-1. Retrovirology. 2010;7:115. doi: 10.1186/1742-4690-7-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.James JA, Kaufman KM, Farris AD, Taylor-Albert E, Lehman TJ, Harley JB. An increased prevalence of Epstein-Barr virus infection in young patients suggests a possible etiology for systemic lupus erythematosus. J Clin Invest. 1997;100:3019–3026. doi: 10.1172/JCI119856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Perl A, Fernandez D, Telarico T, Phillips PE. Endogenous retroviral pathogenesis in lupus. Curr Opin Rheumatol. 2010;22:483–492. doi: 10.1097/BOR.0b013e32833c6297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Burton J, Vera JH, Kapembwa M. HIV and systemic lupus erythematosus: the clinical and diagnostic dilemmas of having dual diagnosis. Int J STD AIDS. 2010;21:845–846. doi: 10.1258/ijsa.2010.010062. [DOI] [PubMed] [Google Scholar]
- 45.Li Y, Liu Y, Strickland FM, Richardson B. Age-dependent decreases in DNA methyltransferase levels and low transmethylation micronutrient levels synergize to promote overexpression of genes implicated in autoimmunity and acute coronary syndromes. Exp Gerontol. 2010;45:312–322. doi: 10.1016/j.exger.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Z, Deng C, Lu Q, Richardson B. Age-dependent DNA methylation changes in the ITGAL (CD11a) promoter. Mech Ageing Dev. 2002;123:1257–1268. doi: 10.1016/s0047-6374(02)00014-3. [DOI] [PubMed] [Google Scholar]
- 47.Fainaru O, Woolf E, Lotem J, Yarmus M, Brenner O, Goldenberg D, et al. Runx3 regulates mouse TGFbeta-mediated dendritic cell function and its absence results in airway inflammation. EMBO J. 2004;23:969–979. doi: 10.1038/sj.emboj.7600085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Domínguez-Soto A, Relloso M, Vega MA, Corbí AL, Puig-Kröger A. RUNX3 regulates the activity of the CD11a and CD49d integrin gene promoters. Immunobiology. 2005;210:133–139. doi: 10.1016/j.imbio.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 49.Ciavatta DJ, Yang J, Preston GA, Badhwar AK, Xiao H, Hewins P, et al. Epigenetic basis for aberrant upregulation of autoantigen genes in humans with ANCA vasculitis. J Clin Invest. 2010;120:3209–3219. doi: 10.1172/JCI40034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Casciola-Rosen LA, Anhalt G, Rosen A. Autoantigens targeted in systemic lupus erythematosus are clustered in two populations of surface structures on apoptotic keratinocytes. J Exp Med. 1994;179:1317–1330. doi: 10.1084/jem.179.4.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Watanabe-Fukunaga R, Brannan CI, Copeland NG, Jenkins NA, Nagata S. Lymphoproliferation disorder in mice explained by defects in Fas antigen that mediates apoptosis. Nature. 1992;356:314–317. doi: 10.1038/356314a0. [DOI] [PubMed] [Google Scholar]
- 52.Strasser A, Whittingham S, Vaux DL, Bath ML, Adams JM, Cory S, et al. Enforced BCL2 expression in B-lymphoid cells prolongs antibody responses and elicits autoimmune disease. Proc Natl Acad Sci USA. 1991;88:8661–8665. doi: 10.1073/pnas.88.19.8661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Emlen W, Niebur J, Kadera R. Accelerated in vitro apoptosis of lymphocytes from patients with systemic lupus erythematosus. J Immunol. 1994;152:3685–3692. [PubMed] [Google Scholar]
- 54.Kalden JR. Defective phagocytosis of apoptotic cells: possible explanation for the induction of autoantibodies in SLE. Lupus. 1997;6:326–327. doi: 10.1177/096120339700600326. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.