Abstract
The oomycete pathogen Phytophthora infestans secretes cystatin-like effector proteins (EPICs) that inhibit secreted host proteases during infection. We recently found that the C14 protease is a relevant target of EPICs and that this protease is under diversifying selection in wild potato species with which P. infestans has coevolved. Here we generated a model of the EPIC-C14 complex based on cystatin-papain crystal structures and discovered three regions where variant residues in C14 might be the result of an arms race between enzyme and inhibitor at the plant-pathogen interface.
Key words: cystatin, papain, arms race, natural variation, variant residues, diversifying selection, structural model, potato, Phytophthora infestans
The plant-pathogen arms race shapes both host and pathogen, and leaves traces in their genomes. It is generally thought that sequence adaptation is particularly strong for genes that are important for ancient, intensive antagonistic plant-pathogen interactions. These arms races occur for instance in inhibitor-enzyme interactions at the plant-pathogen interface.1 The glucanase-inhibitor proteins (GIP) of the oomycete pathogen Phytophthora sojae, for example, inhibit soybean glucanases (EGaseA),2 and natural variation in GIP1 and EGaseA sequences cluster at the surface where GIP1 and EGaseA are thought to interact.3,4
The cystatin-like proteins of P. infestans may be another example of pathogen-derived inhibitors that put selection pressure on their host targets. P. infestans is a devastating oomycete pathogen causing late blight in potato. The species is thought to have co-evolved with wild potato species, Solanum demissum, S. stoliniferum and S. verrucosum in Tolucca Valley, Mexico.5,6 P. infestans secretes cystatin-like proteins during infection, called EPICs. EPIC1 and EPIC2B were originally found to inhibit tomato PIP1 and RCR3, two tomato PLCPs that reside in the apoplast.7,8 We recently identified a third PLCP that forms an even stronger complex with both EPIC1 and EPIC2B.9 This protease is called C14 and was found previously to be transcriptionally induced in tomato upon P. infestans infection.10 Importantly, silencing experiments of C14 in N. benthamiana significantly increase the susceptibility for P. infestans, demonstrating that this host protease contributes to P. infestans resistance, making it a bona fide target for P. infestans effectors.9
In contrast to RCR3 and PIP1, which are under diversifying selection in tomato, C14 is under conservative selection.11 However, tomato is an artificial host for P. infestans and has not been under selection pressure by P. infestans infection. In contrast, we found that C14 is under diversifying selection in wild potato.9 The aim of this work was to map the variant positions on structural models of C14 and EPIC and predict how variant residues may affect the EPIC-C14 interaction.
Variant Residues are Located Around the Substrate Binding Groove of C14
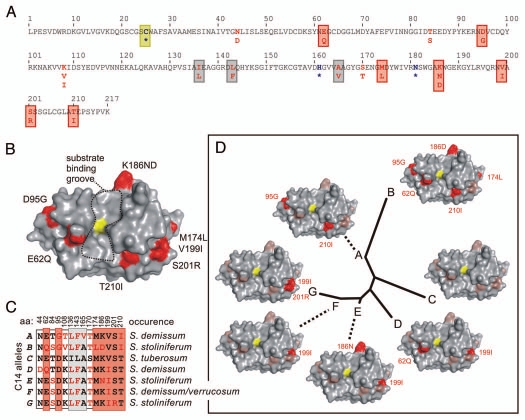
The protein sequence of the protease domain of S. tuberosum C14 (AJ245924) is depicted in Figure 1A. A structural model was generated from potato C14 using SWISS MODEL12 using the crystal structure of the KDEL-tailed Cys protease from Ricinus communis as a template (1S4V13). Ramachandran plot analysis14 showed that the model of potato C14 is reliable, with the exception of S111 which locates at the back surface of C14.
Figure 1.
Structural models of variance in C14 protease domains. (A) Amino acid polymorphism in potato C14 sequences. Only the mature protease domain of S. tuberosum C14 is shown. There are 12 polymorphic amino acids found in wild potato (red), of which seven (boxed red) locate in a ring surrounding the active site and three (boxed grey) locate inside the protein structure. The catalytic residues are indicated in bold blue and with an asterisk (*) and the catalytic Cys is boxed yellow. (B) Model of C14 based on crystal structure of 1S4V. The catalytic Cys is shown in yellow and is in the middle of the substrate binding cleft, which runs from top to bottom. Locations of polymorphic residues are indicated in red and reside in a “ring of fire” around the substrate binding groove. (C) Summary of the occurrence of polymorphic residues in potato C14. There are seven alleles (A–G, left), that differ at 12 positions summarized in the matrix. These were found in wild potato species and cultivated potato (S. tuberosum). (D) Structural polymorphism and phylogenetic relationship of potato C14 proteases. An unrooted tree was generated with the protease domains of C14. The residues indicated in red are different when compared to the protein sequence of cultivated potato (allele C).
We generated surface images of the C14 model using PYMOL and selected the variant residues. Importantly, all variant residues are at surface-exposed positions in the model, with the exception of I136F, L143F and A165V (data not shown). The conservative changes of these internal variant residues are unlikely to affect the overall structure. Variation of the solvent-exposed residues will affect the surface decoration of the C14 proteins, but probably not their overall structure.
Of the 11 solvent-exposed variant residues, seven resides are on the front of the protease around the substrate binding groove with the exposed catalytic C25 (yellow) (Fig. 1B). Interestingly, these variant residues occur in a ring, similar to the “ring of fire” that was observed in EGaseA.3 The variation at these positions is significant and often involves a charge variation, for example for D95G, E62Q, K186ND and S201R.
The occurrence of the variant residues in the different C14 alleles is summarized in Figure 1C. Variance when compared to C14 of S. tuberosum (allele C) is shown in red. Three variant residues that are unique to S. tuberosum (I136, F143 and S170) are located internally or on the back of C14. To visualize the effect of natural variation on protein level we arranged structural models for each allele around a phylogenetic tree that was generated from the protein sequences of the protease domains (Fig. 1D). This figure illustrates that C14 evolved by accumulating mutations and that mutations at positions 62 and 186 may have occurred independently.
Some of the Variant Residues are at the EPIC-C14 Interface
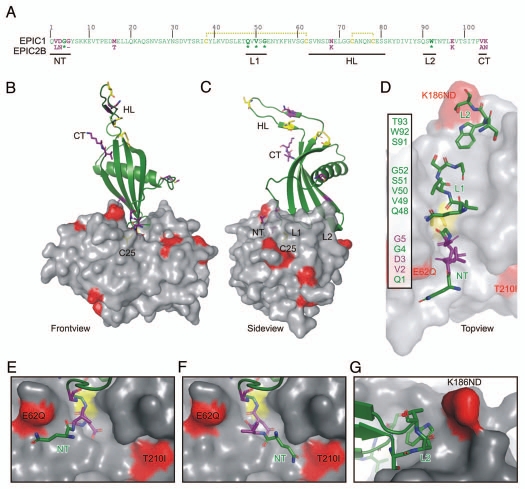
We next generated a structural model for EPIC1 to determine if the variant residues could be in physical contact with EPIC1 and if variant residues between EPIC1 and EPIC2B would interact with the C14 protease. The sequence of EPIC1 without signal peptide is shown in Figure 2A. EPIC2B differs at eight amino acid positions from EPIC2B and the majority of these reside in the termini (Fig. 2A). EPICs were previously shown to be cystatin-like proteins based on their overall alignment with chicken cystatin and the presence of conserved residues at essential positions: the conserved glycine G4 in the N-terminus (NT), the QxVxG motif in loop 2 (L2) and the tryptophane W90 in loop 2 (L2) (Fig. 2A).7
Figure 2.
Model of the EPIC-C14 complex. (A) EPIC 1 protein sequence, without signal peptide. EPIC1 and EPIC2B differ at 8 positions (purple). Residues in the N-terminus (NT), loop-1 (L1) and loop-2 (L2) are directly interacting with the protease. Conserved residues (*) are indicated bold. The hypothetical loop (HT) could not be modelled on the 3IMA template. EPIC1 contains four cysteines (yellow) which are likely connected by disulphide bridges (yellow dashed lines). (C and D) Front-(B), side-(C) and top-(D) views of the model of the EPIC-C14 complex, based on replacement of the tarocystatin-papain crystal structure (3IMA). EPIC 1 is shown as a cartoon (green) and C14 as a semi-transparent surface (gray). The catalytic Cys (C25, yellow) is visible through the transparent surface. As with all cystatins, EPIC s would interact with the substrate binding site of C14 as a wedge consisting of the N-terminus (NT) and two loops (L1 and L2). Polymorphic residues in C14 are indicated in red. Polymorphic sites in EPIC and the cysteins are shown as sticks in purple and yellow, respectively. (D) Model of EPIC residues binding to the substrate binding groove of C14. Residues in the N-terminus (NT) and loops L1 and L2 are shown as sticks. Residues that are polymorphic with EPIC 2B are shown in purple. (E and F) The N-terminus of EPIC may stretch into the left (E) of right (F) cavities of the non-prime substrate binding sites of C14. (G) The L2 loop of EPIC s may interact with the K168ND polymorphic site in C14.
A model for the EPIC1 structure was generated using SWISS MODEL12 using 3IMA as a template. PDB entry 3IMA reports the crystal structure of cystatin from Taro (Colocasia esculenta, hence called tarocystatin), interacting with papain.15 The generated structure contains a hypothetical loop (HL) of residues 63–80 which represents a sequence in EPICs that is absent in tarocystatin. This loop contains two cysteine residues which are unlinked in the model but are likely connected by a disulphide bridge. The Ramachandran plot analysis showed one outlier (S80) in the EPIC1 model which locates at the end of the hypothetical insertion. Loops L1 and L2 which would interact with C14 are on the other side of EPIC1 and are very similar to those in the tarocystatin structure, presumably because the protein sequences align well in these regions.
A model for the EPIC1-C14 complex was generated by replacing papain and tarocystatin in the 3IMA structure with the models of C14 and EPIC1, respectively. As expected for this replacement strategy, EPIC1 docks perfectly into the substrate binding groove of C14 in a way that is typical for cystatins (Fig. 2B–D). The N-terminus occupies the unprimed substrate binding pockets and turns away from the catalytic Cys using the conserved glycine (G4) (Fig. 2C and D). Loop L1 (containing the QxVxG motif) binds above the catalytic Cys and occupies the primed substrate binding pockets, whereas loop L2 contains a conserved tryptophane (W92) that lies flat on the bottom of the hydrophobic substrate binding groove (Fig. 2C and D).
Three variant residues of C14 are able to directly interact with C14. E62Q or T210I might be able to interact with the N-terminus of EPIC proteins. However, the conformation of the EPIC1 N-terminus was not predicted by modelling and therefore two possible conformations are shown (Fig. 2E and F). The N-terminus contains several residues that are polymorphic between EPIC1 and EPIC2B (purple in Fig. 2E and F). K168ND is the other variant residue that may directly interact with loop L2 of EPIC1 (Fig. 2G). Interestingly, this codon was found to be under diversifying selection9 and encodes both basic (K) and acidic (D) residues which may have a severe effect on EPIC binding. The other variant residues in C14 and EPIC1 are not at the interaction interface and therefore unlikely to affect EPIC-C14 binding.
In conclusion, modelling of the EPIC-C14 interactions identified three regions where variant residues in C14 and EPICs may be the result of an arms race at the enzyme-inhibitor interface. The role of these residues can now be addressed using focussed site-directed mutagenesis and inhibition assays. The other variant residues in C14 are less likely to affect the EPIC-C14 interaction, but may affect interactions with other inhibitors. Cladosporium fulvum AVR2, for example, was found to be a weak inhibitor of C14,16 but may be a strong inhibitor for one of the C14 alleles. In addition, C14 was found to physically interact with RxRL effector AvrBlb2 (Bozkurt T, Schornack S, Win J, Shindo T, Oliva R, Cano LM, Jones AME, Huitema E, van der Hoorn RAL, Kamoun S, manuscript submitted) a class of effectors completely different from cystatins or AVR2. Therefore, since C14 is a common target for different effectors that probably share different interaction surfaces with C14, these interactions may have put different regions of C14 under selection pressure, causing the natural variation of C14 that we see in wild potato species today.
References
- 1.Misas-Villamil JC, Van der Hoorn RAL. Enzyme-inhibitor interactions at the plant-pathogen interface. Curr Opin Plant Biol. 2008;11:380–388. doi: 10.1016/j.pbi.2008.04.007. [DOI] [PubMed] [Google Scholar]
- 2.Rose JK, Ham KS, Darvill AG, Albersheim P. Molecular cloning and characterisation of glucanase inhibitor proteins: coevolution of a counter defense mechanism by plant pathogens. Plant Cell. 2002;14:1329–1345. doi: 10.1105/tpc.002253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bishop JG, Ripoll DR, Bashir S, Damasceno CM, Seeds JD, Rose JK. Selection on Glycine beta-1,3-endoglucanase genes differentially inhibited by a Phytophthora glucanase inhibitor protein. Genetics. 2005;169:1009–1019. doi: 10.1534/genetics.103.025098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Damasceno CM, Bishop JG, Ripoll DR, Win J, Kamoun S, Rose JK. Structure of the glucanase inhibitor protein (GIP) family from Phytophthora species suggests coevolution with plant endo-beta-1,3-glucanases. Mol Plant-Microbe Interact. 2008;21:820–830. doi: 10.1094/MPMI-21-6-0820. [DOI] [PubMed] [Google Scholar]
- 5.Debener T, Salamini F, Gebhardt C. Phylogeny of wild and cultivated Solanum species based on nuclear restriction fragment length polymorphisms (RFLPs) Theor Appl Genet. 1990;79:360–368. doi: 10.1007/BF01186080. [DOI] [PubMed] [Google Scholar]
- 6.Grünwald NJ, Flier WG. The biology of Phytophthora infestans at its center of origin. Annu Rev Phytopathol. 2005;43:171–190. doi: 10.1146/annurev.phyto.43.040204.135906. [DOI] [PubMed] [Google Scholar]
- 7.Tian M, Win J, Song J, Van der Hoorn RAL, Van der Knaap E, Kamoun S. A Phytophthora infestans cystatin-like protein interacts with and inhibits a tomato papain-like apoplastic protease. Plant Physiol. 2007;143:364–377. doi: 10.1104/pp.106.090050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Song J, Win J, Tian M, Schornack S, Kaschani F, Muhammad I, et al. Apoplastic effectors secreted by two unrelated eukaryotic plant pathogens target the tomato defense protease Rcr3. Proc Natl Acad Sci USA. 2009;106:1654–1659. doi: 10.1073/pnas.0809201106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaschani F, Shabab M, Bozkurt T, Shindo T, Schornack S, Gu C, et al. An effector-targeted protease contributes to defense against Phytophthora infestans and is under diversifying selection in natural hosts. Plant Physiol. 2010;154:1794–1804. doi: 10.1104/pp.110.158030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Avrova AO, Stewart HE, De Jong WD, Heilbronn J, Lyon GD, Birch PR. A cysteine protease gene is expressed early in resistant potato interactions with Phytophthora infestans. Mol Plant Microbe Interact. 1999;12:1114–1119. doi: 10.1094/MPMI.1999.12.12.1114. [DOI] [PubMed] [Google Scholar]
- 11.Shabab M, Shindo T, Gu C, Kaschani F, Pansuriya T, Chintha R, et al. Fungal effector protein AVR2 targets diversifying defence-related Cys proteases of tomato. Plant Cell. 2008;20:1169–1183. doi: 10.1105/tpc.107.056325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schwede T, Kopp J, Geux N, Peitsch MC. SWISS-MODEL: an automated protein homology-modeling server. Nucl Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Than ME, Helm M, Simpson DJ, Lottspeich F, Huber R, Gietl C. The 2.0 Å crystal structure and substrate specificity of the KDEL-tailed cysteine endopeptidase functioning in programmed cell death of Ricinus communis endosperm. J Mol Biol. 2004;336:1103–1116. doi: 10.1016/j.jmb.2003.12.075. [DOI] [PubMed] [Google Scholar]
- 14.Lovell SC, Davis IW, Arendall WB, III, De Bakker PIW, Word JM, Prisant MG, et al. Structure validation by Calpha geometry: phi, psi and Cbeta deviation. Prot Struct Funct Genet. 2002;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 15.Chu MH, Liu L, Yeh KW, Chen YS. Complex structure of tarocystatin and papain: implications for the inhibition property of group-2 phytocystatins. Planta. 2011 doi: 10.1007/s00425-011-1398-8. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Esse HP, Van't Klooster JW, Bolton MD, Yadeta KA, van Baarlen P, Boeren S, et al. The Cladosporium fulvum virulence protein Avr2 inhibits host proteases required for basal defense. Plant Cell. 2008;20:1948–1963. doi: 10.1105/tpc.108.059394. [DOI] [PMC free article] [PubMed] [Google Scholar]