Abstract
Root architecture is continuously shaped in a manner that helps plants to better adapt to the environment. Gene regulation at the transcriptional or posttranscriptional levels largely controls this environmental response. Recently, RNA silencing has emerged as an important player in gene regulation and is involved in many aspects of plant development, including lateral root formation. In a recent study, we found that FIERY1, a bifunctional abiotic stress and abscisic acid (ABA) signaling regulator and an endogenous RNA silencing suppressor, mediates auxin response during lateral root formation in Arabidopsis. We proposed that FRY1 regulates lateral root development through its activity on adenosine 3′,5′-bisphosphate (PAP), a strong inhibitor of exoribonucleases (XRNs). Interestingly, some of the phenotypes of fry1, such as enhanced response to light in repressing hypocotyl elongation and hypersensitivity to ABA in lateral root growth, are opposite to those of another light- and ABA-signaling mutant, hy5. Here we analyzed the hy5 fry1 double mutant for root and hypocotyl growth. We found that the hy5 mutation can suppress the enhanced light sensitivity in fry1 hypocotyl elongation and restore the lateral root formation. The genetic interaction between HY5 and FRY1 indicates that HY5 and FRY1 may act in overlapping pathways that mediate light signaling and lateral root development.
Key words: lateral root, RNA silencing, light signaling, hypocotyl, XRN4, PAP
Short, double-stranded RNAs repress gene expression by either DNA methylation, mRNA cleavage or mRNA translation blockage.1 The generation of ∼21–24 nucleotide-long RNA silencing triggers requires several conserved protein families, including DICER-LIKE (DCL), ARGONAUTE (AGO) and RNA-dependent RNA polymerase (RDR).2–4 Various developmental defects have been associated with mutations in this RNA silencing pathway.1,4–6 Among them, several miRNA and target mRNA pairs, including miR164 and NAC1,7 tasiRNA-ARF and ARF4,8 and miR160 and ARF17,9 have been shown to be involved in lateral root development. In a recent study,10 we reported that the bifunctional enzyme FRY1 controls lateral root architecture through suppression of RNA silencing. Our work demonstrated that (1) fry1 mutant seedlings generated significantly fewer lateral roots and also displayed a dramatically reduced sensitivity to auxin in lateral root induction; (2) lateral root phenotypes in fry1 may have resulted from loss of the nucleotidase activity, since overexpressing a yeast homolog of FRY1, which possesses only the 3′,5′-bisphosphate nucleotidase activity but not the inositol polyphosphate 1-phosphatase activity, rescues fry1 lateral root defects; (3) knock-out of RNA silencing suppressor XRN4, which is an exoribonuclease inhibited by the substrate of the FRY1 3′,5′-bisphosphate nucleotidase, exhibits similar lateral root defects; and (4) although fry1 mutant seedlings are relatively insensitive to ethylene, their lateral root developmental defects are not caused by the reduced ethylene response.
The FRY1 locus was initially defined in a genetic screen for mutants that exhibit altered induction of a stress- and ABA-responsive reporter gene.11 Null FRY1 mutants are hypersensitive to low temperature, salt and ABA for induction of the reporter gene as well as endogenous stress-responsive genes.11 Interestingly, a different allele isolated in the same screen showed enhanced induction, specifically to cold, of stress-responsive genes.12 The FRY1 gene encodes a bifunctional enzyme that possesses both inositol polyphosphate 1-phosphatase and 3′,(2′)5′-bisphosphatase activities.11 These activities are involved in the catabolism of inositol polyphosphates, such as inositol 1,4,5-trisphosphate (IP3) and 3′-phosphoadenosine-5′-phosphate (PAP), respectively.11–13 The inositol polyphosphate-1-phosphatase activity was initially considered responsible for stress gene regulation.11 A role of the bisphosphate nucleotidase activity in suppressing RNA silencing was later discovered in another genetic screen.14 It was proposed that this nucleotidase activity of FRY1 suppresses RNA silencing through removal of PAP, which is a strong inhibitor of exoribonucleases15 including the ubiquitously expressed XRN2, XRN3 and XRN4 in Arabidopsis.14,16
The mechanism by which enhanced RNA silencing causes defective lateral root formation in fry1 mutants is still unclear, and it may be due to a combination of effects. The enhanced cleavage of mRNA for auxin receptors or NAC1 by miR393 and miR164 in the fry1 mutant might account for some of the lateral root defects.10 It is known that the auxin responsive factor 4 (ARF4) positively regulates lateral root formation and its transcript is targeted by ta-siRNA-ARF.8 It is unclear how much the enhanced silencing of ARF4 could contribute to the lateral root phenotype of fry1, because the generation of ta-siRNA-ARF from TAS3 depends on the intact function of RNA-dependent RNA polymerase RDR6, and the rdr6 mutant has fewer lateral roots than wild type.8 Moreover, in our study, RT-PCR analysis did not detect a clear difference in ARF2 or ARF4 transcript levels between the wild-types and fry1 mutants (data not shown).
Besides their lateral root defects, fry1 mutants also exhibit pleiotropic phenotypes, including serrated leaves, short hypocotyls, retarded phase transition, delayed leaf initiation, increased basal jasmonic acid (JA) content and enhanced light responses.10,17–20 Previously, we reported that HY5, a well-characterized component in the light signal transduction pathway, also mediates ABA response during seed germination and lateral root development.21,22 Using the enhanced light response as a phenotype, we conducted a genetic screen and isolated fry1 suppressors with reduced light sensitivity.20 Map-based cloning of several of these suppressors identified that phyB and hy1 mutant alleles were among the suppressors of fry1 light sensitivity.20 Given the hypersensitivity of fry1 mutant to ABA11 and the enhanced light sensitivity of fry1, we asked whether there is any interaction between HY5-mediated light signaling and FRY1-mediated RNA silencing in modulating lateral root development. To do this, a double mutant of fry1 and hy5 was generated by genetic crossing and was analyzed for response to light and root growth characteristics.
We first examined the light response of fry1, hy5 and hy5 fry1 mutants by growing them on half-strength MS medium under dim light (10 µmols−1m−2). Figure 1A and B shows that fry1 mutants have shorter hypocotyls than the wild type and that the hy5 mutation can restore the hypocotyl length of fry1 to that of the wild.type. The shortened hypocotyl in fry1 may not be due to a general growth inhibition from its hypersensitivity to ABA, since hypocotyl length in dark-grown seedlings is about the same as that of the wild type (Fig. 1B). This is consistent with our discoveries that the light-response mutants such as phyB and hy1 can suppress light sensitivity of fry1.20 It was also recently reported that phyB when crossed into fry1 can rescue the fry1 hypocotyl defect.17
Figure 1.
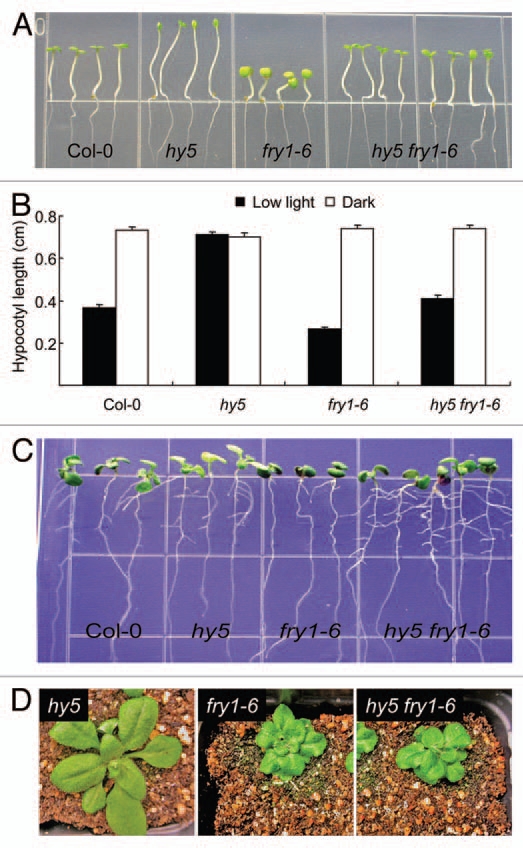
The genetic interaction between HY5 and FRY1 in regulating hypocotyl elongation and lateral root formation. (A) Seedlings of Col-0, hy5, fry1 and hy5 fry1 double mutant grown under dim light. The hy5 (SAL K_096651) and fry1–6 (SAL K_020882, previously incorrectly referred as fry1–4,10) were described in reference 10 and 21. Seeds were planted directly on half-strength MS medium plates and kept at 4°C for two days before incubating under dim light (10 µmols−1m−2, with a 16 h light period) at 22°C. Pictures were taken four days later. (B) Hypocotyl length of Col-0, hy5, fry1 and hy5 fry1 double mutant grown under dim light or dark at 22–24°C for three days. Data represent means and SE (n = 15). (C) Ten day-old seedlings of Col-0, hy5, fry1 and hy5 fry1 double mutant grown on a half-strength MS medium under continuous light at 22–24°C. (D) Morphology of three-week-old seedlings of hy5, fry1 and hy5 fry1 double mutant grown in soil at 22°C with a 16 h light period.
We then checked the lateral root induction in the hy5 fry1 mutant. It was found that the hy5 fry1 double mutant generated more lateral roots than the fry1 single mutant (Fig. 1C). Interestingly, although both phyB and hy1 mutants20 can partially rescue the compacted leaf morphology of fry1 (our unpublished data), hy5 mutation has no obvious effect on the leaf morphology of fry1 (Fig. 1D), indicating that HY5 as a light signaling regulator downstream of the photoreceptor PhyB mediates only a subset of the light signaling cascades that are mis-regulated in the fry1 mutant.
The complementation of fry1 lateral root development by MET22,10 a yeast homolog of FRY1 that cleaves only PAP but not IP3,23 and the resemblance of RNA-silencing suppressor double mutant xrn2 xrn3 to fry1 in shoot morphology14 indicate that the leaf morphological defects of fry1 are caused by its enhanced RNA silencing that arises due to repressed exoribonucleases. However, these exoribonucleases do not seem to function redundantly, since altered shoot morphology and delayed flowering are only observed in xrn3 or xrn2 xrn3 but not in xrn4 mutants,14,17 while the defective lateral root development is only displayed in xrn4 but not xrn2 or xrn3.10 Moreover, the xrn2 xrn3 double mutant has an enhanced response to light in hypocotyl elongation similar to fry1,17 while the xrn4 mutant does not (our unpublished data). It is therefore interesting to note that the hy5 mutant was able to restore both hypocotyl growth and lateral root formation to the fry1 mutant (Fig. 1A–C). These results suggest that HY5 may act downstream of FRY1 in a common or overlapping pathway(s) and that HY5 integrates both the XRN2/XRN3-mediated hypocotyl elongation and the XRN4-mediated lateral root formation pathways seen in fry1 mutant seedlings. Interestingly, a mutation in RNA silencing regulator AGO1 leads to hypersensitivity to light in hypocotyl elongation and reduced adventitious root formation, although there is no change in lateral root formation,24 implicating a diversification of AGO family members in their functionality. In the future, it will be interesting to reveal the detailed circuits and components involved in modulating hypocotyl and lateral root development as observed in fry1 and hy5 mutants.
Acknowledgements
This study was supported by the National Science Foundation grant #0446359 (to L.X.).
References
- 1.Meins F, Jr, Si-Ammour A, Blevins T. RNA silencing systems and their relevance to plant development. Annu Rev Cell Dev Biol. 2005;21:297–318. doi: 10.1146/annurev.cellbio.21.122303.114706. [DOI] [PubMed] [Google Scholar]
- 2.Baulcombe D. RNA silencing in plants. Nature. 2004;431:356–363. doi: 10.1038/nature02874. [DOI] [PubMed] [Google Scholar]
- 3.Matzke MA, Birchler JA. RNAi-mediated pathways in the nucleus. Nat Rev Genet. 2005;6:24–35. doi: 10.1038/nrg1500. [DOI] [PubMed] [Google Scholar]
- 4.Carrington JC, Ambros V. Role of microRNAs in plant and animal development. Science. 2003;301:336–338. doi: 10.1126/science.1085242. [DOI] [PubMed] [Google Scholar]
- 5.Bartel B, Bartel DP. MicroRNAs: at the root of plant development? Plant Physiol. 2003;132:709–717. doi: 10.1104/pp.103.023630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen X. Small RNAs and their roles in plant development. Annu Rev Cell Dev Biol. 2009;25:21–44. doi: 10.1146/annurev.cellbio.042308.113417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo HS, Xie Q, Fei JF, Chua NH. MicroRNA directs mRNA cleavage of the transcription factor NAC1 to downregulate auxin signals for Arabidopsis lateral root development. Plant Cell. 2005;17:1376–1386. doi: 10.1105/tpc.105.030841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yoon EK, Yang JH, Lim J, Kim SH, Kim SK, Lee WS. Auxin regulation of the microRNA390-dependent transacting small interfering RNA pathway in Arabidopsis lateral root development. Nucleic Acids Res. 2010;38:1382–1391. doi: 10.1093/nar/gkp1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mallory AC, Bartel DP, Bartel B. MicroRNA-directed regulation of Arabidopsis AUXIN RESPONSE FACTOR17 is essential for proper development and modulates expression of early auxin response genes. Plant Cell. 2005;17:1360–1375. doi: 10.1105/tpc.105.031716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen H, Xiong L. The bifunctional abiotic stress signaling regulator and endogenous RNA silencing suppressor FIERY1 is required for lateral root formation. Plant Cell Environ. 2010;33:2180–2190. doi: 10.1111/j.1365-3040.2010.02218.x. [DOI] [PubMed] [Google Scholar]
- 11.Xiong L, Lee B, Ishitani M, Lee H, Zhang C, Zhu JK. FIERY1 encoding an inositol polyphosphate 1-phosphatase is a negative regulator of abscisic acid and stress signaling in Arabidopsis. Genes Dev. 2001;15:1971–1984. doi: 10.1101/gad.891901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiong L, Lee H, Huang R, Zhu JK. A single amino acid substitution in the Arabidopsis FIERY1/HOS2 protein confers cold signaling specificity and lithium tolerance. Plant J. 2004;40:536–545. doi: 10.1111/j.1365-313X.2004.02225.x. [DOI] [PubMed] [Google Scholar]
- 13.Quintero FJ, Garciadeblas B, Rodriguez-Navarro A. The SAL1 gene of Arabidopsis, encoding an enzyme with 3′(2′),5′-bisphosphate nucleotidase and inositol polyphosphate 1-phosphatase activities, increases salt tolerance in yeast. Plant Cell. 1996;8:529–537. doi: 10.1105/tpc.8.3.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gy I, Gasciolli V, Lauressergues D, Morel JB, Gombert J, Proux F, et al. Arabidopsis FIERY1, XRN2 and XRN3 are endogenous RNA silencing suppressors. Plant Cell. 2007;19:3451–3461. doi: 10.1105/tpc.107.055319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dichtl B, Stevens A, Tollervey D. Lithium toxicity in yeast is due to the inhibition of RNA processing enzymes. EMBO J. 1997;16:7184–7195. doi: 10.1093/emboj/16.23.7184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gazzani S, Lawrenson T, Woodward C, Headon D, Sablowski R. A link between mRNA turnover and RNA interference in Arabidopsis. Science. 2004;306:1046–1048. doi: 10.1126/science.1101092. [DOI] [PubMed] [Google Scholar]
- 17.Kim BH, von Arnim AG. FIERY1 regulates lightmediated repression of cell elongation and flowering time via its 3′(2′),5′-bisphosphate nucleotidase activity. Plant J. 2009;58:208–219. doi: 10.1111/j.1365-313X.2008.03770.x. [DOI] [PubMed] [Google Scholar]
- 18.Robles P, Fleury D, Candela H, Cnops G, Alonso-Peral MM, Anami S, et al. The RON1/FRY1/SAL1 gene is required for leaf morphogenesis and venation patterning in Arabidopsis. Plant Physiol. 2009;152:1357–1372. doi: 10.1104/pp.109.149369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rodriguez VM, Chetelat A, Majcherczyk P, Farmer EE. Chloroplastic phosphoadenosine phosphosulfate (PAPS) metabolism regulates basal levels of the prohormone jasmonic acid in Arabidopsis leaves. Plant Physiol. 2010;152:1335–1345. doi: 10.1104/pp.109.150474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peterson E, Xiong L. Molecular cloning and characterization of fiery1 suppressors. J Undergrad Res. 2006;5:21–30. [Google Scholar]
- 21.Chen H, Zhang J, Neff MM, Hong SW, Zhang H, Deng XW, et al. Integration of light and abscisic acid signaling during seed germination and early seedling development. Proc Natl Acad Sci. 2008;105:4495–4500. doi: 10.1073/pnas.0710778105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen H, Xiong L. Role of HY5 in abscisic acid response in seeds and seedlings. Plant Signal & Behav. 2008;3:986–988. doi: 10.4161/psb.6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murguia JR, Belles JM, Serrano R. A salt-sensitive 3′(2′),5′-bisphosphate nucleotidase involved in sulfate activation. Science. 1995;267:232–234. doi: 10.1126/science.7809627. [DOI] [PubMed] [Google Scholar]
- 24.Sorin C, Bussell JD, Camus I, Ljung K, Kowalczyk M, Geiss G, et al. Auxin and light control of adventitious rooting in Arabidopsis require ARGONAUTE1. Plant Cell. 2005;17:1343–1359. doi: 10.1105/tpc.105.031625. [DOI] [PMC free article] [PubMed] [Google Scholar]


