Abstract
In this review, we focus on the interaction between the circadian clock of higher plants to that of metabolic and physiological processes that coordinate growth and performance under a predictable, albeit changing environment. In this, the phytochrome and cryptochrome photoreceptors have shown to be important, but not essential for oscillator control under diurnal cycles of light and dark. From this foundation, we will examine how emerging findings have firmly linked the circadian clock, as a central mediator in the coordination of metabolism, to maintain homeostasis. This occurs by oscillator synchronization of global transcription, which leads to a dynamic control of a host of physiological processes. These include the determination of the levels of primary and secondary metabolites, and the anticipation of future environmental stresses, such as mid-day drought and midnight coldness. Interestingly, metabolic and stress cues themselves appear to feedback on oscillator function. In such a way, the circadian clock of plants and abiotic-stress tolerance appear to be firmly interconnected processes.
Key words: circadian clock, Arabidopsis, abiotic stress, metabolism
General Background
Circadian clocks are a time-keeping mechanism used to coordinate the physiology of an organism to its surrounding environment. These clocks anticipate the day to night transitions and consequently have a periodicity of about 24 hours. This speed is not exactly 24 hours, and natural variants and mutants have been isolated, in a wide variety of organisms, that change the speed of this oscillator. This speed has been termed periodicity and periodicity variants/mutants have allowed for a detailed characterization of physiological and developmental processes under circadian control. Several clock-controlled responses include leaf movement, flower-bud opening and stomata aperture in plants, conidiation in fungi, behavioral mannerisms flies and melatonin secretion and sleep cycles in mammals.1 Thus a circadian clock provides fitness by setting physiological events according to the changes between day and night (Fig. 1A). In this way, responses to predicted environmental stresses can be more appropriately alleviated.
Figure 1.
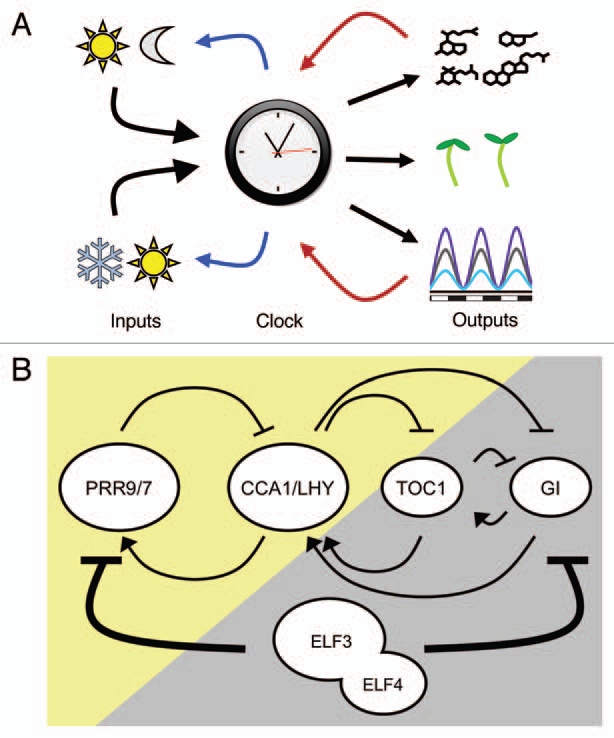
The Plant Circadian System. (A) Crosstalk between the circadian clock with environmental inputs and clock outputs. The clock is entrained by light/dark and warm/cold cycles. In turn, the clock controls multiple output responses, some of which feedback to the clock (red arrows), thus also serving as input signals. Both inputs and some outputs are gated by the clock (lines in blue and red respectively). (B) The current model for the circadian clock. An arrow indicates positive regulation. A stopped arrow indicates negative regulation. Day and night is indicted by the background color (adapted from Kolmos et al.17,18).
The selective advantage provided by an internal pacemaker has been demonstrated in both photosynthetic bacteria and in higher plants. In a competition experiment, strains of cyanobacteria that had circadian clocks with a wild type, short or long period, were subjected to non-24-hour days. These artificial days consisted in equivalent light/dark periods of 11, 12 or 15 hours to thus create a day of 22, 24 and 30 hours, respectively. After 27 days of growth under these conditions, the strain that outgrew the others was the one with a clock period most similar to the subjected artificial day.2 Thus for each of these strains, an artificial day either longer or shorter to their internal rhythm was proposed to cause stress by desynchronizing the external and internal rhythms. In a similar fashion, Dodd et al.3 demonstrated that higher plants with a clock whose periodicity was most similar to the subjected artificial day outperformed those that had a periodicity less matched to the respective environment (Fig. 2). This growth advantage was concluded to be a consequence of higher carbon fixation and improved photosynthesis.3 These studies provided evidence that a clock synchronized with the environment enhances fitness, in both lower and higher organisms. Therefore it was proposed that this fitness benefit was generated by the orchestration of metabolic and stress pathways.
Figure 2.
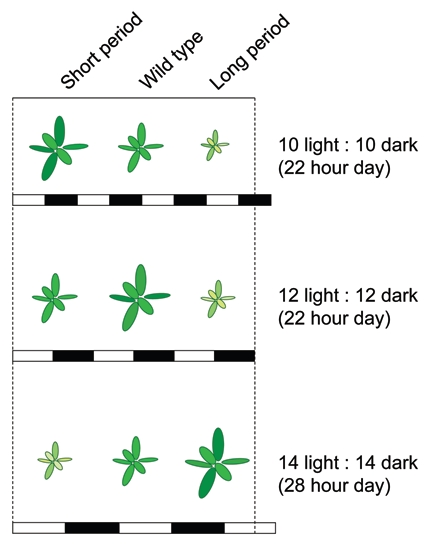
Resonation between the internal oscillator and the external environment enhances plant fitness and performance. An illustration adapted from Figure 4B of (Dodd et al.3). The vigor of wild type, and short- and long-period plants is enhanced when the environment closely matches the ambient environment, in part through improved photosynthesis. Note that the light and dark periods over the depicted days intentionally do not fit the normal diurnal 24 h period (between the horizontal lines). White and black bars denote day and night respectively and their length denotes its duration.
Circadian Clock Generalities
Circadian clocks consist of positive and negative elements that comprise a transcriptional feedback loop.4 In general, the transcription of the positive elements induces the expression of the negative elements, which in turn repress the expression of the formers, thus closing the oscillator. Interestingly, though the clocks of several model organisms share an analogous mechanism, their components are not conserved between them, suggesting that circadian clocks must have appeared several times throughout evolution.
A transcriptional-translational feedback loop is not enough to sustain a ∼24 hour rhythm. To keep such a pace, post-translational processes which govern the activity and stability of both the positive and negative elements. Reversible phosphorylation has emerged as an essential mechanism that drives the timing of the loop by activating, inactivating or providing a targeting signal that ultimately leads to protein degradation.5 Sensing of metabolite homeostasis and internal regulation of stress pathways have numerous control steps that use kinases. One can wonder if metabolic signals could feedback to the clock ensuring proper fine-tuning; this is discussed below.
The oscillator is appropriately set to the correct time of day through the perception of environmental signals, such as light and temperature. These are termed the input pathways to the clock (Fig. 1A). These input signals control responses such as gene expression and metabolic reprogramming, which are termed the outputs of the clock.6 However these clock-driven outputs can also feedback to the clock serving as an input. The oscillator modulates the perception of the input signals in a process called gating, which sets the clock susceptibility to certain inputs to a particular time of day.7,8 Thus one ultimate function of the circadian clock is to coordinate physiological and metabolic processes in order to take place at the time when they are needed. For instance, priming light harvesting at dawn.
The clock framework of transcription and translation is fine tuned in a daily basis by environmental signals, which consist mainly of light and temperature.6,7 Both of these have been considered as the key input signals to the clock. As the clock is sensitive to the dark to light transitions and changes in temperature at dawn, the clock is reset, thus providing and adjustment to the environment on a daily basis.6,7 Once the clock is set to its environment, it coordinates numerous processes, such as transcription, hormone production, metabolic reactions (see below), which have been considered the primary outputs of the clock. As these clock-driven outputs can also feedback to the clock serving as an input, this view of input and output signals has recently been changed to an intricate network [see below and reviewed in ref. 9]. As the outputs of a clock can serve as input signals and the input signals are gated to specific times of the day, the picture that arises is that of a continuous crosstalk between the input/environmental sensing pathways and the circadian clock that sets a rhythmic time window of environmental perception.
The Arabidopsis thaliana Circadian Clock
The core of the A. thaliana oscillator was first proposed to comprise of a loop between two morning components and one evening component (Fig. 1B). The formers are two Myb-related transcription factors, CIRCADIAN CLOCK ASSOCIATED (CCA1) and LATE ELONGATED HYPOCOTYL (LHY),10,11 whose encoded proteins bind to the Evening Element (EE) in the promoter of TIMING OF CAB EXPRESSION/PSEUDORESPONSE REGULATOR1 (TOC1/PRR1), the evening component.12 This binding represses TOC1 transcription, but at dusk the protein levels of CCA1 and LHY diminish and the repression of TOC1 is relieved. As a consequence, TOC1 mRNA reaches its maximum around dusk, and by an unknown mechanism, TOC1 protein activates the transcription of CCA1 and LHY,12,13 and it has been proposed that this is direct.14,15 The role of CCA1/LHY and TOC1 as core elements has been confirmed by genetic experiments.16 Though these three elements have been considered the core of the plant clock, this model cannot fully explain all experimental data.17–21
Mathematical models have been formalized to comprise a clock with three interconnected feedback loops, which harbors unknown components within them. In a three-loop model,20–22 besides their described role in the core oscillator, CCA1 and LHY also participate in a morning loop to induce the expression of the PSEUDO RESPONSE REGULATOR 7 and 9 (PRR7 and PPR9), whose encoded proteins in turn bind and repress the transcription of the formers.23 A so-called evening loop places TOC1 as a repressor of an unidentified ‘Y’ component, which feeds back by inducing TOC1 expression. GIGANTEA (GI) was proposed to be the Y component, but this factor only partially fulfils this role.20 Within this model, the identity of an X factor, which serves as the component between TOC1 expression and CCA1/LHY was mathematically required. A gene with these characteristics has not yet been found. An element acting after TOC1 was identified and termed CCA1 HIKING EXPEDITION (CHE).15 This factor cannot be X, as its protein only binds the promoter of CCA1 and not that of LHY. Furthermore CHE binding to CCA1 promoter represses CCA1 transcription. This is opposed to the expected X function of an activator.20 Thus the circadian clock model still has pieces to be discovered and fit into the current model of the circadian framework. Research efforts in this direction have placed EARLY FLOWERING 4 (ELF4) within the oscillator as a key circadian repressor.17,18,24,25
Other studies have shown that post-translational modifications, including protein targeted degradation and protein phosphorylation are essential for the function of the plant circadian clock. As an example, it has been shown that ZEITLUPE (ZTL), a light-sensitive protein, binds to TOC1 during the night, directing TOC1 to degradation by the proteasome.26,27 Furthermore ZTL was proposed to be stabilized by GI when plants are under blue light.28 In contrast to the action of ZTL on TOC1 instability, PRR3 protects TOC1 from degradation.29 Targeted protein degradation also takes place in the morning phase of the oscillator. As an example, LHY proteolysis was shown to be regulated by DEETIOLATED 1 (DET1). There it was demonstrated that DET1 inhibited LHY degradation in a light-independent process.30 Besides protein degradation, the localization and import of clock components to the nucleus have been shown to be crucial for clock function.31 In that seminal work, the authors showed that TOC1 interacts with PRR5 in the cytosol and that this interaction directs TOC1 nuclear localization as well as its phosphorylation.31 Taken together, protein modification and stability of clock components are essential for the oscillator function.
Light Perception and Signal Transduction to the Clock
Light has at least three roles in a plant's life. Firstly light provides the energy for photosynthesis. Secondly though light is a source for energy, it also produces cellular damage and cell death through the production of Reactive oxygen species (ROS). Consequently plants are subjected on a daily basis to both events and therefore adjust their metabolism to the duration and intensity of the light source. Finally light also serves an environmental signal by directing photomorphogenesis.
Plants perceive light through different classes of photoreceptors. Phytochromes are the dominant sensors for red and far-red light, whereas the cryptochromes perceive predominantly blue light.32 These chromic molecules are essential for plant development, as mutations in photoreceptor genes impair the de-etiolation process, development and fitness.32
As light is the most dominant input signal to the clock, the role of the photoreceptors in the circadian clock has been studied. Photoreceptors gene transcription was reported to be under circadian control.33 Photoreceptor mutants were used to establish their role within the circadian clock. It was shown that the photoreceptor function within the clock was wavelength and intensity specific as for photomorphogenesis: phytochrome B (phyB) was shown to be the main red light photoreceptor, cryptochrome 1 (cry1) the blue light photoreceptor and phytochrome A (phyA) the low-fluence photoreceptor.34 It was shown that photoreceptors mutants such as that phyA and phyB were not necessary for sustaining circadian responses.35 Also photoreceptors mutants displayed a light intensity dependent effect on clock periodicity. In a fluence rate and light-quality dependent manner, photoreceptor mutants exhibited a lengthened period.34 Interestingly Devlin et al.36 demonstrated that red-light signaling through phyA required a functional cry1. In parallel, Más et al.37 showed that phy and cry2 proteins interacted, forming foci in a light-dependent manner. Thus phytochromes and cryptochromes interaction coordinate light input to the clock. Recently Palágyi et al. showed that phyB amino terminus is sufficient for the red light signaling to the clock; however under white light, the carboxy terminus was shown to be required.38
Further research showed that even a quadruple photoreceptor mutant (phyA/phyB/cry1/cry2),39 as did a quintuple phytochrome mutant (phyA/phyB/phyC/phyD/phyE), displayed robust circadian rhythms.40 Consequently though light input is essential for clock periodicity, photic-entrainment can still take place without canonical photoreceptors. This result could suggest that other molecules could be responsible of transducing the light signals to the oscillator.
If photoreceptors are not required to transduce light signals to the clock, other light perceiving molecules could act as circadian photoreceptors. As mentioned before, ZTL has been proposed as a blue-light photoreceptor. ZTL structure contains a Light-Oxygen-Voltage (LOV) domain that is essential for the light dependent binding of ZTL with GI.28 ztl was described as a clock mutant that altered period in a fluence-dependent manner.41 Two other proteins exist in the A. thaliana genome with a similar structure to ZTL. These are the LOV-KELCH PROTEIN 2 (LKP2) and FLAVIN BINDING KELCH REPEAT F-BOX 1 (FKF1), which both posses a LOV domain, though no clear role in light perception has yet been established for these products. lkp2 and fkf1 single mutants were found to display subtle effects on circadian rhythms, but a ztl/lkp2/fkf1 triple mutant became essentially arrhythmic.42 Thus ZTL had a major role in the circadian clock. Consequently ZTL remains a likely candidate for light transduction to the clock.
Several other loci have been genetically shown to affect light input to the clock. These include genes such as ELF3,43 ELF4,25 TIME FOR COFFEE,44,45 SENSITIVITY TO RED LIGHT REDUCED (SRR1),46 LIGHT INSENSITIVE PERIOD 1 (LIP1),47,48 and XAP5 CIRCADIAN TIMEKEEPER (XCT).49 None of these genes encode a protein with apparent chromophore interactions, required as a canonical domain involved in light perception, which suggests that their protein products act as signaling components in a step downstream of the light signal perception. As an example, lip1 mutant, which encodes for an atypical plant GTPase, is almost insensitive to changes in light intensity.47,48 In summary, light input to the clock, in particular to entrainment, is far from resolved. Finding the missing pieces, as well as fully understanding the input function of known light signaling mutants may help elucidate photoentrainment.
Genome Transcriptional Control by the Circadian Clock
Microarray expression analysis demonstrated the pervasiveness of the circadian clock in the plant transcriptome.50 Estimates of the percentage of genes under circadian control ranged between 6% up to 15%, but more thorough experimentation concluded that at least 30% of the transcriptome is under clock control.22,51,52 Within this percentage of genes, it was found that a number of transcription factors including the MYB, bHLH and bZIP families were overrepresented as being under circadian control.53 This is suggestive of a mechanism by which rhythms in several transcription factors drive a genome wide transcriptional rhythm on a daily basis. Besides transcription factors, phytohormone production and sensitization,51,54 as well as the expression of cold responses55 appeared under clock control. These transcript profile analyses demonstrated how the circadian clock drives the expression of different biological processes in anticipation of physiological events.
Other transcriptomic analysis took into consideration not only circadian microarray datasets, but also diurnal data and concluded that at least 89% of the transcribed genome is rhythmic under natural, diurnal conditions.56,57 This analysis suggested that such an extensive control was achieved by dispersed promoter sequences throughout the plant genome that provide a particular time frame of expression.56,57 These authors also found that at least in one of the conditions tested, more than 10,000 genes were under circadian control, while more than 16,000 were diurnally controlled. Interestingly, an overlap of 8,500 between them was found, providing the possibility that a substantial fraction of diurnal rhythms are not under clock control. Furthermore gene-expression peaks mainly tended towards being either at dawn or dusk. Thus, it seems that either clock or diurnal controlled, most of the responses to the environment are tightly time coordinated using the light/dark transitions as the main environmental signal.
Covington et al.51 found a significant set of genes involved in hormonal metabolism to be circadian regulated. In particular ABA, cytokinines, methyl jasmonate, salicylic acid and auxin were strongly circadian controlled. This exerted control by the circadian clock on the transcription of hormones is not a surprise as hormones are outputs of the clock. Interestingly it has also been shown that hormones can feedback to the clock, being capable of affecting period length, amplitude or phase. Furthermore the physiological responses to hormones have been shown to be gated by the circadian clock. For example stomatal opening is dictated by the clock in well-watered plants, opening at dawn and closing before dusk.3 Similarly, treatment with the hormone ABA, which promotes stomatal closure, is less effective early in the day than at the end of it.58 Additionally, many auxin genes are clock regulated and the later gates the sensitivity to the hormone depending on the time of day.59,60 The gated response to auxin explains the observed growth of plants around dawn in a diurnal photoperiod.61 At this time of the day, auxin levels and plant responsiveness coincide with increased water turgor pressure and renewal of carbon supply, promoting growth. In summary, the plethora of genes under circadian control and the capacity the clock has on modulating multiple responses through a gating mechanism, highlights the importance of the circadian clock in adjusting the plant to its environment.
ABA and Non-ABA Stress Responses: Drought, Cold and Osmotic Stress
From the available microarray expression profiles, circadian datasets were found to extensively overlap mainly with ABA62,63 and cADPR64 datasets. The relationship between the clock and ABA is interesting because ABA controls many environmental stress responses, such as water use and responses to drought, as well as to priming for frost tolerance. Recently it was found that TOC1 expression is induced by ABA.65 These authors found that the central oscillator component TOC1 was implicated in plant responses to drought by controlling stomatal aperture through the circadian and diurnal regulation of the H subunit of the magnesium-protoporphyrin IX chelatase, also known as GENOME UNCOUPLED 5 (ABAR/GUN5).65 Overexpression of TOC1 led to drought hypersensitivity, as it impeded stomatal closure.65 Therefore, the reciprocal link between the circadian clock and ABA-related responses could define how the clock prepares and deals with stress responses to enhance fitness.
The overlap between ABA and clock microarray datasets arose from the circadian control of many key genes involved in ABA biosynthesis and signal transduction. Genes such as EARLY RESPONSE TO DEHYDRATION 10 (ERD10) and 7 (ERD7), COLD REGULATED 15 B (COR15B) and A (COR15A) and RESPONSE TO DISSECATION (RD29A) were found to be transcriptionally induced during the day.63 All of these genes are known to be involved in the responses to drought and water deprivation, and to the responses to osmotic stress. A detailed examination by Covington et al.59 showed not only that ABA inducible genes are expressed during the day, but that key enzymes in ABA precursors and biosynthesis are also clock controlled. These included CLOROPLASTOS ALTERADOS 1 (CLA1), PHYTOENO SYNTHASE (PSY), 9-CIS'EPOXYCAROTENOID DIOXYGENASE (NCED3) and ABA DEFICIENT 2 (ABA2), which participate in isoprenoid precursors synthesis, carotenoid synthesis and ABA biosynthesis, respectively. Recalling that carotenoids participate in the xanthophyll cycle in chloroplast to avoid excess of solar energy absorption,66 as well as the circadian control of stomatal opening,58 the clock seems to link the daytime heat from solar irradiation with stress and water loss, and consequently prepares in advance to them.
Given the relationship between the circadian clock and ABA, it is perhaps not surprising that microarray datasets from osmotic, salt and water deprivation stress also have a high number of genes under circadian control.51 Through the use of genome tilling arrays, it was shown that both annotated and unannotated regions responded to a diversity of abiotic stress.67 In the same report the similarity between salt and osmotic stress profiles was the highest between all the treatments. ABA treated samples showed certain similarity with the previous stresses, whereas cold and heat profiles shared less identity.67
Dusk plays a role as an environmental signal for circadian control. At dusk, genes involved in starch remobilization and lipid modification reach their peak expression.50,68,69 The later has been interpreted as a correlation with the anticipation of cold nights and consequently freezing tolerance.70 In fact, the circadian clock controls the expression of cold responsive genes through the C-REPEAT BINDING FACTOR 1/DEHYDRATION RESPONSIVE ELEMENT BINDING 1 (CBF1/DREB1) family of transcription factors. Expression of CBF1/2/3 is gated by the clock to take place in the light phase, but the signal transduction to CBFs targets delays the freezing tolerance after dusk, a time in which the plant faces chilling.55 Furthermore the expression of the more than 100 targets known as the CBF/DREB regulon not only provides freezing tolerance, but also resistance to salt and drought.71,72 Though the later share the resistance profile with the ABA induced genes, the CBF/DREB transcription factors are independent of ABA signals.73 Interestingly Franklin et al. demonstrated that CBF1, CBF2 and CBF3 expression was increased under a low R/FR ratio (increased far-red light), which is typically present under a canopy or other shading. After experiencing a low R/FR illumination, plants acquired freezing tolerance. An enriched far-red light quality is mainly found during dawn and dusk and its duration is particularly longer in higher latitudes. Then the requirement of a low R/FR for CBF expression together with the gating of cold responses to dusk by the circadian clock55 prepares the plant for the oncoming night. Thus the circadian clock, ABA and light signals coordinate the transcriptional cold response and interconnects the clock input and output pathways.
Links between the Plant Circadian Clock and Metabolism
A key piece of molecular-genetic evidence mediating between the plant circadian clock and metabolism was provided by Dodd et al.64 These authors found that cyclic adenosine diphosphate ribose (cADPR), which is synthesized from NAD by the ADP ribosyl cyclase, peaked early in the morning and affected the oscillator. A decrease in the concentration of cADPR lengthened the period of clock-controlled genes, whereas nicotinamide inhibited the ADP ribosyl cyclase and weakened circadian calcium oscillations.64
Previously it was demonstrated that ADPR cyclase activity was induced by ABA and that 30% of all ABA responsive genes were expressed in a similar pattern that those from cADPR.75 Recalling that circadian clock microarray datasets overlapped with ABA transcriptomic profiles,63 a link between the circadian clock and metabolism in plants through cADPR has been suggested.64
What could emerge as a common factor between the clock, ABA and energy is the status of carbon availability throughout the day. Previously several screens for altered sugar responses led to the description of glucose insensitive (gin), sugar insensitive (sis) and sucrose uncoupled (sun) mutants. Many of these mutants also displayed ABA response mutant phenotypes, and some were allelic to ABA INSENSITIVE 4 (ABI4) and ABA DEFICIENT 2 (ABA2).76 Thus forward-genetic screens have implicated a direct link from carbon availability to ABA signaling.
As the circadian clock has a tight crosstalk with ABA gene expression profiles, the influence of carbohydrates on the expression of clock responsive genes is of particular interest. Blasing et al. reported that half of the circadian-controlled genes could respond to sugar. Similarly cellular sugar levels showed a major contribution in the establishment of diurnal gene expression patterns.77 Interestingly these authors described that sugar-controlled gene expression was sensitive and responsive to low sugar levels, but not to high sugar. Through the study of the phosphoglucomutase (pgm) mutant, which impaired starch synthesis, they observed that when endogenous sugars levels diminished, sugar responsive genes were rapidly induced. In contrast, when high sugar levels were present during the light period, gene expression did not change. Therefore in the wild type, transcriptional reprogramming to declining levels of sugars occurs at the end of the night.77 It seems that the circadian clock and diurnal changes in carbon availability through photosynthesis are tightly linked and are responsible for many of the cyclic patterns of gene expression under natural day lengths.
Recently the results obtained by Graf et al. indicated that plant fitness and performance are obtained through the adequate consumption of carbon resources. Starch is broken during the night phase and its consumption rate lasts until the end of the dark period.68 Then, can carbon sources can be replenished by photosynthesis at dawn. These authors demonstrated that a short-period mutant consumed its starch before the end of the night and consequently triggered a starvation response, which lead to diminished growth. Therefore Graf et al. concluded that the anticipation of the light and dark cycles provides an advantage by adjusting carbon supplies and not through affecting the photosynthesis rate, as suggested by Dodd et al.3 It remains to be seen if this principle also applies to clock mutants that cause a long period and/or starch accumulation.
Light-Induced Oxidative Stress
Photosynthesis is the primary and most important metabolic process in plants. Prime metabolites in the photosynthetic process are NADPH and ATP. Though photosynthesis is essential, the process of light absorption creates oxidative stress due to the formation ROS species, such as singlet oxygen (1O2), superoxide (O2•−) and hydrogen peroxide (H2O2).79 Furthermore, under high light, the electron flow through the photosynthethic chain overcomes the passage of electrons from ferredoxin to several reductases, and this causes an over-reduction of the plastoquinone and cytochrome b complex.79 Thus, during a day with high irradiance, plants are under constant oxidative stress.
As ROS generation is concomitant to photosynthesis and respiration, it was surprising that ROS responsive genes were not found to be under clock control.51 This could be explained by three characteristics of ROS responsive genes. Firstly, the transcription of these genes is induced upon stress. Thus, if a ROS responsive gene is expressed at a low constitutive level through the day or at a particular time, its expression will increase significantly only after being subjected to stress. Secondly, the activity of many ROS quenching enzymes depends on the redox state,79,80 which bypasses the need for regulated transcription. Finally, the rate of transcription of genes involved in ROS quenching is environment dependent. It has been observed that the longer the photoperiod or the higher the light intensity, ROS antioxidant genes are higher expressed and the ascorbate pool increases (reviewed in ref. 81 and 82, respectively). In summary, plants have a complex response system to cope with ROS generation and the role of the circadian clock in ROS transcriptional control is still unclear.
Stress and Energy as a Metabolic Input to the Clock
A link between energy metabolism, ROS production and environmental responses, could be provided by the changes in NAD and poly-ADPribosylation in response to stress. NAD synthesis has been shown to be rhythmic in mammals.83,84 As photosynthesis is under circadian control, pyridine nucleotide levels could also oscillate through the day in plants. It has been reported that the degree of poly ADP ribose (PAR) synthesis by the PAR polymerases (PARP) is increased in proportion to stress severity.85 PARP reaction consumes NAD and ATP, therefore affecting energy homeostasis.86 These authors observed that the downregulation of PARP enhanced stress tolerance probably due to a reduced consumption of NAD and ATP. However Vanderauwera et al. suggested an alternative hypothesis in which the downregulation of PARP activity caused plants to display abiotic stress tolerance due to induction of ABA responsive and ABA signaling genes and not through the direct consumption of energy (NAD and ATP). PAR changes also occur in response to ROS and DNA damage.88 Thus it seems that a tight correlation in the appropriate use of energy is essential to acquire resistance against ROS, regardless of their origin.
Polyribosylation of proteins by PARPs is overcome by the antagonistic reaction of the PAR glycohydrolases (PARG). Interestingly one A. thaliana clock mutant, tej (from the Sanskrit word for “bright”) was identified as a PARG.89 tej displayed higher luciferase activity that traduced into brighter luminescence (thus the gene name). Also the periodicty of circadian rhythms in tej was lengthened independent of light quality and quantity. Furthermore, the mutant was found to display alterations in the transcript accumulation of clock-regulated genes, and the mutant flowered earlier than the wild type, independent of the photoperiod.89 Therefore it could be plausible that PARP/PARG activity modifies ABA signaling through changes in NAD and cADPR levels. Consequently the changes in cADPR and ABA would alter the pace of the clock.64 Conceptually, any of these molecules could provide signals of stress to the oscillator. Taken together, these results suggest that a link exists between energy homeostasis and the plant circadian clock, though this interaction has not been completely defined. Intriguingly, the signaling interactions between the metabolic and stress signaling networks centrally require the sucrose non-fermenting-1 (SNF1)-related protein kinases.90 Whether these kinases participate in integrating abiotic stress signals to the oscillator is not known, but deserves investigation.
The Circadian Clock Beyond Transcriptional Control: Clock Mutants with Abiotic Stress Phenotypes
Recently the effects on metabolism and plant performance as a consequence of disrupted rhythms in circadian clock mutants have been reported in reference 65 and 91. Mutation in toc1, displayed altered plant responses to drought by controlling stomata aperture (see above) and thus gas exchange. TOC1 effect on stomata aperture requires a functional ABAR as the former binds the promoter of ABAR/GUN5.65 Related to TOC1 effects on ABA responses, Kant et al. used a functional genomic pipeline to search for genes involved in multiple abiotic stresses. These authors found that the mutation in cca1 and lhy, the other two components of the central loop, displayed hypersensitivity to salt, osmotic and heat stress. Thus the core of the plant clock is involved in modulating abiotic responses.
In a comprehensive work, Fukushima et al.91 demonstrated that the triple mutant prr9/prr7/prr5 had several metabolic defects that were different to those produced as a consequence of CCA1 overexpression. The authors found through a metabolic profile of the triple prr9/prr7/prr5 mutant was altered. This was particularly rue in examinations of primary metabolism, in particular the tricarboxilic acid cycle (TCA). This mutant-combination also displayed defects in biosynthethic pathways involved in chlorophyll, carotenoid, tocopherol and ABA. The later was found to result in an increased content of ABA. Furthermore this triple mutant displayed drought resistance, higher freezing tolerance, as well as upregulation of cold responsive genes.93 The extensive analysis of the prr9/prr7/prr5 triple mutant provided evidence, for the first time in plants, of a molecular link between metabolism and the circadian clock. In particular, this study also showed that respiration is imbalanced by disruption of the circadian clock.91 It is tempting to speculate that the circadian clock is tightly linked to primary metabolism associated with mitochondria respiration and photosynthesis.
A prime example of an interconnection between transcriptomic and metabolic pathways is seen in the mutant gigantea. The GI locus, which encodes for a protein with uncharacterized domains, is involved in rosette development94 and flowering time,95 starch metabolism,96,97 correct circadian clock function,95,98 and resistance to oxidative stress.99 While the role of GI in clock control and timing of flower development are starting to emerge, its role in starch accumulation and oxidative stress is still obscure. GI protein is not thought to be an enzyme directly involved in starch metabolism, but the gi mutant was found to display a starch-excess phenotype.96 Not only does gi accumulate starch, it also has higher proportions of simple carbohydrates,97 suggesting that a higher rate of carbon fixation must take place to sustain such a carbohydrate metabolic imbalance. The oxidative-stress resistant phenotype observed by Kurepa et al.99 was partly explained by a constitutive higher expression of ascorbate peroxidase (APX1) and Cd/Zn and Fe superoxide dismutases (CSD2 and FeSOD, respectively).100 Furthermore, when oxidative stress was created by methylviologen application, gi showed lower increase in H2O2 and superoxide production compared to wild type,99,101 as well as reduced lipid peroxidation.100 However in both cases, the mechanism that leads to starch accumulation and oxidative stress resistance in gi is still unknown. Given the pleiotropy of gi phenotypes, one could speculate that either these are a consequence of the disruption of the clock in gi, or perhaps more likely that GI is involved in several protein complexes in different cellular compartments with different and distinct biochemical/enzymatic functions.
It is still unclear if the mutations in the clock genes cause the transcriptomic and metabolic reprogramming so far observed through circadian processes. It is alternatively plausible that clock components have functions beyond their role in generating the oscillator. For example, it is a possibility that circadian-clock components are directly linked to primary metabolic functions. A continuous crosstalk through signaling between the clock and the organelles, and more generally, cellular metabolism could be responsible of providing a selective advantage upon a changing environment.
Perspectives
In the last decade, the plant circadian clock has essentially been elucidated at a molecular-genetic level of understanding. Research efforts established a coherent model of transcription and translational control.7,8,102 However the biochemical and mechanistic functions of the clock components beyond its clock control are broadly unknown. As the biochemistry of clock proteins is characterized, we may gain insight into their more specific mechanistic function. Simultaneously, these analyses, together with transcriptomic, proteomic and metabolic studies could uncover how clock components are linked to cellular metabolism. After all, the circadian clock evolved to enhance fitness in response to a perpetually and predictably changing environment. Thus “solving” the clock could be a key to understand how environmental signals are integrated by an organism in its response toward its environment.
Taking into consideration the abiotic-stress phenotypes displayed by clock mutants, a scenario results in which the clock and abiotic stress responses are tightly linked. However, the relationship between the clock components and their phenotypes is not straightforward. We hypothesize that the various described abiotic/metabolic phenotypes seen in circadian mutants cannot solely be explained by the given alterations of transcript levels of other clock genes. Gene overexpression or null mutant mutations do not correlate with a particular phenotypes. As examples: we note that the increase in TOC1 in the cca1/lhy does not explain drought/osmotic stress phenotypes,65,92 nor the increase in CCA1 in the prr9/ppr7/prr5 triple mutant explain alterations in TCA metabolites.91 Thus, the alteration of transcript levels of clock components are not the sole cause of a metabolic imbalance nor are the only cause of their abiotic stress phenotype. Thus, it seems that the general disruption of the circadian clock affects plant homeostasis and development. This could take place by desynchronizing the transcriptional control of the clock from its 24-hour environment.
The biological significance of the circadian clock is to enhance fitness over intervals that range from the daily to the life-span.8 Recent reports by numerous groups have firmly implicated the circadian clock as an integral component of plant homeostasis. As the chronobiological community expands its knowledge on the biochemistry and domain structure of clock components, it should emerge how the specificities of clock-protein action mediate in the convergence of circadian signaling to that of ABA, osmotic stress and primary metabolism signaling-responses (Fig. 3). Similarly as research fields take a more integrative approach towards studying the interaction between the clock, stress/ABA responses/cADPR and primary metabolism, we should gain a clearer picture of how homeostasis, growth and fitness are a product of correct timing to the prevailing ambient environment.
Figure 3.
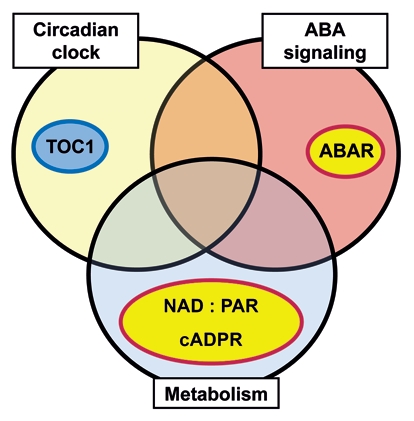
Interconnections between stress, metabolism and the circadian clock. An illustration that highlights the strong over lap in transcriptomic and molecular genetic data sets from the clock, from ABA, and from cADPR revealed extensive inter-connections between the systems that generate the circadian oscillator, abiotic stress signaling and control of basal metabolism.
References
- 1.Dunlap JC, Loros JJ, DeCoursey PJ. Chronobiology: Biological Timekeeping. Sunderland: Sinauer Associates Inc., Publishers; 2009. [Google Scholar]
- 2.Ouyang Y, Andersson CR, Kondo T, Golden SS, Johnson CH. Resonating circadian clocks enhance fitness in cyanobacteria. Proc Natl Acad Sci USA. 1998;95:8660–8664. doi: 10.1073/pnas.95.15.8660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dodd AN, Salathia N, Hall A, Kevei E, Toth R, Nagy F, et al. Plant circadian clocks increase photosynthesis, growth, survival and competitive advantage. Science. 2005;309:630–633. doi: 10.1126/science.1115581. [DOI] [PubMed] [Google Scholar]
- 4.Bell-Pedersen D, Cassone VM, Earnest DJ, Golden SS, Hardin PE, Thomas TL, et al. Circadian rhythms from multiple oscillators: lessons from diverse organisms. Nat Rev Genet. 2005;6:544–556. doi: 10.1038/nrg1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gallego M, Virshup DM. Post-translational modifications regulate the ticking of the circadian clock. Nat Rev Mol Cell Biol. 2007;8:139–148. doi: 10.1038/nrm2106. [DOI] [PubMed] [Google Scholar]
- 6.Salome PA, McClung CR. The Arabidopsis thaliana clock. J Biol Rhythms. 2004;19:425–435. doi: 10.1177/0748730404268112. [DOI] [PubMed] [Google Scholar]
- 7.Boikoglou E, Davis SJ. Signaling in the Circadian Clock. In: Mancuso FBaS., editor. Signaling in Plants. Berlin: Springer Publishers; 2009. pp. 261–285. [Google Scholar]
- 8.Harmer SL. The circadian system in higher plants. Annu Rev Plant Biol. 2009;60:357–377. doi: 10.1146/annurev.arplant.043008.092054. [DOI] [PubMed] [Google Scholar]
- 9.Pruneda-Paz JL, Kay SA. An expanding universe of circadian networks in higher plants. Trends Plant Sci. 2010;15:259–265. doi: 10.1016/j.tplants.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schaffer R, Ramsay N, Samach A, Corden S, Putterill J, Carre IA, et al. The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell. 1998;93:1219–1229. doi: 10.1016/s0092-8674(00)81465-8. [DOI] [PubMed] [Google Scholar]
- 11.Wang ZY, Tobin EM. Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell. 1998;93:1207–1217. doi: 10.1016/s0092-8674(00)81464-6. [DOI] [PubMed] [Google Scholar]
- 12.Alabadi D, Oyama T, Yanovsky MJ, Harmon FG, Mas P, Kay SA. Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science. 2001;293:880–883. doi: 10.1126/science.1061320. [DOI] [PubMed] [Google Scholar]
- 13.Mizoguchi T, Wheatley K, Hanzawa Y, Wright L, Mizoguchi M, Song HR, et al. LHY and CCA1 are partially redundant genes required to maintain circadian rhythms in Arabidopsis. Dev Cell. 2002;2:629–641. doi: 10.1016/s1534-5807(02)00170-3. [DOI] [PubMed] [Google Scholar]
- 14.Kolmos E, Schoof H, Plumer M, Davis SJ. Structural insights into the function of the core-circadian factor TIMING OF CAB2 EXPRESSION 1 (TOC1) J Circadian Rhythms. 2008;6:3. doi: 10.1186/1740-3391-6-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pruneda-Paz JL, Breton G, Para A, Kay SA. A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock. Science. 2009;323:1481–1485. doi: 10.1126/science.1167206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ding Z, Doyle MR, Amasino R, Davis SJ. A complex genetic interaction between Arabidopsis thaliana TOC1 and CCA1/LHY in driving the circadian clock and in output regulation. Genetics. 2007;176:1501–1510. doi: 10.1534/genetics.107.072769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kolmos E, Davis SJ. ELF4 as a central gene in the circadian clock. Plant Signal Behav. 2007;2:370–372. doi: 10.4161/psb.2.5.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kolmos E, Nowak M, Werner M, Fischer K, Schwarz G, Mathews S, et al. Integrating ELF4 into the circadian system through combined structural and functional studies. HFSP J. 2009;3:350–366. doi: 10.2976/1.3218766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Locke JC, Millar AJ, Turner MS. Modelling genetic networks with noisy and varied experimental data: the circadian clock in Arabidopsis thaliana. J Theor Biol. 2005;234:383–393. doi: 10.1016/j.jtbi.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 20.Locke JCW, Kozma-Bognar L, Gould PD, Feher B, Kevei E, Nagy F, et al. Experimental validation of a predicted feedback loop in the multi-oscillator clock of Arabidopsis thaliana. Mol Syst Biol. 2006;2:59. doi: 10.1038/msb4100102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zeilinger MN, Farre EM, Taylor SR, Kay SA, Doyle FJ., 3rd A novel computational model of the circadian clock in Arabidopsis that incorporates PRR7 and PRR9. Mol Syst Biol. 2006;2:58. doi: 10.1038/msb4100101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shin J, Davis SJ. Recent advances in computational modeling as a conduit to understand the plant circadian clock. F1000 Biol Rep. 2010;2:49. doi: 10.3410/B2-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakamichi N, Kiba T, Henriques R, Mizuno T, Chua NH, Sakakibara H. PSEUDO-RESPONSE REGULATORS 9, 7 and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell. 2010;22:594–605. doi: 10.1105/tpc.109.072892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adeyemo OS, Kolmos E, Tohme J, Chavariaga P, Fregene M, Davis SJ. Identification and characterization of the cassava core-clock gene EARLY FLOWERING 4. Tropical Plant Biol. (In press) [Google Scholar]
- 25.McWatters HG, Kolmos E, Hall A, Doyle MR, Amasino RM, Gyula P, et al. ELF4 is required for oscillatory properties of the circadian clock. Plant Physiol. 2007;144:391–401. doi: 10.1104/pp.107.096206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kevei E, Gyula P, Hall A, Kozma-Bognar L, Kim WY, Eriksson ME, et al. Forward genetic analysis of the circadian clock separates the multiple functions of ZEITLUPE. Plant Physiol. 2006;140:933–945. doi: 10.1104/pp.105.074864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mas P, Kim WY, Somers DE, Kay SA. Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana. Nature. 2003;426:567–570. doi: 10.1038/nature02163. [DOI] [PubMed] [Google Scholar]
- 28.Kim WY, Fujiwara S, Suh SS, Kim J, Kim Y, Han L, et al. ZEITLUPE is a circadian photoreceptor stabilized by GIGANTEA in blue light. Nature. 2007;449:356–360. doi: 10.1038/nature06132. [DOI] [PubMed] [Google Scholar]
- 29.Para A, Farre EM, Imaizumi T, Pruneda-Paz JL, Harmon FG, Kay SA. PRR3 Is a vascular regulator of TOC1 stability in the Arabidopsis circadian clock. Plant Cell. 2007;19:3462–3473. doi: 10.1105/tpc.107.054775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Song HR, Carre IA. DET1 regulates the proteasomal degradation of LHY, a component of the Arabidopsis circadian clock. Plant Mol Biol. 2005;57:761–771. doi: 10.1007/s11103-005-3096-z. [DOI] [PubMed] [Google Scholar]
- 31.Wang L, Fujiwara S, Somers DE. PRR5 regulates phosphorylation, nuclear import and subnuclear localization of TOC1 in the Arabidopsis circadian clock. EMBO J. 2010;29:1903–1915. doi: 10.1038/emboj.2010.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kami C, Lorrain S, Hornitschek P, Fankhauser C. Light-regulated plant growth and development. Curr Top Dev Biol. 2010;91:29–66. doi: 10.1016/S0070-2153(10)91002-8. [DOI] [PubMed] [Google Scholar]
- 33.Toth R, Kevei E, Hall A, Millar AJ, Nagy F, Kozma-Bognar L. Circadian clock-regulated expression of phytochrome and cryptochrome genes in Arabidopsis. Plant Physiol. 2001;127:1607–1616. doi: 10.1104/pp.010467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Somers DE, Devlin PF, Kay SA. Phytochromes and cryptochromes in the entrainment of the Arabidopsis circadian clock. Science. 1998;282:1488–1490. doi: 10.1126/science.282.5393.1488. [DOI] [PubMed] [Google Scholar]
- 35.Anderson SL, Somers DE, Millar AJ, Hanson K, Chory J, Kay SA. Attenuation of phytochrome A and B signaling pathways by the Arabidopsis circadian clock. Plant Cell. 1997;9:1727–1743. doi: 10.1105/tpc.9.10.1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Devlin PF, Kay SA. Cryptochromes are required for phytochrome signaling to the circadian clock but not for rhythmicity. Plant Cell. 2000;12:2499–2510. doi: 10.1105/tpc.12.12.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mas P, Devlin PF, Panda S, Kay SA. Functional interaction of phytochrome B and cryptochrome 2. Nature. 2000;408:207–211. doi: 10.1038/35041583. [DOI] [PubMed] [Google Scholar]
- 38.Palagyi A, Terecskei K, Adam E, Kevei E, Kircher S, Merai Z, et al. Functional analysis of amino-terminal domains of the photoreceptor phytochrome B. Plant Physiol. 2010;153:1834–1845. doi: 10.1104/pp.110.153031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yanovsky MJ, Mazzella MA, Casal JJ. A quadruple photoreceptor mutant still keeps track of time. Curr Biol. 2000;10:1013–1015. doi: 10.1016/s0960-9822(00)00651-5. [DOI] [PubMed] [Google Scholar]
- 40.Strasser B, Sanchez-Lamas M, Yanovsky MJ, Casal JJ, Cerdan PD. Arabidopsis thaliana life without phytochromes. Proc Natl Acad Sci USA. 2010;107:4776–4781. doi: 10.1073/pnas.0910446107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Somers DE, Schultz TF, Milnamow M, Kay SA. ZEITLUPE encodes a novel clock-associated PAS protein from Arabidopsis. Cell. 2000;101:319–329. doi: 10.1016/s0092-8674(00)80841-7. [DOI] [PubMed] [Google Scholar]
- 42.Baudry A, Ito S, Song YH, Strait AA, Kiba T, Lu S, et al. F-box proteins FKF1 and LKP2 act in concert with ZEITLUPE to control Arabidopsis clock progression. Plant Cell. 2010;22:606–622. doi: 10.1105/tpc.109.072843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hicks KA, Millar AJ, Carre IA, Somers DE, Straume M, Meeks-Wagner DR, et al. Conditional circadian dysfunction of the Arabidopsis early-flowering 3 mutant. Science. 1996;274:790–792. doi: 10.1126/science.274.5288.790. [DOI] [PubMed] [Google Scholar]
- 44.Hall A, Bastow RM, Davis SJ, Hanano S, McWatters HG, Hibberd V, et al. The TIME FOR COFFEE gene maintains the amplitude and timing of Arabidopsis circadian clocks. Plant Cell. 2003;15:2719–2729. doi: 10.1105/tpc.013730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ding Z, Millar AJ, Davis AM, Davis SJ. TIME FOR COFFEE encodes a nuclear regulator in the Arabidopsis thaliana circadian clock. Plant Cell. 2007;19:1522–1536. doi: 10.1105/tpc.106.047241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Staiger D, Allenbach L, Salathia N, Fiechter V, Davis SJ, Millar AJ, et al. The Arabidopsis SRR1 gene mediates phyB signaling and is required for normal circadian clock function. Genes Dev. 2003;17:256–268. doi: 10.1101/gad.244103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kevei E, Gyula P, Feher B, Toth R, Viczian A, Kircher S, et al. Regulation of the Arabidopsis thaliana circadian clock by the small GTPase LIP1. Curr Biol. 2007;17:1456–1464. doi: 10.1016/j.cub.2007.07.018. [DOI] [PubMed] [Google Scholar]
- 48.Kolmos E, Davis SJ. Rho-related signals in time-specific light perception. Curr Biol. 2007;17:808–810. doi: 10.1016/j.cub.2007.07.031. [DOI] [PubMed] [Google Scholar]
- 49.Martin-Tryon EL, Harmer SL. XAP5 CIRCADIAN TIMEKEEPER coordinates light signals for proper timing of photomorphogenesis and the circadian clock in Arabidopsis. Plant Cell. 2008;20:1244–1259. doi: 10.1105/tpc.107.056655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Harmer SL, Hogenesch JB, Straume M, Chang HS, Han B, Zhu T, et al. Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science. 2000;290:2110–2113. doi: 10.1126/science.290.5499.2110. [DOI] [PubMed] [Google Scholar]
- 51.Covington MF, Maloof JN, Straume M, Kay SA, Harmer SL. Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Biology. 2008;9:130. doi: 10.1186/gb-2008-9-8-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hubbard KE, Robertson FC, Dalchau N, Webb AA. Systems analyses of circadian networks. Mol Biosyst. 2009;5:1502–1511. doi: 10.1039/B907714f. [DOI] [PubMed] [Google Scholar]
- 53.Hanano S, Stracke R, Jakoby M, Merkle T, Domagalska MA, Weisshaar B, et al. A systematic survey in Arabidopsis thaliana of transcription factors that modulate circadian parameters. BMC Genomics. 2008;9:182. doi: 10.1186/1471-2164-9-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hanano S, Domagalska MA, Nagy F, Davis SJ. Multiple phytohormones influence distinct parameters of the plant circadian clock. Genes Cells. 2006;11:1381–1392. doi: 10.1111/j.1365-2443.2006.01026.x. [DOI] [PubMed] [Google Scholar]
- 55.Fowler SG, Cook D, Thomashow MF. Low temperature induction of Arabidopsis CBF1, 2 and 3 is gated by the circadian clock. Plant Physiol. 2005;137:961–968. doi: 10.1104/pp.104.058354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Michael TP, Breton G, Hazen SP, Priest H, Mockler TC, Kay SA, et al. A morning-specific phytohormone gene expression program underlying rhythmic plant growth. PLoS Biol. 2008;6:225. doi: 10.1371/journal.pbio.0060225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Michael TP, Mockler TC, Breton G, McEntee C, Byer A, Trout JD, et al. Network discovery pipeline elucidates conserved time-of-day-specific cis-regulatory modules. PLoS Genet. 2008;4:14. doi: 10.1371/journal.pgen.0040014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Robertson FC, Skeffington AW, Gardner MJ, Webb AA. Interactions between circadian and hormonal signalling in plants. Plant Mol Biol. 2009;69:419–427. doi: 10.1007/s11103-008-9407-4. [DOI] [PubMed] [Google Scholar]
- 59.Covington MF, Harmer SL. The circadian clock regulates auxin signaling and responses in Arabidopsis. PLoS Biol. 2007;5:222. doi: 10.1371/journal.pbio.0050222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rawat R, Schwartz J, Jones MA, Sairanen I, Cheng Y, Andersson CR, et al. REVEILLE1, a Myb-like transcription factor, integrates the circadian clock and auxin pathways. Proc Natl Acad Sci USA. 2009;106:16883–16888. doi: 10.1073/pnas.0813035106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nozue K, Covington MF, Duek PD, Lorrain S, Fankhauser C, Harmer SL, et al. Rhythmic growth explained by coincidence between internal and external cues. Nature. 2007;448:358–361. doi: 10.1038/nature05946. [DOI] [PubMed] [Google Scholar]
- 62.Matsui A, Ishida J, Morosawa T, Mochizuki Y, Kaminuma E, Endo TA, et al. Arabidopsis transcriptome analysis under drought, cold, high-salinity and ABA treatment conditions using a tiling array. Plant Cell Physiol. 2008;49:1135–1149. doi: 10.1093/pcp/pcn101. [DOI] [PubMed] [Google Scholar]
- 63.Mizuno T, Yamashino T. Comparative transcriptome of diurnally oscillating genes and hormone-responsive genes in Arabidopsis thaliana: insight into circadian clock-controlled daily responses to common ambient stresses in plants. Plant Cell Physiol. 2008;49:481–487. doi: 10.1093/pcp/pcn008. [DOI] [PubMed] [Google Scholar]
- 64.Dodd AN, Gardner MJ, Hotta CT, Hubbard KE, Dalchau N, Love J, et al. The Arabidopsis circadian clock incorporates a cADPR-based feedback loop. Science. 2007;318:1789–1792. doi: 10.1126/science.1146757. [DOI] [PubMed] [Google Scholar]
- 65.Legnaioli T, Cuevas J, Mas P. TOC1 functions as a molecular switch connecting the circadian clock with plant responses to drought. EMBO J. 2009;28:3745–3757. doi: 10.1038/emboj.2009.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Young AJ, Frank HA. Energy transfer reactions involving carotenoids: quenching of chlorophyll fluorescence. J Photochem Photobiol B. 1996;36:3–15. doi: 10.1016/S1011-1344(96)07397-6. [DOI] [PubMed] [Google Scholar]
- 67.Zeller G, Henz SR, Widmer CK, Sachsenberg T, Ratsch G, Weigel D, et al. Stress-induced changes in the Arabidopsis thaliana transcriptome analyzed using whole-genome tiling arrays. Plant J. 2009;58:1068–1082. doi: 10.1111/j.1365-313X.2009.03835.x. [DOI] [PubMed] [Google Scholar]
- 68.Smith AM, Stitt M. Coordination of carbon supply and plant growth. Plant Cell Environ. 2007;30:1126–1149. doi: 10.1111/j.1365-3040.2007.01708.x. [DOI] [PubMed] [Google Scholar]
- 69.Smith SM, Fulton DC, Chia T, Thorneycroft D, Chapple A, Dunstan H, et al. Diurnal changes in the transcriptome encoding enzymes of starch metabolism provide evidence for both transcriptional and post-transcriptional regulation of starch metabolism in Arabidopsis leaves. Plant Physiol. 2004;136:2687–2699. doi: 10.1104/pp.104.044347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.McClung CR, Davis SJ. Ambient Thermometers in Plants: From Physiological Outputs towards Mechanisms of Thermal Sensing. Curr Biol. 2010;20:1086–1092. doi: 10.1016/j.cub.2010.10.035. [DOI] [PubMed] [Google Scholar]
- 71.Fowler S, Thomashow MF. Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell. 2002;14:1675–1690. doi: 10.1105/tpc.003483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki Y, et al. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell. 1998;10:1391–1406. doi: 10.1105/tpc.10.8.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Urano K, Kurihara Y, Seki M, Shinozaki K. ‘Omics’ analyses of regulatory networks in plant abiotic stress responses. Curr Opin Plant Biol. 2010;13:132–138. doi: 10.1016/j.pbi.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 74.Franklin KA, Whitelam GC. Light-quality regulation of freezing tolerance in Arabidopsis thaliana. Nat Genet. 2007;39:1410–1413. doi: 10.1038/ng.2007.3. [DOI] [PubMed] [Google Scholar]
- 75.Sanchez JP, Duque P, Chua NH. ABA activates ADPR cyclase and cADPR induces a subset of ABA-responsive genes in Arabidopsis. Plant J. 2004;38:381–395. doi: 10.1111/j.1365-313X.2004.02055.x. [DOI] [PubMed] [Google Scholar]
- 76.Rook F, Hadingham SA, Li Y, Bevan MW. Sugar and ABA response pathways and the control of gene expression. Plant Cell Environ. 2006;29:426–434. doi: 10.1111/j.1365-3040.2005.01477.x. [DOI] [PubMed] [Google Scholar]
- 77.Blasing OE, Gibon Y, Gunther M, Hohne M, Morcuende R, Osuna D, et al. Sugars and circadian regulation make major contributions to the global regulation of diurnal gene expression in Arabidopsis. Plant Cell. 2005;17:3257–3281. doi: 10.1105/tpc.105.035261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Graf A, Schlereth A, Stitt M, Smith AM. Circadian control of carbohydrate availability for growth in Arabidopsis plants at night. Proc Natl Acad Sci USA. 2010;107:9458–9463. doi: 10.1073/pnas.0914299107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Oelze ML, Kandlbinder A, Dietz KJ. Redox regulation and overreduction control in the photosynthesizing cell: complexity in redox regulatory networks. Biochim Biophys Acta. 2008;1780:1261–1272. doi: 10.1016/j.bbagen.2008.03.015. [DOI] [PubMed] [Google Scholar]
- 80.Wormuth D, Heiber I, Shaikali J, Kandlbinder A, Baier M, Dietz KJ. Redox regulation and antioxidative defence in Arabidopsis leaves viewed from a systems biology perspective. J Biotechnol. 2007;129:229–248. doi: 10.1016/j.jbiotec.2006.12.006. [DOI] [PubMed] [Google Scholar]
- 81.Bartoli CG, Yu J, Gomez F, Fernandez L, McIntosh L, Foyer CH. Inter-relationships between light and respiration in the control of ascorbic acid synthesis and accumulation in Arabidopsis thaliana leaves. J Exp Bot. 2006;57:1621–1631. doi: 10.1093/jxb/erl005. [DOI] [PubMed] [Google Scholar]
- 82.Becker B, Holtgrefe S, Jung S, Wunrau C, Kandlbinder A, Baier M, et al. Influence of the photoperiod on redox regulation and stress responses in Arabidopsis thaliana L. (Heynh.) plants under long- and short-day conditions. Planta. 2006;224:380–393. doi: 10.1007/s00425-006-0222-3. [DOI] [PubMed] [Google Scholar]
- 83.Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, Chen D, et al. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008;134:329–340. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ramsey KM, Yoshino J, Brace CS, Abrassart D, Kobayashi Y, Marcheva B, et al. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science. 2009;324:651–654. doi: 10.1126/science.1171641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hashida SN, Takahashi H, Uchimiya H. The role of NAD biosynthesis in plant development and stress responses. Ann Bot. 2009;103:819–824. doi: 10.1093/aob/mcp019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.De Block M, Verduyn C, De Brouwer D, Cornelissen M. Poly(ADP-ribose) polymerase in plants affects energy homeostasis, cell death and stress tolerance. Plant J. 2005;41:95–106. doi: 10.1111/j.1365-313X.2004.02277.x. [DOI] [PubMed] [Google Scholar]
- 87.Vanderauwera S, De Block M, Van de Steene N, van de Cotte B, Metzlaff M, Van Breusegem F. Silencing of poly(ADP-ribose) polymerase in plants alters abiotic stress signal transduction. Proc Natl Acad Sci USA. 2007;104:15150–15155. doi: 10.1073/pnas.0706668104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Qin XJ, Hudson LG, Liu W, Timmins GS, Liu KJ. Low concentration of arsenite exacerbates UVR-induced DNA strand breaks by inhibiting PARP-1 activity. Toxicol Appl Pharmacol. 2008;232:41–50. doi: 10.1016/j.taap.2008.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Panda S, Poirier GG, Kay SA. tej defines a role for poly(ADP-ribosyl)ation in establishing period length of the arabidopsis circadian oscillator. Dev Cell. 2002;3:51–61. doi: 10.1016/s1534-5807(02)00200-9. [DOI] [PubMed] [Google Scholar]
- 90.Baena-Gonzalez E, Sheen J. Convergent energy and stress signaling. Trends Plant Sci. 2008;13:474–482. doi: 10.1016/j.tplants.2008.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fukushima A, Kusano M, Nakamichi N, Kobayashi M, Hayashi N, Sakakibara H, et al. Impact of clock-associated Arabidopsis pseudo-response regulators in metabolic coordination. Proc Natl Acad Sci USA. 2009;106:7251–7256. doi: 10.1073/pnas.0900952106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kant P, Gordon M, Kant S, Zolla G, Davydov O, Heimer YM, et al. Functional-genomics-based identification of genes that regulate Arabidopsis responses to multiple abiotic stresses. Plant Cell Environ. 2008;31:697–714. doi: 10.1111/j.1365-3040.2008.01779.x. [DOI] [PubMed] [Google Scholar]
- 93.Nakamichi N, Kusano M, Fukushima A, Kita M, Ito S, Yamashino T, et al. Transcript profiling of an Arabidopsis PSEUDO RESPONSE REGULATOR arrhythmic triple mutant reveals a role for the circadian clock in cold stress response. Plant Cell Physiol. 2009;50:447–462. doi: 10.1093/pcp/pcp004. [DOI] [PubMed] [Google Scholar]
- 94.Redei GP. Supervital Mutants of Arabidopsis. Genetics. 1962;47:443–460. doi: 10.1093/genetics/47.4.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Fowler S, Lee K, Onouchi H, Samach A, Richardson K, Morris B, et al. GIGANTEA: a circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J. 1999;18:4679–4688. doi: 10.1093/emboj/18.17.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Eimert K, Wang SM, Lue WI, Chen J. Monogenic Recessive Mutations Causing Both Late Floral Initiation and Excess Starch Accumulation in Arabidopsis. Plant Cell. 1995;7:1703–1712. doi: 10.1105/tpc.7.10.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Messerli G, Partovi Nia V, Trevisan M, Kolbe A, Schauer N, Geigenberger P, et al. Rapid classification of phenotypic mutants of Arabidopsis via metabolite fingerprinting. Plant Physiol. 2007;143:1484–1492. doi: 10.1104/pp.106.090795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Park DH, Somers DE, Kim YS, Choy YH, Lim HK, Soh MS, et al. Control of circadian rhythms and photoperiodic flowering by the Arabidopsis GIGANTEA gene. Science. 1999;285:1579–1582. doi: 10.1126/science.285.5433.1579. [DOI] [PubMed] [Google Scholar]
- 99.Kurepa J, Smalle J, Van Montagu M, Inze D. Oxidative stress tolerance and longevity in Arabidopsis: the late-flowering mutant gigantea is tolerant to paraquat. Plant J. 1998;14:759–764. doi: 10.1046/j.1365-313x.1998.00168.x. [DOI] [PubMed] [Google Scholar]
- 100.Cao S, Jiang S, Zhang R. The Role of GIGANTEA Gene in Mediating the Oxidative Stress Response and in Arabidopsis. Plant Growth Regulation. 2006;48:261–270. [Google Scholar]
- 101.Kurepa J, Smalle J, Van Montagu M, Inze D. Polyamines and paraquat toxicity in Arabidopsis thaliana. Plant Cell Physiol. 1998;39:987–992. doi: 10.1093/oxfordjournals.pcp.a029463. [DOI] [PubMed] [Google Scholar]
- 102.Imaizumi T. Arabidopsis circadian clock and photoperiodism: time to think about location. Curr Opin Plant Biol. 2010;13:83–89. doi: 10.1016/j.pbi.2009.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]


