Abstract
Molecular chaperones and foldases are a diverse group of proteins that in vivo bind to misfolded or unfolded proteins (non-native or unstable proteins) and play important role in their proper folding. Stress conditions compel altered and heightened chaperone and foldase expression activity in the endoplasmic reticulum (ER), which highlights the role of these proteins, due to which several of the proteins under these classes were identified as heat shock proteins. Different chaperones and foldases are active in different cellular compartment performing specific tasks. The review will discuss the role of ER chaperones and foldases under stress conditions, to maintain proper protein folding dynamics in the plant cells and recent advances in the field. The ER chaperones and foldases, which are described in article, are binding protein (BiP), glucose regulated protein (GRP94), protein-disulfide isomerase (PDI), peptidyl-prolyl isomerases (PPI) or immunophilins, calnexin and calreticulin.
Key words: Abiotic stress, chaperones, endoplasmic reticulum, foldases, immunophilins, protein folding, signal transduction
Introduction
Protein folding often requires molecular protein folding machineries like chaperones and foldases.1,2 Foldases include two isomerases, namely, protein disulfide isomerase and peptidyl prolyl isomerase. The two foldases catalyze the formation of disulfide bonds or isomerization of peptide bonds proximal to Pro residues, respectively. Molecular chaperones are rather diverse group of proteins, sharing a common property to bind to unstable substrate proteins in non-native and intermediate structural states. Intriguingly, homologs of almost all the proteins involved in protein folding are conserved across eukaryotes, plant or animal kingdom.
The chaperones increase the overall efficiency of protein foldings by recognizing and stabilizing the partially folded intermediates during polypeptide folding, assembly and disassembly. On the other hand foldases influence the protein folding by affecting the rate limiting steps. There are two types of foldases, using different modes of action, namely Peptidyl-prolyl cis or trans isomerase (PPI) (immunophilins), and protein-disulfide isomerase (PDI). The cyclophilin and FK506 binding protein are also the ER foldases. The foldases facilitate the folding of every protein by catalyzing isomerization of prolyl peptide bonds or formation and isomerization of disulfide bonds for proper folding.
Folding of the nascent secretory proteins, tagged and delivered by ribosomes into ER lumen is assisted and corrected by a host of chaperones and foldases. The ER lumen is a specialized organelle compartment dedicated primarily to protein folding as proteins enter it in an unfolded conformation and leaves it fully folded.3 ER located protein folding mediators include BiP (binding protein) or GRP78 (glucose-regulated protein 78), calnexin, calreticulin, GRP94 (endoplasmin or glucose-regulated protein 94), and PDI (protein disulfide isomerase). These proteins were initially studied in yeast and mammalian systems and later homologs of these ER molecular chaperones have been also discovered in higher plants.4,5 BiP and GRP94 are ER isoforms of cytosolic proteins, whereas others are unique ER proteins. The conservation of the proteins across different species and kingdom indicates existence of broadly common protein folding pathway. Plant stress induces enhance expression of ER chaperones and foldases. For example enhanced expression of chaperones is observed in tunicamycin-induced stress in plant cells or tissues. The antibiotic is known to inhibit N-linked glycosylation, which hinders proper protein folding and slows down assembly of oligomers.6–8 A variety of other stress conditions both artificial as well as natural like addition of plant growth regulators, infection etc. are also known to enhance chaperone production.9 Recently, microarray expression analysis of a beta peptide (expressed in Alzheimer's disease) expressing transgenic rice endosperm cells affected ER response in the cells, accompanied with changes in expression of several several BiPs, PDIs and OsbZIP60 and an opaque and shrunken phenotype.10 Disruption or enhanced demand for protein folding causes ER stress- designated the Unfolded Protein Response (UPR), activates signaling cascades leading to restoration or enhancement in protein folding capacity.
Binding protein (BiP).
Binding protein (BiP), a HSP70 molecular chaperone, is an important and most well studied ER protein implicated in stress response of cells.11–13 Alvim, Carolino et al. found that under progressive drought, the leaf BiP levels correlated with the maintenance of the shoot turgidity and water content. The protective effect of BiP overexpression against water stress was disrupted by expression of an antisense BiP cDNA construct. Although overexpression of BiP prevented cellular dehydration, the stomatal conductance and transpiration rate in droughted sense leaves were higher than in control and antisense leaves. Their experiments for the first time demonstrated the role of BiP in multicellular organisms, namely transgenic tobacco Nicotiana tabacum L. cv Havana. BiP is also an essential component of the protein translocation machinery, as well as playing a role in retrograde transport across the ER membrane of aberrant proteins destined for degradation by the proteasome.12
It was discovered that the molecular chaperone BiP purified from bovine liver (bBiP) exhibits a low basal level of ATPase activity that can be enhanced 3-to 6-fold by synthetic peptides.14 This and several other experiments also confirmed that there is no consensus binding recognition sites or motif but the fact that BiP interacts more tightly with non-native protein domains.15 It has been established that BiP binds stabilizes partially folded intermediates, then releases and rebinds to sites that remain in non-native conformations.16 Amongst the higher plant ER chaperones that have been identified, BiP is most likely involved in the early stages of protein folding. BiP binds nascent polypeptides, and is vital in the ER translocation and corrects folding of newly synthesized proteins. The role of plant BiP was also established by experiments in which a chimeric gene containing the coding region of one of the tobacco BiP genes is able to complement a mutation in the Saccharomyces cerevisiae BiP gene 6. Although the complementation data do not directly infer the role of BiP in plants, they do show that amino acid residues important for BiP function in yeast have been evolutionarily conserved between yeast and higher plants and that the His-Asp-Glu-Leu peptide that may form the signal for retention in the endoplasmic reticulum. It is also known that BiP gene is encoded as multigene family in higher plants and other organisms. For illustration- yeast and spinach have only one BiP gene,17,18 where as maize, tobacco and soybean have more than one BiP gene. The BiP RNA level is enhanced in stress conditions in soybeans (Glycine max L. Merr.) like external stimuli or the agents that perturb protein folding.19
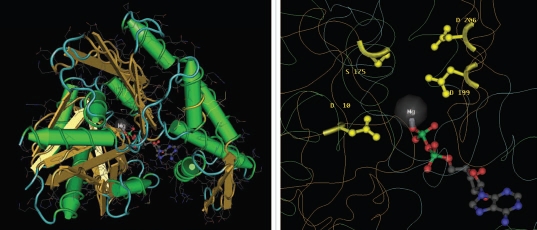
It has been demonstrated that plant and animal BiP proteins behave differently in concentrations of magnesium or calcium. Maize BiP ATPase activity is enhanced in low concentrations of calcium but inhibited by high concentrations of calcium or magnesium, whereas bovine BiP ATPase activity is independent of calcium or magnesium concentrations.20 The workers also proved that four acidic residues of the ATPase fragment which might participate in catalysis (Asp-10 and Asp-199, which are Mg2+ ion ligands; Glu-175 and Asp-206, which are candidates for a role as catalytic base) have been individually mutated to both the cognate amide residue (aspartate to asparagine, glutamate to glutamine) and to serine, and the effects of the mutations on the kinetics and structure have been determined.21 For details, see Figure 1, a Cn3D22 image of a Bovine HSP 70 structure, corresponding to the Protein Data Bank (PDB) entry 1NGA. BiP has been shown to be associated with nascent prolamine storage proteins in the ER during protein body formation in rice seeds.23
Figure 1.
Structural basis of the 70-kilodalton heat shock cognate protein ATP hydrolytic activity site (PDB: 1NGA, MMDB: 56948): The Cn3D image on the left shows overall tertiary crystal structure of the 44 KDa amino-terminal mutant fragment of Hsc70. The image on the right is zoomed image of the binding site: showing Asp-10 and Asp-199, which are Mg2+ ion ligands; Glu-175 (mutated to Ser in this mutant) and Asp-206, which are candidates for a role as catalytic base are marked in the figure.
BiP plays a significant role in folding and assembly of newly synthesized seed storage proteins. The role of BiP in the folding and assembly of trimeric phaseolin in developing bean cotyledons has been demonstrated.24 They demonstrated that even in the presence of a significant pool of phaseolin trimers in the ER, only monomeric phaseolin is detected in association with BiP. A similar stable association of BiP with unassembled protein subunits was observed in tobacco protoplasts expressing an assembly-defective bean phaseolin mutant.25 The authors found that association of BiP with normal, assembly-competent phaseolin was not observed in the tobacco protoplast system and provided evidence for the presence of a quality control mechanism in the ER of plant cells that avoids intracellular trafficking of severely defective proteins and hence promotes their degradation.
BiP mediates an increase in drought tolerance in soybean and delays drought-induced leaf senescence in soybean and tobacco.26 The authors reported that the leaves in their transgenic BiP overexpressing lines did not wilt and exhibited only a small decrease in water potential and delayed senescence. Further, they concluded that BiP overexpression confers resistance to drought, through an as yet unknown mechanism that is related to ER functioning. Recently, it has been shown that, BiP predominantly expressed in maturing rice endosperm, acts as not only a chaperone but also a stress sensor protein for ER quality control.27
GRP94.
GRP94 (Glucose Regulated Protein), also known as endoplasmin, is a HSP90 protein family member found in the ER lumen. The expression of the protein was found to be upregulated in the cases of powdery mildew fungal infection in barley.28 Mammalian homologs of GRP94 have implicated function in cancer progression, autoimmune disease, Alzheimer's disease and other stress conditions.29,30 Based on immunoglobulin (Ig) binding studies, it was found that BiP preferentially binds an early disulphide intermediate of light chains and dissociates within a few minutes, GRP94 exclusively binds fully oxidized molecules and dissociates with a half-time of 50 min, indicating that GRP94 acts after BiP.31
Foldases in the ER: PDI and PPI.
Protein disulfide isomerase (PDI) contains thioredoxin (TRX) domains and act as a catalyst of disulfide bond formation in the oxidizing environment of the ER,3,32 hence stabilizing the tertiary and quaternary structures of protein folding. Plant homologs of PDI when compared with those in vertebrates, have been found to have conserved active site (Ala-Pro-Trp-Cys-Gly-His-Cys-Lys) and ER retention signal (Lys-Asp-Glu-Leu).33–36 The altered expression profiles in stress conditions have now been reported in Arabidopsis.37 They have studied the AtPDI gene expression in different tissues, in response to chemically induced UPR, and in null mutants of UPR signaling mediators (AtIRE1-2 and AtbZIP60). Their experiments revealed that expression of six AtPDI genes was significantly upregulated by UPR and sharply attenuated by the transcription inhibitor, actinomycin D, indicating UPR induced AtPDI gene transcription. Similarly, PDI is induced in alfalfa and tobacco cells challenged with tunicamycin. In Soybean too, the PDI expression was found to be upregulated in ER stress conditions, expressed ubiquitously in tissues like cotyledon.38 Soybean PDI sequence showed significant conservation of the exon/intron structure with that of Arabidopsis thaliana and Oryza sativa. Their results reconfirmed that PDI may play a role in the folding of storage proteins and functions not only as a thiol-oxidoredactase, but also as molecular chaperone. Multiple sequence alignments and phylogenetic analysis of PDI sequences have revealed 10 classes of PDI in plants.39 The structure of several non plant PDI homologs has been solved. The yeast PDI40 (Figure 2) consists of two catalytically inactive thioredoxin domains inserted between two catalytically active thioredoxin domains and an acidic C-terminal. The structure reveals that the four thioredoxin domains are arranged in the shape of a twisted “U” with the active sites facing each other across the long sides of the “U.” The inside surface of the “U” is enriched in hydrophobic residues, thereby facilitating interactions with misfolded proteins.
Figure 2.

The crystal structure of yeast protein disulfide isomerase (PDB: 2B5E, MMDB: 37123). The Cn3D image features two catalytically inactive thioredoxin domains inserted between two catalytically active thioredoxin domains and an acidic C-terminal tail. The reveals that the four thioredoxin domains are arranged in the shape of a twisted “U” with the active sites facing each other across the long sides of the “U.” The inside surface of the “U” is enriched with hydrophobic residues, facilitating interactions with misfolded proteins.
Peptidyl-prolyl isomerases (PPI), PPIases or immunophilins are also been reported in plant ER for example in Vicia faba41 and Arabidopsis.42 Immunophilins are intracellular receptors for the immunosuppressants like cyclosporin A, FK506, and rapamycin. PPIases are involved in folding and trafficking of proteins, particularly those with signal transducing functions and susceptibility to immunosuppressant drugs. The immunophilins have been used to investigate signaling pathways in yeast, plant, and mammalian cells. Luan et al.41 have identified a 15 kDa FK506 and rapamycin-binding protein from V. faba (VfFKBP15). The amino acid sequence of the protein starts with a signal peptide of 22 hydrophobic amino acids. The core region of VfFKBP15 is highly similar to that of yeast and mammalian FKBP13, localized in the ER. VfFKBP15 has a carboxyl-terminal sequence ending with SSEL, a putative ER retention signal. The mRNA of VfFKBP15 is ubiquitously expressed in various plant tissues including leaves, stems, and roots, consistent with the ER localization of the protein. Levels of VfFKBP15 mRNA are elevated by heat shock, suggesting a possible role for this FKBP member under stress conditions. Wu et al.42 demonstrated that Mutations in TWD1, a arabidopsis immunophilin, caused mislocalization of ABC transporters like ABCB1, ABCB4, and ABCB19 to the ER instead of the plasma membrane as shown by confocal microscopy of fluorescently tagged fusion proteins and transmission electron microscopy of immunogold-labeled samples in the case of ABCB19. However, a very little work has been done on these proteins in plants.
Calnexin and calreticulin.
Calnexins and calreticulins (CRT) are two closely related calcium-binding molecular chaperones localized in ER.43 Calcium functions as a central node in the overall “signaling web” and play important role in stress tolerance in plant.43–47 In response to the stress the cytosolic calcium concentration was found to increased, which initiates the stress signal transduction pathways for stress tolerance.47 Calnexin along with calreticulin are quality control systems of protein folding machinery that promotes correct folding of proteins that enter the secretory pathway and targets misfolded proteins for degradation.48 Calreticulin and calnexin are present in all the green plants and possibly share a common origin. In green plants the calreticulin founder gene are known to duplicated in early tracheophytes while the calnexin founder gene was inherited from basal green algae during evolution in a very conservative copy number.49 The evolutionary history of calreticulin and calnexin genes in green plants has recently been described.49 Calnexins are generally 90 kDa ER protein consisting of a large N-terminal calcium-binding luminal domain, a single transmembrane helix and a short acidic cytoplasmic tail.48 The luminal domain shares similarity with calreticulin, hence calnexin was initially identified as membrane anchored homolog of calreticulin. In plants, Calnexin was first characterized in Arabidopsis. It has been established that AtCRT1a is an alleviator of the tunicamycin-induced unfolded protein response.50 After Arabidopsis, the calnexin was also cloned and well characterized from pea.51 Plant calreticulins have been identified in Spinach,52 barley,53 tobacco54 and other plant species. Recently, it has been demonstrated that plant CRTs have a number of specific properties different from their animal counterparts, highlighting the significance of CRTs in plants growth and development as well as biotic and abiotic stress responses. There are at least two distinct groups of calreticulin isoforms in higher plants.55 Recent work on Arabidopsis CRT have revealed that the physiological functions of the two CRT subgroups in Arabidopsis have diverged, resulting in a role for AtCRT3 in PAMP (Pathogen-Associated Molecular Pattern) associated responses, and possibly more general chaperone functions for AtCRT1a and CRT1b.56
Conclusions
The involvement of ER chaperones and foldases in protein folding processes in plant cells might have general implications. They also play an important role in stress signaling and management in plants. Knowledge about the proteins is of immense importance to crop scientists working towards development of stress resistance plants. Few regulatory elements controlling expression of these important proteins have been identified and very little is know about how these proteins act selectively on proteins. Results from new generation high throughput techniques will help reconstruct the system networks and protein interactions, controlling expression of these proteins and further our understanding. More emphasis should also be laid on solving the crystal structures of these proteins. Overall, the ER chaperones and foldases have now also emerged as important molecules to understand stress signaling and tolerance in plants.
Acknowledgements
Work is partially supported by Department of Biotechnology (DBT), and Department of Science and Technology (DST), Government of India.
References
- 1.Nagradova N. Enzymes catalyzing protein folding and their cellular functions. Curr Protein Pept Sci. 2007;8:273–282. doi: 10.2174/138920307780831866. [DOI] [PubMed] [Google Scholar]
- 2.Boston RS, Viitanen PV, Vierling E. Molecular chaperones and protein folding in plants. Plant Mol Biol. 1996;32:191–222. doi: 10.1007/BF00039383. [DOI] [PubMed] [Google Scholar]
- 3.Rowling PJ, Freedman RB. Folding, assembly, and post-translational modification of proteins within the lumen of the endoplasmic reticulum. Subcell Biochem. 1993;21:41–80. doi: 10.1007/978-1-4615-2912-5_3. [DOI] [PubMed] [Google Scholar]
- 4.Boston RS, Fontes EB, Shank BB, Wrobel RL. Increased expression of the maize immunoglobulin binding protein homolog b-70 in three zein regulatory mutants. Plant Cell. 1991;3:497–505. doi: 10.1105/tpc.3.5.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vitale A, Boston RS. Endoplasmic reticulum quality control and the unfolded protein response: insights from plants. Traffic. 2008;9:1581–1588. doi: 10.1111/j.1600-0854.2008.00780.x. [DOI] [PubMed] [Google Scholar]
- 6.Denecke J, Goldman MH, Demolder J, Seurinck J, Botterman J. The tobacco luminal binding protein is encoded by a multigene family. Plant Cell. 1991;3:1025–1035. doi: 10.1105/tpc.3.9.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.D'Amico L, Valsasina B, Daminati MG, Fabbrini MS, Nitti G, Bollini R, et al. Bean homologs of the mammalian glucose-regulated proteins: induction by tunicamycin and interaction with newly synthesized seed storage proteins in the endoplasmic reticulum. Plant J. 1992;2:443–455. doi: 10.1111/j.1365-313x.1992.00443.x. [DOI] [PubMed] [Google Scholar]
- 8.Dwek RA. Glycobiology: more functions for oligosaccharides. Science. 1995;269:1234–1235. doi: 10.1126/science.7652569. [DOI] [PubMed] [Google Scholar]
- 9.Vasquez-Robinet C, Watkinson JI, Sioson AA, Ramakrishnan N, Heath LS, Grene R. Differential expression of heat shock protein genes in preconditioning for photosynthetic acclimation in water-stressed loblolly pine. Plant Physiol Biochem. 2010;48:256–264. doi: 10.1016/j.plaphy.2009.12.005. [DOI] [PubMed] [Google Scholar]
- 10.Oono Y, Wakasa Y, Hirose S, Yang L, Sakuta C, Takaiwa F. Analysis of ER stress in developing rice endosperm accumulating beta-amyloid peptide. Plant Biotechnol J. 2010;8:691–718. doi: 10.1111/j.1467-7652.2010.00502.x. [DOI] [PubMed] [Google Scholar]
- 11.Alvim FC, Carolino SM, Cascardo JC, Nunes CC, Martinez CA, Otoni WC, et al. Enhanced accumulation of BiP in transgenic plants confers tolerance to water stress. Plant Physiol. 2001;126:1042–1054. doi: 10.1104/pp.126.3.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gething MJ. Role and regulation of the ER chaperone BiP. Semin Cell Dev Biol. 1999;10:465–472. doi: 10.1006/scdb.1999.0318. [DOI] [PubMed] [Google Scholar]
- 13.Muench DG, Wu Y, Zhang Y, Li X, Boston RS, Okita TW. Molecular cloning, expression and subcellular localization of a BiP homolog from rice endosperm tissue. Plant Cell Physiol. 1997;38:404–412. doi: 10.1093/oxfordjournals.pcp.a029183. [DOI] [PubMed] [Google Scholar]
- 14.Flynn GC, Chappell TG, Rothman JE. Peptide binding and release by proteins implicated as catalysts of protein assembly. Science. 1989;245:385–390. doi: 10.1126/science.2756425. [DOI] [PubMed] [Google Scholar]
- 15.Gething MJ, Sambrook J. Protein folding in the cell. Nature. 1992;355:33–45. doi: 10.1038/355033a0. [DOI] [PubMed] [Google Scholar]
- 16.Knittler MR, Haas IG. Interaction of BiP with newly synthesized immunoglobulin light chain molecules: cycles of sequential binding and release. EMBO J. 1992;11:1573–1581. doi: 10.1002/j.1460-2075.1992.tb05202.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Normington K, Kohno K, Kozutsumi Y, Gething MJ, Sambrook J. S. cerevisiae encodes an essential protein homologous in sequence and function to mammalian BiP. Cell. 1989;57:1223–1236. doi: 10.1016/0092-8674(89)90059-7. [DOI] [PubMed] [Google Scholar]
- 18.Anderson JV, Li QB, Haskell DW, Guy CL. Structural organization of the spinach endoplasmic reticulum-luminal 70-kilodalton heat-shock cognate gene and expression of 70-kilodalton heat-shock genes during cold acclimation. Plant Physiol. 1994;104:1359–1370. doi: 10.1104/pp.104.4.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kalinski A, Rowley DL, Loer DS, Foley C, Buta G, Herman EM. Binding-protein expression is subject to temporal, developmental and stress-induced regulation in terminally differentiated soybean organs. Planta. 1995;195:611–621. doi: 10.1007/BF00195722. [DOI] [PubMed] [Google Scholar]
- 20.Wilbanks SM, DeLuca-Flaherty C, McKay DB. Structural basis of the 70-kilodalton heat shock cognate protein ATP hydrolytic activity. I. Kinetic analyses of active site mutants. J Biol Chem. 1994;269:12893–12898. [PubMed] [Google Scholar]
- 21.Flaherty KM, Wilbanks SM, DeLuca-Flaherty C, McKay DB. Structural basis of the 70-kilodalton heat shock cognate protein ATP hydrolytic activity. II. Structure of the active site with ADP or ATP bound to wild type and mutant ATPase fragment. J Biol Chem. 1994;269:12899–12907. [PubMed] [Google Scholar]
- 22.Wang Y, Geer LY, Chappey C, Kans JA, Bryant SH. Cn3D: sequence and structure views for Entrez. Trends Biochem Sci. 2000;25:300–302. doi: 10.1016/s0968-0004(00)01561-9. [DOI] [PubMed] [Google Scholar]
- 23.Li X, Wu Y, Zhang DZ, Gillikin JW, Boston RS, Franceschi VR, et al. Rice prolamine protein body biogenesis: a BiP-mediated process. Science. 1993;262:1054–1056. doi: 10.1126/science.8235623. [DOI] [PubMed] [Google Scholar]
- 24.Vitale A, Bielli A, Ceriotti A. The binding protein associates with monomeric phaseolin. Plant Physiol. 1995;107:1411–1418. doi: 10.1104/pp.107.4.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pedrazzini E, Giovinazzo G, Bielli A, de Virgilio M, Frigerio L, Pesca M, et al. Protein quality control along the route to the plant vacuole. Plant Cell. 1997;9:1869–1880. doi: 10.1105/tpc.9.10.1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Valente MA, Faria JA, Soares-Ramos JR, Reis PA, Pinheiro GL, Piovesan ND, et al. The ER luminal binding protein (BiP) mediates an increase in drought tolerance in soybean and delays drought-induced leaf senescence in soybean and tobacco. J Exp Bot. 2009;60:533–546. doi: 10.1093/jxb/ern296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wakasa Y, Yasuda H, Oono Y, Kawakatsu T, Hirose S, Takahashi H, et al. Expression of ER quality control-related genes in response to changes in BiP1 levels in developing rice endosperm. Plant J. 2011;65:675–689. doi: 10.1111/j.1365-313X.2010.04453.x. [DOI] [PubMed] [Google Scholar]
- 28.Walther-Larsen H, Brandt J, Collinge DB, Thordal-Christensen H. A pathogen-induced gene of barley encodes a HSP90 homologue showing striking similarity to vertebrate forms resident in the endoplasmic reticulum. Plant Mol Biol. 1993;21:1097–1108. doi: 10.1007/BF00023606. [DOI] [PubMed] [Google Scholar]
- 29.Mao C, Wang M, Luo B, Wey S, Dong D, Wesselschmidt R, et al. Targeted mutation of the mouse Grp94 gene disrupts development and perturbs endoplasmic reticulum stress signaling. PLoS One. 2010;5:e10852. doi: 10.1371/journal.pone.0010852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kudo T. [Therapeutic strategies for Alzheimer disease based on endoplasmic reticulum stress] Nihon Shinkei Seishin Yakurigaku Zasshi. 2010;30:163–168. [PubMed] [Google Scholar]
- 31.Melnick J, Dul JL, Argon Y. Sequential interaction of the chaperones BiP and GRP94 with immunoglobulin chains in the endoplasmic reticulum. Nature. 1994;370:373–375. doi: 10.1038/370373a0. [DOI] [PubMed] [Google Scholar]
- 32.Gruber CW, Cemazar M, Mechler A, Martin LL, Craik DJ. Biochemical and biophysical characterization of a novel plant protein disulfide isomerase. Biopolymers. 2009;92:35–43. doi: 10.1002/bip.21113. [DOI] [PubMed] [Google Scholar]
- 33.Gruber CW, Cemazar M, Clark RJ, Horibe T, Renda RF, Anderson MA, et al. A novel plant protein-disulfide isomerase involved in the oxidative folding of cystine knot defense proteins. J Biol Chem. 2007;282:20435–20446. doi: 10.1074/jbc.M700018200. [DOI] [PubMed] [Google Scholar]
- 34.Coughlan SJ, Hastings C, Winfrey RJ., Jr Molecular characterisation of plant endoplasmic reticulum. Identification of protein disulfide-isomerase as the major reticuloplasmin. Eur J Biochem. 1996;235:215–224. doi: 10.1111/j.1432-1033.1996.00215.x. [DOI] [PubMed] [Google Scholar]
- 35.Shorrosh BS, Subramaniam J, Schubert KR, Dixon RA. Expression and localization of plant protein disulfide isomerase. Plant Physiol. 1993;103:719–726. doi: 10.1104/pp.103.3.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shorrosh BS, Dixon RA. Molecular cloning of a putative plant endomembrane protein resembling vertebrate protein disulfide-isomerase and a phosphatidylinositol-specific phospholipase C. Proc Natl Acad Sci U S A. 1991;88:10941–10945. doi: 10.1073/pnas.88.23.10941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu DP, Christopher DA. Endoplasmic reticulum stress activates the expression of a sub-group of protein disulfide isomerase genes and AtbZIP60 modulates the response in Arabidopsis thaliana. Mol Genet Genomics. 2008;280:199–210. doi: 10.1007/s00438-008-0356-z. [DOI] [PubMed] [Google Scholar]
- 38.Wadahama H, Kamauchi S, Nakamoto Y, Nishizawa K, Ishimoto M, Kawada T, et al. A novel plant protein disulfide isomerase family homologous to animal P5 - molecular cloning and characterization as a functional protein for folding of soybean seed-storage proteins. FEBS J. 2008;275:399–410. doi: 10.1111/j.1742-4658.2007.06199.x. [DOI] [PubMed] [Google Scholar]
- 39.Houston NL, Fan C, Xiang JQ, Schulze JM, Jung R, Boston RS. Phylogenetic analyses identify 10 classes of the protein disulfide isomerase family in plants, including single-domain protein disulfide isomerase-related proteins. Plant Physiol. 2005;137:762–778. doi: 10.1104/pp.104.056507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tian G, Xiang S, Noiva R, Lennarz WJ, Schindelin H. The crystal structure of yeast protein disulfide isomerase suggests cooperativity between its active sites. Cell. 2006;124:61–73. doi: 10.1016/j.cell.2005.10.044. [DOI] [PubMed] [Google Scholar]
- 41.Luan S, Kudla J, Gruissem W, Schreiber SL. Molecular characterization of a FKBP-type immunophilin from higher plants. Proc Natl Acad Sci USA. 1996;93:6964–6969. doi: 10.1073/pnas.93.14.6964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu G, Otegui MS, Spalding EP. The ER-localized TWD1 immunophilin is necessary for localization of multidrug resistance-like proteins required for polar auxin transport in Arabidopsis roots. Plant Cell. 2010;22:3295–3304. doi: 10.1105/tpc.110.078360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Crofts AJ, Denecke J. Calreticulin and calnexin in plants. Trends Plant Sci. 1998;3:4. [Google Scholar]
- 44.Mahajan S, Pandey GK, Tuteja N. Calcium- and salt-stress signaling in plants: shedding light on SOS pathway. Arch Biochem Biophys. 2008;471:146–158. doi: 10.1016/j.abb.2008.01.010. [DOI] [PubMed] [Google Scholar]
- 45.Mahajan S, Tuteja N. Cold, salinity and drought stresses: an overview. Arch Biochem Biophys. 2005;444:139–158. doi: 10.1016/j.abb.2005.10.018. [DOI] [PubMed] [Google Scholar]
- 46.Tuteja N. Mechanisms of high salinity tolerance in plants. Methods Enzymol. 2007;428:419–438. doi: 10.1016/S0076-6879(07)28024-3. [DOI] [PubMed] [Google Scholar]
- 47.Tuteja N, Mahajan S. Calcium signaling network in plants: an overview. Plant Signal Behav. 2007;2:79–85. doi: 10.4161/psb.2.2.4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sarwat M, Tuteja N. Calnexin: a versatile calcium binding integral membrane-bound chaperone of endoplasmic reticulum. Calcium Binding Proteins. 2007;2:36–50. [Google Scholar]
- 49.Del Bem LE. The evolutionary history of calreticulin and calnexin genes in green plants. Genetica. 2011;139:255–259. doi: 10.1007/s10709-010-9544-y. [DOI] [PubMed] [Google Scholar]
- 50.Christensen A, Svensson K, Persson S, Jung J, Michalak M, Widell S, et al. Functional characterization of Arabidopsis calreticulin1a: a key alleviator of endoplasmic reticulum stress. Plant Cell Physiol. 2008;49:912–924. doi: 10.1093/pcp/pcn065. [DOI] [PubMed] [Google Scholar]
- 51.Ehtesham NZ, Phan TN, Gaikwad A, Sopory SK, Tuteja N. Calnexin from Pisum sativum: cloning of the cDNA and characterization of the encoded protein. DNA Cell Biol. 1999;18:853–862. doi: 10.1089/104454999314854. [DOI] [PubMed] [Google Scholar]
- 52.Menegazzi P, Guzzo F, Baldan B, Mariani P, Treves S. Purification of calreticulin-like protein(s) from spinach leaves. Biochem Biophys Res Commun. 1993;190:1130–1135. doi: 10.1006/bbrc.1993.1167. [DOI] [PubMed] [Google Scholar]
- 53.Chen F, Hayes PM, Mulrooney DM, Pan A. Identification and characterization of cDNA clones encoding plant calreticulin in barley. Plant Cell. 1994;6:835–843. doi: 10.1105/tpc.6.6.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Denecke J, Carlsson LE, Vidal S, Hoglund AS, Ek B, van Zeijl MJ, et al. The tobacco homolog of mammalian calreticulin is present in protein complexes in vivo. Plant Cell. 1995;7:391–406. doi: 10.1105/tpc.7.4.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jia XY, He LH, Jing RL, Li RZ. Calreticulin: conserved protein and diverse functions in plants. Physiol Plant. 2009;136:127–138. doi: 10.1111/j.1399-3054.2009.1223.x. [DOI] [PubMed] [Google Scholar]
- 56.Christensen A, Svensson K, Thelin L, Zhang W, Tintor N, Prins D, et al. Higher plant calreticulins have acquired specialized functions in Arabidopsis. PLoS One. 2010;28:e11342. doi: 10.1371/journal.pone.0011342. [DOI] [PMC free article] [PubMed] [Google Scholar]