Abstract
Communication between the cytoplasm and the nucleus is a fundamental feature shared by both plant and animal cells. Cellular factors involved in the transport of macromolecules through the nuclear envelope, including nucleoporins, importins and Ran-GTP related components, are conserved among a variety of eukaryotic systems. Interestingly, mutations in these nuclear components compromise resistance signaling, illustrating the importance of nucleocytoplasmic trafficking in plant innate immunity. Indeed, spatial restriction of defense regulators by the nuclear envelope and stimulus-induced nuclear translocation constitute an important level of defense-associated gene regulation in plants. A significant number of effectors from different microbial pathogens are targeted to the plant cell nucleus. In addition, key host factors, including resistance proteins, immunity components, transcription factors and transcriptional regulators shuttle between the cytoplasm and the nucleus, and their level of nuclear accumulation determines the output of the defense response, further confirming the crucial role played by the nucleus during the interaction between plants and pathogens. Here, we discuss recent findings that situate the nucleus at the frontline of the mutual recognition between plants and invading microbes.
Key words: NES, NLS, nuclear pore complex, plant immunity, protein translocation
Plant resistance to disease is mediated by a mutilayered innate immune system that shares features with animal immunity.1 The first line of active defense relies on the recognition of pathogen- (or microbe-) associated molecular patterns (PAMPs or MAMPs), which are conserved among a diversity of microbes.2 PAMP recognition is mediated by specific pattern-recognition receptors (PRRs) and leads to the so-called PTI (pathogen-triggered immunity), which involves the induction of basal defenses.3–5 A second layer of defense involves molecular recognition of pathogen-produced virulence effectors that evolved to suppress PTI and promote pathogen growth.6 This defense layer is also known as effector-triggered immunity (ETI),4 and involves additional receptors, mostly nucleotide-binding leucine-rich repeat (NB-LRR) resistance (R) proteins, which recognize specific pathogen effectors or their action(s) on host components. R proteins of the NB-LRR type have been classified in two groups based on the presence of the N-terminal coiled-coil (CC) or Toll/interleukin-1 receptor (TIR) domains. NB-LRR R proteins work as molecular switches whose effector-triggered activation results in the transcriptional reprogramming of defense-associated genes and promotion of host defenses. ETI is often associated with the hypersensitive response (HR), a form of programmed cell death that restricts the growth of the pathogen at the infection sites.7,8
Plant innate immunity against infection by microbial pathogens is a highly dynamic process that involves spatial and temporal regulation of a variety of defense components. Studies on the localization of R proteins, pathogen effectors and key host factors during plant innate immunity have revealed the implication of multiple organelles in the recognition and signaling mechanisms associated to defense responses.9 In particular, a growing body of evidence shows the importance of protein trafficking to the nucleus to regulate gene expression related to plant defense.10–13 Various nucleocytoplasmic trafficking pathways, including mRNA export, nuclear localization signal (NLS)-dependent nuclear protein import and nuclear export signal (NES)-mediated nuclear protein export, have been involved in plant innate immunity.14–17 A significant number of effector proteins from viruses, bacteria and oomycetes are targeted to the nucleus where they may directly target nuclear host components (or functions) essential for the immune response.18–21 Alternatively, nuclear translocation of effectors may affect subcellular localization of their cognate R proteins.22,23 Indeed, dynamic trafficking of different R proteins between the cytoplasm and the nucleus is crucial for plant immune responses.10–12,24,25 Together, these findings situate the nucleus at the forefront of the interaction between plants and pathogens. Moreover, essential components of the plant immune response shuttle between the cytoplasm and the nucleus,26,27 providing a possible framework for defense signal trafficking between the two compartments. Finally, cytoplasmic retention of inactive transcription factors or transcriptional regulators, followed by their activation and signal-dependent translocation into the nucleus, allows plants to rapidly connect signal perception at the cell surface and cytoplasmic signal transduction to defense gene activation in a stimulus-dependent manner.28–31
In this review, we provide an overview of recent reports that highlight the importance of cellular dynamics, and particularly protein translocation to the nucleus, for defense-associated gene regulation in plants.
Plant-Pathogen Recognition: “Love will Tear us Apart”
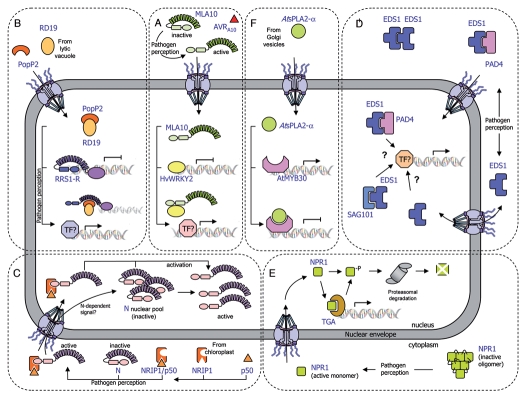
Recent studies reveal that several cytoplasmic NB-LRR receptors partially localize to the nucleus where they function to trigger innate immune responses. The barley Mla resistance locus that conditions isolate-specific immunity to the powdery mildew fungus Blumeria graminis f. sp. hordei encodes CC-NB-LRR receptor proteins.32–35 Shen and colleagues23 showed that the intracellular MLA10 protein is localized both in the cytoplasm and in the nucleus of plant cells. Importantly, activation of the MLA10-mediated immune response requires nuclear accumulation of MLA10, which is triggered by its cognate effector AVRA10. Indeed, the fusion of a nuclear export signal (NES) to MLA10 prevents its nuclear accumulation and leads to the loss of MLA10 function. Interestingly, the CC domain of MLA10 was shown to associate with two closely related barley WKRY transcription factors, HvWRKY1 and HvWRKY2, which act as repressors of PAMP-triggered basal defense responses.23 In planta interaction between nuclear MLA10 and HvWRKY2 occurs in an AVRA10- dependent manner. MLA receptor activation appears to allow rapid transcriptional reprogramming in response to pathogen perception by interfering with HvWRKY1/2 repressor function. Therefore, direct targeting of PAMP-activated HvWRKY1/2 repressors by MLA receptors represents a molecular shortcut to rapidly connect pathogen perception with regulation of defense-related gene expression independently of the action of R gene-specific signaling components (Fig. 1A).
Figure 1.
Schematic representation of the transport of plant immune receptors and regulatory components across the nuclear envelope. (A) The MLA10-mediated immune response is activated by the AVRA10 effector, which promotes nuclear accumulation of CC-NB-LRR resistance protein MLA10. In resting cells, HvWRKY2 acts as a transcriptional repressor of basal defense responses. After pathogen perception, MLA10 binds HvWRKY2 through its CC domain, thereby releasing the negative regulation of the immune response. (B) The PopP2 effector, which triggers the RRS1-R-mediated resistance response, promotes nuclear accumulation of the TIR-NB-LRR-WRKY immune receptor RRS1-R and the vacuolar cysteine protease RD19. In unchallenged cells, RRS1-R is hypothesized to act as a transcriptional repressor of basal defense responses. The RD19/PopP2 protein complex would be recognized by RRS1-R leading to either modification of RRS1-R transcriptional activity or transcriptional activation by additional transcription factors, thereby derepressing defense responses. (C) The tobacco TIR-NB-LRR immune receptor N resides in the cytoplasm and the nucleus of non-infected cells. After pathogen inoculation, the tobacco rhodanase sulfurtransferase NRIP1, which localizes to the stroma of chloroplast in resting cells, is recruited to the cytoplasm of the plant cell by the p50 effector, to form a pre-recognition complex. This NRIP1/p50 complex interacts with the N receptor thanks to NRIP1, which binds to the TIR domain of N, leading to the activation of cytoplasmic N. Once activated, cytoplasmic N either enters the nucleus or sends a signal that activates the N nuclear pool, resulting in the activation of a successful defense response. (D) Basal and TIR-NB-LRR resistance protein-mediated defense responses depend on the regulatory immune complexes formed by the EDS1, PAD4 and SAG101 proteins. EDS1 can exist in a complex with PAD4 in both the cytoplasm and nucleus, and with SAG101 in the nucleus, resulting in the regulation of defense gene transcription. The role of EDS1 homodimers in the cytoplasm is still unknown. Dynamic changes in the binding affinities and concentrations of these complexes modulate nuclear accumulation of defense-related components, which allows the plant to mount an appropriate immune response. (E) The transcriptional regulator NPR1 primarily exists as inactive oligomers in the cytoplasm of non-elicited cells. Pathogen recognition leads to a change in the cell redox potential, resulting in the formation of reduced active NPR1 monomers that are translocated into the nucleus. Within the nucleus, NPR1 associates with TGA transcription factors to promote defense gene expression. Levels of nuclear NPR1 accumulation are controlled by protein degradation through the 26S proteasome. (F) The secreted phospholipase AtsPLA2-α is partially relocalized, via a yet undescribed mechanism, from Golgi vesicles to the nucleus where it interacts with the transcription factor AtMYB30, leading to transcriptional repression of defense responses.
A more condensed version of a typical defense signaling pathway is illustrated by the Arabidopsis RRS1-R protein whose modular architecture brings together the NB-LRR domains and a WKRY motif in the same protein.22,36 RRS1-R was shown to interact in yeast cells with its cognate Ralstonia solanacearum Type III effector PopP2. Both protein partners co-localize in the plant cell nucleus where they physically interact.22,37 Upon association with PopP2, RRS1-R may regulate defense-related gene expression either directly via its WKRY domain or through the action of additional transcription factors (Fig. 1B). Interestingly, PopP2 induces nuclear targeting of the Arabidopsis cysteine protease RESPONSIVE TO DEHYDRATATION19 (RD19), otherwise localized to mobile vacuole-associated vesicles and destined to the lytic vacuole.38 The physical interaction between RD19, whose expression is induced by Ralstonia infection, and PopP2 was demonstrated in living plant cells. Since Arabidopsis rd19 mutant plants are compromised in resistance to Ralstonia, it was proposed that RD19 associates with PopP2 to form a nuclear complex that is required for activation of the plant resistance response (Fig. 1B). Unlike RRS1-R, RD19 does not present an obvious NLS and details about the molecular mechanism that activates RD19 nuclear relocalization remain unknown. Several hypotheses about how RD19 nuclear targeting may occur are discussed below.
Recent data reveal a more sophisticated mechanism behind the RRS1-R-mediated resistance response. Indeed, the Arabidopsis RPS4 protein, previously described as conferring resistance to Pseudomonas syringae pv. tomato strain DC3000 expressing AvrRps4,39 was recently shown to function cooperatively with RRS1 allelic forms to prevent infection against at least three different pathogens (Ralstonia solanacearum, Pseudomonas syringae and Colletotrichum higginsianum).40,41 It is not yet known how RRS1 and RPS4 act together into the nucleus to trigger plant immunity. Interestingly, the AvrRps4-triggered immune response requires nuclear accumulation of RPS4 that is distributed between endomembranes and nuclei in uninfected plants. However, RPS4 subcellular localization is not influenced by AvrRps4,42 whereas RRS1-R nuclear accumulation depends on the presence of PopP2.22,37 It is conceivable that a threshold concentration of both R proteins in the nucleus controls their respective functions for efficient activation of immune responses.
The tobacco TIR-NB-LRR immune receptor N is an additional R protein that has been shown to function in the nucleus. In non-inoculated cells, N resides in the cytoplasm and the nucleus. Recognition of the 50 kDa helicase domain of Tobacco mosaic virus (TMV) replicase (p50) occurs in the cytoplasm through its indirect association with the TIR domain of the N protein through NRIP1, a tobacco rhodanase sulfurtransferase, which normally localizes to the stroma of chloroplasts and is recruited to the cytoplasm (and nucleus) of the plant cell by the p50 effector.27 In this scenario, NRIP1 has been proposed to form a cytoplasmic pre-recognition complex with p50 and to recognize the TIR domain of N to activate a successful defense response. Once activated, cytoplasmic N is either translocated into the nucleus or able to send a signal that activates the N nuclear pool, resulting in the activation of a successful defense response (Fig. 1C). Shuttling of p50-activated N from the cytoplasm to the nucleus may be required for N function, since nuclear accumulation of N is needed to trigger an efficient defense response.43 Nuclear relocalization of NRIP1 is required to provide full resistance to TMV infection.27 Different hypotheses were formulated to explain the molecular mechanism of p50-mediated NRIP1 nuclear relocalization. First, p50 may intercept cytosolic NRIP1 on its way to the chloroplast, in a process that would mask NRIP1 chloroplast targeting signal and allow its nuclear recruitment. Alternatively, p50 may indirectly disrupt global chloroplast import affecting the translocation of NRIP1. Otherwise, similar to the mitochondrial release of pro-cell death components, such as cytochrome c,44 chloroplast-localized NRIP1 may be released into the cytoplasm and the nucleus by p50-induced permeabilization of the outer membrane. Finally, the involvement of stromules in NRIP1 nuclear import was also suggested. Stromules have been associated with defense reactions in response to both biotic and abiotic stresses.27,45 Although there is not reported evidence demonstrating that stromules directly connect nuclei and chloroplasts, close contact between stromules and nuclei has been observed27,45,46 and this close association may enhance the nuclear import of chloroplastic factors, such as NRIP1.
Immunity Components: “With a Little Help from My Friends”
In addition to R proteins, essential components of the plant immune system have been shown to dynamically translocate across the nuclear envelope. The plant immune regulator EDS1 (enhanced disease susceptibility1) is a master component of plant defense47,48 that transiently associates in the cytoplasm and in the nucleus with its co-regulator partners PAD4 (Phytoalexin Deficient4) and SAG101 (Senescence-Associated Gene101).48,49 EDS1-, PAD4-, SAG101-containing complexes constitute an essential regulatory node that mediates both effector-triggered activation of several TIR-NB-LRR and basal defense responses48–50 (Fig. 1D). Although EDS1 was found not to be necessary for accumulation or nuclear accessibility of RPS4, EDS1 is strictly required for RPS4-mediated signaling and functions somewhere between the activation of the R protein and the transcriptional reprogramming in the cell nucleus.42 Recently, Garcia and colleagues showed that RPS4 recognition of AvrRps4 triggers nuclear accumulation of EDS1, which directs transcriptional reprogramming.51 The Arabidopsis snc1 (suppressor of npr1-1, constitutive 1) mutant carries a gain of function mutation in a TIR-NB-LRR-type R gene and shows constitutive defense activation and increased resistance to virulent pathogens.52,53 EDS1-dependent constitutive resistance displayed by the snc1 mutant was shown to be associated with changes in relative amounts of cytoplasmic and nuclear EDS1. Thus, EDS1 functions through balanced nuclear and cytoplasmic activities to mediate the induction and repression of particular defense-related genes thereby allowing the plant to mount an appropriate immune response.51
Transcriptional Regulators: “The Winner Takes it All”
Spatial restriction of transcription factors and regulators of transcription by the nuclear envelope and their stimulus-induced nuclear translocation constitute an important level of defense-associated gene regulation in plants. NPR1 (Non expressor of Pathogenesis-Related genes1, also known as NIM1) is a key regulator of systemic acquired resistance (SAR), an inducible form of plant defense conferring broad-spectrum immunity to secondary infection.54–56 In the absence of infection, NPR1, which contains a functional bipartite NLS, is predominantly sequestered in the cytoplasm as an oligomeric complex.57 Upon pathogen challenge, a change in the redox potential of the cell is induced so that the NPR1 oligomer is partially reduced to a monomeric state and translocates to the nucleus. Once in the nucleus, NPR1 functions as a co-activator of gene transcription through its interaction with TGA-bZIP transcription factors.58–60 Interestingly, proteasomal-mediated turnover of NPR1 in the nucleus is necessary for its function as a transcriptional co-activator in SAR61 (Fig. 1E).
Similarly to the Arabidopsis vacuolar cysteine protease RD19, the tomato LeCp vacuolar protease is relocalized to the nucleus following treatment with the ethylene-inducing xylanase (EIX) elicitor.62 In the nucleus, LeCp acts as a transcription factor to activate the expression of the ACC synthase gene via a yet unexplained mechanism.62 As RD19, LeCp does not contain a consensus NLS, and since LeCp interacts with SUMO in yeast, the authors proposed that SUMO modification of LeCp generates the signal required for LeCp to enter the nucleus. In this context, LeCp would generally function as a Cys protease, but upon modification by SUMO, it would enter the nucleus and induce expression of the ACC synthase gene.
An additional example of the importance of dynamic protein trafficking to the nucleus for the regulation of transcription is provided by the secreted phospholipase AtsPLA2-α. Surprisingly, AtsPLA2-α was identified in a yeast two-hybrid screen as an interactor of the nuclear Arabidopsis MYB transcription factor AtMYB30, a positive regulator of bacterial-induced HR and defense responses.28,63 AtsPLA2-α has been implicated in the control of auxin transport protein trafficking to the plasma membrane.64 Consistent with the presence of a signal peptide in the N-terminus of AtsPLA2-α, YFP-tagged AtsPLA2-α was localized intracellularly in Golgi-associated vesicles and later secreted to the extracellular space.28,64 However, in the presence of AtMYB30, targeting of AtsPLA2-α is partially modified from cytoplasmic vesicles to the plant cell nucleus, where both proteins interact. This protein interaction leads to repression of the AtMYB30-mediated transcriptional activity and negative regulation of plant HR. Furthermore, an Arabidopsis Atspla2-α mutant is more resistant to bacterial inoculation, whereas AtsPLA2-α overexpression leads to decreased resistance, confirming the role of AtsPLA2-α as a negative regulator of AtMYB30-mediated defense (Fig. 1F). As in the previously mentioned examples, the mechanism behind the nuclear recruitment of AtsPLA2-α is still unknown. In the case of LeCp and RD19, EIX and PopP2 were proposed to induce a membrane permeabilization process, triggering the collapse of the vacuolar membrane. This would lead to the respective release of LeCp and RD19 from vacuole-associated compartments into the cytoplasm, where they may become available for SUMOylation, thereby allowing their nuclear targeting.38,62 Similarly, AtMYB30 might release AtsPLA2-α from vesicle-associated compartments to the nucleus through an unknown mechanism that may involve AtsPLA2-α SUMOylation. Indeed, lysine residues with high probability of being SUMOylated are present in LeCp, RD19 and AtsPLA2-α. Alternatively, we cannot rule out that AtMYB30 may intercept AtsPLA2-α on its way to the extracellular compartment via retrograde transport from the endomembrane system, which has some continuity with the nuclear envelope, as it was previously proposed for PopP2-induced nuclear translocation of RD19. Finally, binding of AtMYB30 to AtsPLA2-α may mask AtsPLA2-α vesicle targeting signal or AtMYB30 may indirectly disrupt global vesicle sorting affecting the translocation of AtsPLA2-α.
The Nuclear Import-Export Machinery: “I Like to Move it”
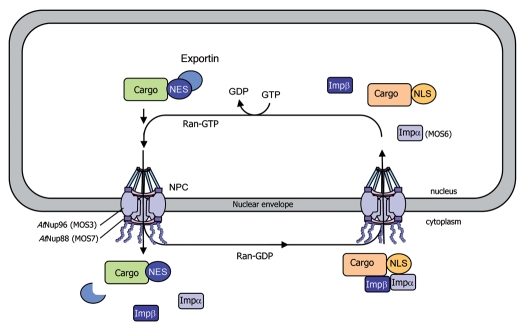
In eukaryotic cells, protein and RNA trafficking between the cytoplasm and the nucleus occurs through nuclear pore complexes (NPCs), which are composed of nucleoporins (Nups) and span the double membrane of the nuclear envelope. Nucleocytoplasmic transport of macromolecules typically depends on import and export receptors (importins and exportins), which translocate cargo through the nuclear pore.65,66 Importins and exportins respectively recognize exposed NLSs and NESs that label their cargo proteins.67 α importins are adapter proteins that bind to NLS-containing cargo proteins and bridge their interaction with importin β. The trimeric complex importin α/importin β/cargo protein goes across the NPC, via the importin β-mediated interaction with Nups, and is translocated into the nucleus. The directionality of the nuclear transport is maintained by the ratio of Ran-GDP (cytoplasmic side) and Ran-GTP (nuclear side). Upon binding of Ran-GTP in the nucleus, the complex is disassembled and importin α and β shuttle back to the cytoplasm to allow additional rounds of transport65,66 (Fig. 2).
Figure 2.
Model for nucleocytoplasmic transport of macromolecules through the nuclear pore complex. Cytoplasmic proteins with a nuclear localization signal (NLS) are translocated into the nucleus through nuclear pore complexes (NPCs), which are composed of nucleoporins (Nups) and span the double membrane of the nuclear envelope. The NLS in cargo proteins is recognized and bound by importin receptors (importin α and β) promoting their transport across the NPC, via the importin β-mediated interaction with Nups. The Ras-related nuclear (Ran) protein provides the directionality of transport via its binding to GDP (cytoplasmic side) or GTP (nuclear side). Export receptors (exportins) recognize nuclear export signals (NES) in cargo proteins and promote their nuclear export to the cytoplasm. Mutations in AtNup96 (MOS3), AtNup88 (MOS7) and importin α3 (MOS6) have been shown to impair the nuclear transport of immunity components, resulting in altered defense responses.
The Arabidopsis snc1 mutant shows increased resistance to several pathogens.52,53 The identification of MOS3, MOS6 and MOS7 (for MODIFIER OF SNC1) in a snc1 suppressor screen underlines the importance of nucleocytoplasmic trafficking for both basal resistance and resistance mediated by several R genes in addition of snc1. Arabidopsis Nup96 (MOS3/SAR3), which is involved in mRNA export, and Nup88 (MOS7) are localized in the nuclear envelope and required for basal resistance as well as R gene-mediated resistance to avirulent pathogens.14,17 However, it is unclear how MOS proteins regulate snc1-mediated defense and basal resistance responses. MOS3 may be involved in the nuclear export of an RNA encoding a positive regulator essential for activation of disease resistance.14 MOS7 is required for specific nuclear accumulation of the autoactivated R protein SNC1, as well as the downstream defense signaling components EDS1 and NPR1,17 indicating that MOS7 acts a nuclear chaperone for NB-LRR proteins. Furthermore, these data suggest that specific modulation of the nuclear concentration of a set of defense regulators is crucial for the fine-tuning of plant immune responses. In Arabidopsis, importin α4 is preferentially involved in Agrobacterium-mediated transformation68 and importin α3 (MOS6) is required for plant defense against virulent pathogens.15 Finally, the implication of Ran-regulated elements of the nuclear pore in plant immune responses was confirmed by the finding that RanGap2 (Ran-GTPase-activating protein2) interacts with the NB-LRR protein Rx, and silencing of RanGap2 impairs Rx-mediated resistance.16,69 Therefore, it is likely that RanGap2 directs Rx into the nucleus, either serving as a carrier to promote Rx nuclear transport or modifying Rx folding so that it may interact with an additional carrier protein that may facilitate Rx nuclear translocation.
Together, these findings strongly suggest that components of the NPC may specifically mediate nuclear translocation of cargo proteins, including NB-LRR R proteins, immunity components (such as the EDS1/PAD4/SAG101 complex), TGA or bZIP transcription factors, transcriptional regulators (including NPR1 and AtsPLA2-α) or mitogen-activated protein kinases, which are implicated in disease resistance and translocate to the nucleus upon activation of the defense response.13,29,30,70–72 Specific modulation of the nuclear concentration of these host components would thus allow appropriately mounting an efficient immune response to pathogen attack.
Conclusions and Perspectives
Exciting progress has been made during the last few years in uncovering the previously unexpected dynamic translocation of a great number of defense-related proteins across the nuclear envelope. However, we are still far from comprehending the molecular mechanisms controlling the dynamic changes in protein subcellular localization that are necessary to achieve resistance. It is becoming increasingly evident that, to coordinate appropriate defense outputs, plant cells rely on the integrated translocation of defense-related proteins through different organelles, including the chloroplast, mitochondria, nucleus or endoplasmic reticulum. In the view of the recent findings in our field, unravelling the flexible and intricate mechanisms behind the establishment of plant immune responses is an exciting challenge for future research.
References
- 1.Ausubel FM. Are innate immune signaling pathways in plants and animals conserved? Nat Immunol. 2005;6:973–979. doi: 10.1038/ni1253. [DOI] [PubMed] [Google Scholar]
- 2.Zipfel C. Pattern-recognition receptors in plant innate immunity. Curr Opin Immunol. 2008;20:10–16. doi: 10.1016/j.coi.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 3.Boller T, Felix G. A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol. 2009;60:379–406. doi: 10.1146/annurev.arplant.57.032905.105346. [DOI] [PubMed] [Google Scholar]
- 4.Jones JD, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 5.Meylan E, Tschopp J, Karin M. Intracellular pattern recognition receptors in the host response. Nature. 2006;442:39–44. doi: 10.1038/nature04946. [DOI] [PubMed] [Google Scholar]
- 6.Gohre V, Robatzek S. Breaking the barriers: microbial effector molecules subvert plant immunity. Annu Rev Phytopathol. 2008;46:189–215. doi: 10.1146/annurev.phyto.46.120407.110050. [DOI] [PubMed] [Google Scholar]
- 7.Tao Y, Xie Z, Chen W, Glazebrook J, Chang HS, Han B, et al. Quantitative nature of Arabidopsis responses during compatible and incompatible interactions with the bacterial pathogen Pseudomonas syringae. Plant Cell. 2003;15:317–330. doi: 10.1105/tpc.007591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Greenberg JT, Yao N. The role and regulation of programmed cell death in plant-pathogen interactions. Cell Microbiol. 2004;6:201–211. doi: 10.1111/j.1462-5822.2004.00361.x. [DOI] [PubMed] [Google Scholar]
- 9.Padmanabhan MS, Dinesh-Kumar SP. All hands on deck: The role of chloroplasts, endoplasmic reticulum and the nucleus in driving plant innate immunity. Mol Plant Microbe Interact. 2010;23:1368–1380. doi: 10.1094/MPMI-05-10-0113. [DOI] [PubMed] [Google Scholar]
- 10.Caplan J, Padmanabhan M, Dinesh-Kumar SP. Plant NB-LRR immune receptors: from recognition to transcriptional reprogramming. Cell Host Microbe. 2008;3:126–135. doi: 10.1016/j.chom.2008.02.010. [DOI] [PubMed] [Google Scholar]
- 11.Liu J, Coaker G. Nuclear trafficking during plant innate immunity. Mol Plant. 2008;1:411–422. doi: 10.1093/mp/ssn010. [DOI] [PubMed] [Google Scholar]
- 12.Wiermer M, Palma K, Zhang Y, Li X. Should I stay or should I go? Nucleocytoplasmic trafficking in plant innate immunity. Cell Microbiol. 2007;9:1880–1890. doi: 10.1111/j.1462-5822.2007.00962.x. [DOI] [PubMed] [Google Scholar]
- 13.Garcia AV, Parker JE. Heaven's Gate: nuclear accessibility and activities of plant immune regulators. Trends Plant Sci. 2009;14:479–487. doi: 10.1016/j.tplants.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 14.Zhang Y, Li X. A putative nucleoporin 96 Is required for both basal defense and constitutive resistance responses mediated by suppressor of npr1-1, constitutive 1. Plant Cell. 2005;17:1306–1316. doi: 10.1105/tpc.104.029926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Palma K, Zhang Y, Li X. An importin alpha homolog, MOS6, plays an important role in plant innate immunity. Curr Biol. 2005;15:1129–1135. doi: 10.1016/j.cub.2005.05.022. [DOI] [PubMed] [Google Scholar]
- 16.Tameling WI, Baulcombe DC. Physical association of the NB-LRR resistance protein Rx with a Ran GTPase-activating protein is required for extreme resistance to Potato virus X. Plant Cell. 2007;19:1682–1694. doi: 10.1105/tpc.107.050880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng YT, Germain H, Wiermer M, Bi D, Xu F, Garcia AV, et al. Nuclear pore complex component MOS7/Nup88 is required for innate immunity and nuclear accumulation of defense regulators in Arabidopsis. Plant Cell. 2009;21:2503–2516. doi: 10.1105/tpc.108.064519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vetter G, Hily JM, Klein E, Schmidlin L, Haas M, Merkle T, Gilmer D. Nucleo-cytoplasmic shuttling of the beet necrotic yellow vein virus RNA-3-encoded p25 protein. J Gen Virol. 2004;85:2459–2469. doi: 10.1099/vir.0.80142-0. [DOI] [PubMed] [Google Scholar]
- 19.Szurek B, Rossier O, Hause G, Bonas U. Type III-dependent translocation of the Xanthomonas AvrBs3 protein into the plant cell. Mol Microbiol. 2002;46:13–23. doi: 10.1046/j.1365-2958.2002.03139.x. [DOI] [PubMed] [Google Scholar]
- 20.Kanneganti TD, Bai X, Tsai CW, Win J, Meulia T, Goodin M, et al. A functional genetic assay for nuclear trafficking in plants. Plant J. 2007;50:149–158. doi: 10.1111/j.1365-313X.2007.03029.x. [DOI] [PubMed] [Google Scholar]
- 21.Schornack S, van Damme M, Bozkurt TO, Cano LM, Smoker M, Thines M, et al. Ancient class of translocated oomycete effectors targets the host nucleus. Proc Natl Acad Sci USA. 2010;107:17421–17426. doi: 10.1073/pnas.1008491107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deslandes L, Olivier J, Peeters N, Feng DX, Khounlotham M, Boucher C, et al. Physical interaction between RRS1-R, a protein conferring resistance to bacterial wilt and PopP2, a type III effector targeted to the plant nucleus. Proc Natl Acad Sci USA. 2003;100:8024–8029. doi: 10.1073/pnas.1230660100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shen QH, Saijo Y, Mauch S, Biskup C, Bieri S, Keller B, et al. Nuclear activity of MLA immune receptors links isolate-specific and basal disease-resistance responses. Science. 2007;315:1098–1103. doi: 10.1126/science.1136372. [DOI] [PubMed] [Google Scholar]
- 24.Burch-Smith TM, Dinesh-Kumar SP. The functions of plant TIR domains. Sci STKE. 2007;2007:46. doi: 10.1126/stke.4012007pe46. [DOI] [PubMed] [Google Scholar]
- 25.Shen QH, Schulze-Lefert P. Rumble in the nuclear jungle: compartmentalization, trafficking and nuclear action of plant immune receptors. EMBO J. 2007;26:4293–4301. doi: 10.1038/sj.emboj.7601854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wiermer M, Feys BJ, Parker JE. Plant immunity: the EDS1 regulatory node. Curr Opin Plant Biol. 2005;8:383–389. doi: 10.1016/j.pbi.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 27.Caplan JL, Mamillapalli P, Burch-Smith TM, Czymmek K, Dinesh-Kumar SP. Chloroplastic protein NRIP1 mediates innate immune receptor recognition of a viral effector. Cell. 2008;132:449–462. doi: 10.1016/j.cell.2007.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Froidure S, Canonne J, Daniel X, Jauneau A, Briere C, Roby D, Rivas S. AtsPLA2-alpha nuclear relocalization by the Arabidopsis transcription factor AtMYB30 leads to repression of the plant defense response. Proc Natl Acad Sci USA. 2010;107:15281–15286. doi: 10.1073/pnas.1009056107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dong X. NPR1, all things considered. Curr Opin Plant Biol. 2004;7:547–552. doi: 10.1016/j.pbi.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 30.Kaminaka H, Nake C, Epple P, Dittgen J, Schutze K, Chaban C, et al. bZIP10-LSD1 antagonism modulates basal defense and cell death in Arabidopsis following infection. EMBO J. 2006;25:4400–4411. doi: 10.1038/sj.emboj.7601312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rochon A, Boyle P, Wignes T, Fobert PR, Despres C. The coactivator function of Arabidopsis NPR1 requires the core of its BTB/POZ domain and the oxidation of C-terminal cysteines. Plant Cell. 2006;18:3670–3685. doi: 10.1105/tpc.106.046953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Halterman D, Zhou F, Wei F, Wise RP, Schulze-Lefert P. The MLA6 coiled-coil, NBS-LRR protein confers AvrMla6-dependent resistance specificity to Blumeria graminis f. sp. hordei in barley and wheat. Plant J. 2001;25:335–348. doi: 10.1046/j.1365-313x.2001.00982.x. [DOI] [PubMed] [Google Scholar]
- 33.Zhou F, Kurth J, Wei F, Elliott C, Vale G, Yahiaoui N, et al. Cell-autonomous expression of barley mla1 confers race-specific resistance to the powdery mildew fungus via a rar1-independent signaling pathway. Plant Cell. 2001;13:337–350. doi: 10.1105/tpc.13.2.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shen QH, Zhou F, Bieri S, Haizel T, Shirasu K, Schulze-Lefert P. Recognition specificity and RAR1/SGT1 dependence in barley Mla disease resistance genes to the powdery mildew fungus. Plant Cell. 2003;15:732–744. doi: 10.1105/tpc.009258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Halterman DA, Wise RP. A single-amino acid substitution in the sixth leucine-rich repeat of barley MLA6 and MLA13 alleviates dependence on RAR1 for disease resistance signaling. Plant J. 2004;38:215–226. doi: 10.1111/j.1365-313X.2004.02032.x. [DOI] [PubMed] [Google Scholar]
- 36.Deslandes L, Olivier J, Theulieres F, Hirsch J, Feng DX, Bittner-Eddy P, et al. Resistance to Ralstonia solanacearum in Arabidopsis thaliana is conferred by the recessive RRS1-R gene, a member of a novel family of resistance genes. Proc Natl Acad Sci USA. 2002;99:2404–2409. doi: 10.1073/pnas.032485099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tasset C, Bernoux M, Jauneau A, Pouzet C, Briíre C, Kieffer-Jacquinode S, et al. Autoacetylation of the Ralstonia solanacearum effector PopP2 targets a lysine residue essential for RRS1-R-mediated immunity in Arabidopsis. PloS Pathog. 2010 doi: 10.1371/journal.ppat.1001202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bernoux M, Timmers T, Jauneau A, Briere C, de Wit PJ, Marco Y, Deslandes L. RD19, an Arabidopsis cysteine protease required for RRS1-R-mediated resistance, is relocalized to the nucleus by the Ralstonia solanacearum PopP2 effector. Plant Cell. 2008;20:2252–2264. doi: 10.1105/tpc.108.058685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gassmann W, Hinsch ME, Staskawicz BJ. The Arabidopsis RPS4 bacterial-resistance gene is a member of the TIR-NBS-LRR family of disease-resistance genes. Plant J. 1999;20:265–277. doi: 10.1046/j.1365-313x.1999.t01-1-00600.x. [DOI] [PubMed] [Google Scholar]
- 40.Birker D, Heidrich K, Takahara H, Narusaka M, Deslandes L, Narusaka Y, et al. A locus conferring resistance to Colletotrichum higginsianum is shared by four geographically distinct Arabidopsis accessions. Plant J. 2009;60:602–613. doi: 10.1111/j.1365-313X.2009.03984.x. [DOI] [PubMed] [Google Scholar]
- 41.Narusaka M, Shirasu K, Noutoshi Y, Kubo Y, Shiraishi T, Iwabuchi M, Narusaka Y. RRS1 and RPS4 provide a dual Resistance-gene system against fungal and bacterial pathogens. Plant J. 2009;60:218–226. doi: 10.1111/j.1365-313X.2009.03949.x. [DOI] [PubMed] [Google Scholar]
- 42.Wirthmueller L, Zhang Y, Jones JD, Parker JE. Nuclear accumulation of the Arabidopsis immune receptor RPS4 is necessary for triggering EDS1-dependent defense. Curr Biol. 2007;17:2023–2029. doi: 10.1016/j.cub.2007.10.042. [DOI] [PubMed] [Google Scholar]
- 43.Burch-Smith TM, Schiff M, Caplan JL, Tsao J, Czymmek K, Dinesh-Kumar SP. A novel role for the TIR domain in association with pathogen-derived elicitors. PLoS Biol. 2007;5:68. doi: 10.1371/journal.pbio.0050068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gogvadze V, Orrenius S, Zhivotovsky B. Multiple pathways of cytochrome c release from mitochondria in apoptosis. Biochim Biophys Acta. 2006;1757:639–647. doi: 10.1016/j.bbabio.2006.03.016. [DOI] [PubMed] [Google Scholar]
- 45.Holzinger A, Buchner O, Lutz C, Hanson MR. Temperature-sensitive formation of chloroplast protrusions and stromules in mesophyll cells of Arabidopsis thaliana. Protoplasma. 2007;230:23–30. doi: 10.1007/s00709-006-0222-y. [DOI] [PubMed] [Google Scholar]
- 46.Kwok EY, Hanson MR. Plastids and stromules interact with the nucleus and cell membrane in vascular plants. Plant Cell Rep. 2004;23:188–195. doi: 10.1007/s00299-004-0824-9. [DOI] [PubMed] [Google Scholar]
- 47.Falk A, Feys BJ, Frost LN, Jones JDG, Daniels MJ, Parker JE. EDS1, an essential component of R gene-mediated disease resistance in Arabidopsis has homology to eukaryotic lipases. Proc Natl Acad Sci USA. 1999;96:3292–3297. doi: 10.1073/pnas.96.6.3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Feys BJ, Moisan LJ, Newman MA, Parker JE. Direct interaction between the Arabidopsis disease resistance signaling proteins, EDS1 and PAD4. EMBO J. 2001;20:5400–5411. doi: 10.1093/emboj/20.19.5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Feys BJ, Wiermer M, Bhat RA, Moisan LJ, Medina-Escobar N, Neu C, et al. Arabidopsis SENESCENCE-ASSOCIATED GENE101 stabilizes and signals within an ENHANCED DISEASE SUSCEPTIBILITY1 complex in plant innate immunity. Plant Cell. 2005;17:2601–2613. doi: 10.1105/tpc.105.033910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Venugopal SC, Jeong RD, Mandal MK, Zhu S, Chandra-Shekara AC, Xia Y, et al. Enhanced disease susceptibility 1 and salicylic acid act redundantly to regulate resistance gene-mediated signaling. PLoS Genet. 2009;5:1000545. doi: 10.1371/journal.pgen.1000545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Garcia AV, Blanvillain-Baufume S, Huibers RP, Wiermer M, Li G, Gobbato E, et al. Balanced nuclear and cytoplasmic activities of EDS1 are required for a complete plant innate immune response. PLoS Pathog. 2010;6:1000970. doi: 10.1371/journal.ppat.1000970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li X, Clarke JD, Zhang Y, Dong X. Activation of an EDS1-mediated R-gene pathway in the snc1 mutant leads to constitutive, NPR1-independent pathogen resistance. Mol Plant Microbe Interact. 2001;14:1131–1139. doi: 10.1094/MPMI.2001.14.10.1131. [DOI] [PubMed] [Google Scholar]
- 53.Zhang Y, Goritschnig S, Dong X, Li X. A gain-of-function mutation in a plant disease resistance gene leads to constitutive activation of downstream signal transduction pathways in suppressor of npr1-1, constitutive 1. Plant Cell. 2003;15:2636–2646. doi: 10.1105/tpc.015842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cao H, Bowling SA, Gordon SA, Dong X. Characterization of an Arabidopsis mutant that is non-responsive to inducers of systemic acquired resistance. Plant Cell. 1994;6:1583–1592. doi: 10.1105/tpc.6.11.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Delaney TP, Friedrich L, Ryals JA. Arabidopsis signal transduction mutant defective in chemically and biologically induced disease resistance. Proc Natl Acad Sci USA. 1995;92:6602–6606. doi: 10.1073/pnas.92.14.6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang D, Amornsiripanitch N, Dong X. A genomic approach to identify regulatory nodes in the transcriptional network of systemic acquired resistance in plants. PLoS Pathog. 2006;2:123. doi: 10.1371/journal.ppat.0020123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mou Z, Fan W, Dong X. Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell. 2003;113:935–944. doi: 10.1016/s0092-8674(03)00429-x. [DOI] [PubMed] [Google Scholar]
- 58.Zhang Y, Fan W, Kinkema M, Li X, Dong X. Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR-1 gene. Proc Natl Acad Sci USA. 1999;96:6523–6528. doi: 10.1073/pnas.96.11.6523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Després C, DeLong C, Glaze S, Liu E, Fobert PR. The Arabidopsis NPR1/NIM1 protein enhances the DNA binding activity of a subgroup of the TGA family of bZIP transcription factors. Plant Cell. 2000;12:279–290. [PMC free article] [PubMed] [Google Scholar]
- 60.Zhou JM, Trifa Y, Silva H, Pontier D, Lam E, Shah J, Klessig DF. NPR1 differentially interacts with members of the TGA/OBF family of transcription factors that bind an element of the PR-1 gene required for induction by salicylic acid. Molecular Plant-Microbe Interact. 2000;13:191–202. doi: 10.1094/MPMI.2000.13.2.191. [DOI] [PubMed] [Google Scholar]
- 61.Spoel SH, Mou Z, Tada Y, Spivey NW, Genschik P, Dong X. Proteasome-mediated turnover of the transcription coactivator NPR1 plays dual roles in regulating plant immunity. Cell. 2009;137:860–872. doi: 10.1016/j.cell.2009.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Matarasso N, Schuster S, Avni A. A novel plant cysteine protease has a dual function as a regulator of 1-aminocyclopropane-1-carboxylic acid synthase gene expression. Plant Cell. 2005;17:1205–1216. doi: 10.1105/tpc.105.030775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vailleau F, Daniel X, Tronchet M, Montillet JL, Triantaphylides C, Roby D. A R2R3-MYB gene, AtMYB30, acts as a positive regulator of the hypersensitive cell death program in plants in response to pathogen attack. Proc Natl Acad Sci USA. 2002;99:10179–10184. doi: 10.1073/pnas.152047199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee OR, Kim SJ, Kim HJ, Hong JK, Ryu SB, Lee SH, et al. Phospholipase A(2) is required for PIN-FORMED protein trafficking to the plasma membrane in the Arabidopsis root. Plant Cell. 2010;22:1812–1825. doi: 10.1105/tpc.110.074211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Merkle T. Nuclear import and export of proteins in plants: a tool for the regulation of signaling. Planta. 2001;213:499–517. doi: 10.1007/s004250100621. [DOI] [PubMed] [Google Scholar]
- 66.Meier I. Composition of the plant nuclear envelope: theme and variations. J Exp Bot. 2007;58:27–34. doi: 10.1093/jxb/erl009. [DOI] [PubMed] [Google Scholar]
- 67.Patel SS, Belmont BJ, Sante JM, Rexach MF. Natively unfolded nucleoporins gate protein diffusion across the nuclear pore complex. Cell. 2007;129:83–96. doi: 10.1016/j.cell.2007.01.044. [DOI] [PubMed] [Google Scholar]
- 68.Bhattacharjee S, Lee LY, Oltmanns H, Cao H, Veena, Cuperus J, Gelvin SB. IMPa-4, an Arabidopsis importin alpha isoform, is preferentially involved in agrobacterium-mediated plant transformation. Plant Cell. 2008;20:2661–2680. doi: 10.1105/tpc.108.060467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sacco MA, Mansoor S, Moffett P. A RanGAP protein physically interacts with the NB-LRR protein Rx, and is required for Rx-mediated viral resistance. Plant J. 2007;52:82–93. doi: 10.1111/j.1365-313X.2007.03213.x. [DOI] [PubMed] [Google Scholar]
- 70.Pedley KF, Martin GB. Role of mitogen-activated protein kinases in plant immunity. Curr Opin Plant Biol. 2005;8:541–547. doi: 10.1016/j.pbi.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 71.Cheong YH, Moon BC, Kim JK, Kim CY, Kim MC, Kim IH, et al. BWMK1, a rice mitogen-activated protein kinase, locates in the nucleus and mediates pathogenesis-related gene expression by activation of a transcription factor. Plant Physiol. 2003;132:1961–1972. doi: 10.1104/pp.103.023176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lee J, Rudd JJ, Macioszek VK, Scheel D. Dynamic changes in the localization of MAPK cascade components controlling pathogenesis-related (PR) gene expression during innate immunity in parsley. J Biol Chem. 2004;279:22440–22448. doi: 10.1074/jbc.M401099200. [DOI] [PubMed] [Google Scholar]