Abstract
The covalent addition of Small Ubiquitin-Like Modifier (SUMO) to various intracellular proteins is an essential regulatory step in most eukaryotes. Due to its necessity and the large number of putative targets, SUMO is thought to be second only to ubiquitin (Ub) among Ub-fold proteins in terms of regulatory influence. Whereas, ubiquitylation (i.e., the attachment of Ub) is generally associated with protein degradation, SUMOylation appears to have more diverse consequences, including the regulation of transcription, chromatin structure/accessibility, nuclear import and various protein-protein interactions, and even appears to block the action of Ub by competing for the same binding sites on targets.1–3 Paramount to understanding SUMO function(s) is knowing the complete catalog of SUMO targets. In the following addendum we review our recent publication4 describing the proteomic identification of SUMO substrates in the model plant, Arabidopsis thaliana, and expand our analyses with regard to the changes in SUMOylation patterns that are induced by heat stress. Collectively, our data indicate that SUMOylation is highly dynamic with evidence that SUMO addition globally modifies transcription and chromatin accessibility, especially during stress.
Key words: SUMO, Arabidopsis, proteomics, transcription, chromatin, ubiquitin
SUMOylation of accessible lysines on target proteins is mediated by a sequential three-enzyme conjugation pathway, which in plants is comprised of an E1 activating enzyme heterodimer (SAE1a or b combined with SAE2), a single E2 conjugating enzyme (SCE1), and at least two E3 ligases (SIZ1 and MMS21/HPY2).5–8 Often but not always, SUMO becomes bound within a SUMO-acceptor motif—ΨKxE, where Ψ is a large hydrophobic residue, K is the modified lysine, x is any residue and E is a glutamic acid.9,10 In plants, the SUMO pathway is essential,11 and has been shown genetically to control multiple cellular processes, including the response to various stresses, cell division, flowering time, pathogen defense and signaling by the hormones ABA and salicylic acid.1,7,12,13 Of special interest are the observations that various abiotic stresses induce a substantial and reversible rise in SUMO1/2 conjugates, presumably as a way to protect plants from environmental insults.5,11,14,15
Prior to our proteomic analyses, only a few SUMO substrates were described in plants (e.g., ABI5, PHR1, ICE1 and FLD1,12). To expand this list, we took a proteomic approach, similar to those using yeast and mammalian cell cultures,16,17 to identify SUMO substrates both before and during stress. Key to our success was the creation of an Arabidopsis line in which the essential SUMO1/2 isoforms were replaced with a tagged variant (6His-SUMO1-H89R) designed for faithful rescue, stringent purification, and the ability to more easily map SUMO attachment sites by mass spectrometry (MS).4 SUMO conjugates were then purified from these 6His-SUMO1-H89R sum1-1 sum2-1 plants by a three-step affinity procedure that employed strong denaturants (8 M urea/7 M guanidine HCl) to minimize contaminants and proteins that bind non-covalently to SUMO or to SUMOylated proteins. The enriched samples from non-stressed plants or plants subjected to heat or oxidative stress were then analyzed by tandem MS to identify SUMO substrates generated in planta.
In total, we identified 357 high probability SUMO substrates in Arabidopsis, which upon inspection provide a number of important insights into SUMO functions in plants.4 First, in agreement with cell fractionation studies,11 most of the SUMO substrates identified reside in the nucleus. Consistent with this location, many are known or predicted to regulate nuclear activities, including roles in transcription, chromatin remodeling, DNA/RNA modifications, DNA repair and nuclear pore shuttling. Second, many of these Arabidopsis targets overlap with those identified in yeast and metazoans with the list similarly enriched for protiens harboring domains common among DNA/RNA interactors and chromatin modifiers.16 This connection strongly suggests that SUMOylation modifies a conserved set of targets and their associated processes in eukaryotes. Third, the SUMO pathway itself is a main target of SUMOylation, which may have important regulatory consequences.18,19 The detection of SUMO footprints on SUMO1 also unequivocally demonstrates that poly-SUMO chains are assembled in plants. Fourth, we found that the Arabidopsis conjugation machinery likely uses the same SUMO-acceptor sites as in other eukaryotes.9,10 Our substrate list was similarly enriched for proteins with predicted ΨKxE motifs. And for sites mapped via our engineered SUMO MS footprint, ∼60% conformed to the ΨKxE sequence.4 Sequences surrounding the remaining SUMO acceptors sites were highly divergent, indicating that a variety of other recognition motifs are possible. And fifth, our detection of proteins that are both SUMOylated and ubiquitylated revealed an intriguing crosstalk between the SUMO and Ub pathways as has been seen in other eukaryotes.20–22 These SUMO+Ub-modified proteins dramatically increase upon heat stress, with our mapping studies suggesting that some Ubs are directly attached to the SUMO moieties.4 An intriguing possibility is that this Ub addition directs the degradation of a subset of SUMOylated proteins with the SUMO moiety providing the signal for ubiquitylation.
Arabidopsis like other eukaryotes examined,16,23,24 dramatically and reversibly increase the levels of SUMOylated proteins upon various abiotic stresses.4,5,11 This increase involves the SUMO1/2 isoforms specifically and requires the SIZ1 E3,11 which may explain the pleiotropic phenotype of siz1 null mutants and their sensitivity to stress.8,11,12,14,25 One goal of our study was to identify the changes in Arabidopsis SUMOylation patterns during heat stress. As can be seen in Figure 1, a 37°C heat stress for 30 minutes increases the level of SUMO1/2 conjugates by 3 fold (measure 30 minutes later at the peak), which is paralleled by a commensurate drop in the level of free SUMO1/2.
Figure 1.
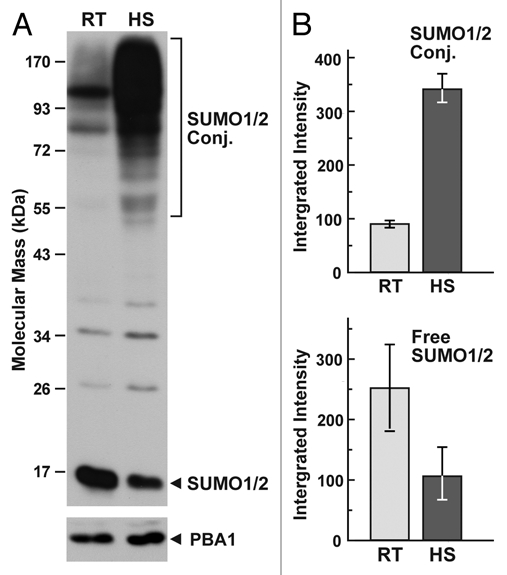
Effect of heat stress on the levels of free SUMO and SUMO conjugates in Arabidopsis. (A) Profile of SUMO conjugates in seedlings before and after heat stress. Seven-day-old seedlings were grown at 24°C, exposed to 37°C for 30 min, and then returned to 24°C for 30 min before harvest. Crude protein extracts were subjected to SDS-PAGE and immunoblot analysis with anti-SUMO1 antibodies. Levels of the proteasome subunit PBA1 were used to confirm equal loading. HS, heat-stressed seedlings. RT, seedlings not exposed to heat stress. (B) Quantification of high molecular mass SUMO conjugates and free SUMO. Levels of each fraction were quantified by densitometric scanning of the immunoblot membranes probed with anti-SUMO1 antibodies. Error bars represent the standard deviation of three independent heat-stress experiments.
Semi-quantitative analysis of individual SUMO conjugates indicates that heat stress can dynamically affect their levels. Here, we compared the spectral counts obtained from MS precursor scans of specific peptides from non-stressed and heat-stressed seedling, which were normalized by the number of total spectral counts. In total, the analyses combined datasets from five independent MS experiments (three heat-stress and two non-stress datasets). Given the likely role of SUMO in regulating transcription,26 we expected to see a significant over-representation of transcription factors and chromatin remolding complexes in the heat-stress versus nonstress datasets. Surprisingly, most transcription factors, including some HSFs were present in both. However, a few were significantly increased upon heat stress. For example, the TOPLESS (TPL) and SEUSS (SEU) corepressors and several components of the SWI/SNF chromatin modification complex (PICKLE, SWI3C and CHR11), which have important regulatory roles in plant development,27,28 were substantially more SUMOylated during heat stress than expected (Fig. 2A). We also discovered a collection of DNA repair components that were over-represented in the heat-stress MS datasets. Of the 12 SUMO targets we detected that have been directly or indirectly connected to DNA repair, seven were found exclusively in the heat-stress dataset (LIG1, DRT111, POLD3, RPA1a, RFC1, ATTRP1 and RPA70d) and another (KU80) had substantially higher spectral counts than the average after heat stress (Fig. 2A). It is worth noting that despite a global increase in SUMOylation during heat stress, the SUMOylation levels of some targets decreased (e.g., histone H2b, SUVH9 and ANACO51) (Fig. 2B). The underpinning mechanism(s) behind the changes in SUMOylation patterns remains to be determined. Increased SUMOylation of specific targets could be driven by activation of the SUMO conjugation machinery and/or inhibition of the de-SUMOylation machinery, whereas decreased SUMOylation could be driven by inverse changes as well as by conjugate breakdown as is possible for SUMO+Ub conjugates.
Figure 2.
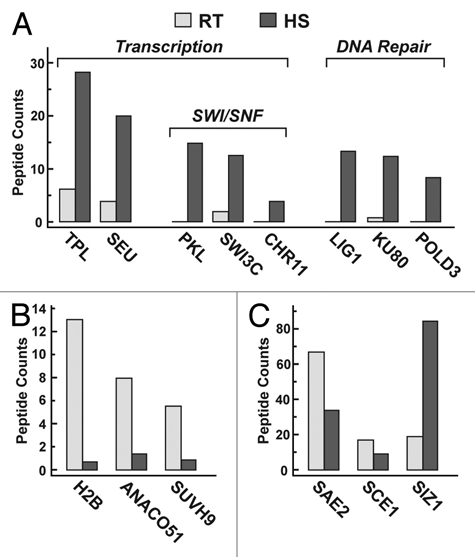
Effect of heat stress on the SUMOylation levels of representative Arabidopsis targets. Seven-day-old seedlings were subjected to heat stress as in Figure 1. HS, heat-stressed seedlings. RT, seedlings not exposed to heat stress. Levels of each protein in the RT and HS MS datasets were estimated by the number of spectral counts in the MS precursor scans that were obtained for each target; these numbers were then normalized by the total number of spectral counts for each MS run. (A) SUMOylated transcription and DNA repair components that increase in abundance during heat stress. (B) SUMOylated proteins that decrease in abundance during heat stress. (C) Members of the SUMO conjugation pathway.
Finally, our data identify a potentially interesting autoregulatory loop within the SUMO pathway itself. Whereas the SUMOylation status of the SAE2 E1 subunit and the SCE1 E2 decreases during heat stress, the SUMOylation status of the SIZ1 E3 rises substantially (Fig. 2C). Given the observation that the target preference of the mammalian SCE1 ortholog—UBC9 can be dramatically altered by SUMOylation,19 it is possible that this auto-SUMOyation of the Arabidopsis E1-E2-E3 cascade represents a mechanism to alter the profile of SUMO addition during plant stress.
In conclusion, our proteomic studies revealed that SUMOyation is a remarkably dynamic process that affects a wide range of nuclear targets in Arabidopsis. Its ability to modify many components important for transcription and chromatin assembly suggests that it represents an important epigenetic mark to globally affect gene expression and chromatin stability. Moreover, our discovery of proteins containing both SUMO and Ub identifies an intriguing interplay between these two post-translational modifiers. Certainly, a more complete understanding of the dynamics of SUMOylation, especially during stress, will be essential to fully understand the exact role(s) of SUMO. While MS spectra counts can provided a rudimentary picture, more quantitative MS methods will be needed to better clarify the role of stress-induced SUMOylation on specific substrates.
Acknowledgements
We thank Gregory Barrett-Wilt for help with the MS analysis. This work was supported by a grant (MCB 0929395) from the US National Science Foundation-Arabidopsis 2010 Program to R.D.V.
References
- 1.Miura K, Jin JB, Hasegawa PM. SUMOylation, a post-translational regulatory process in plants. Curr Opin Plant Biol. 2007;10:495–502. doi: 10.1016/j.pbi.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 2.Geiss-Friedlander R, Melchior F. Concepts in SUMOylation: a decade on. Nat Rev Mol Cell Biol. 2007;8:947–956. doi: 10.1038/nrm2293. [DOI] [PubMed] [Google Scholar]
- 3.Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol. 2006;22:159–180. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- 4.Miller MJ, Barrett-Wilt GA, Hua Z, Vierstra RD. Proteomic analyses identify a diverse array of nuclear processes affected by small ubiquitin-like modifier conjugation in Arabidopsis. Proc Natl Acad Sci USA. 2010;107:16512–16517. doi: 10.1073/pnas.1004181107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kurepa J, Walker JM, Smalle J, Gosink MM, Davis SJ, Durham TL, et al. The small ubiquitin-like modifier (SUMO) protein modification system in Arabidopsis. Accumulation of SUMO1 and -2 conjugates is increased by stress. J Biol Chem. 2003;278:6862–6872. doi: 10.1074/jbc.M209694200. [DOI] [PubMed] [Google Scholar]
- 6.Huang L, Yang S, Zhang S, Liu M, Lai J, Qi Y, et al. The Arabidopsis SUMO E3 ligase AtMMS21, a homologue of NSE2/MMS21, regulates cell proliferation in the root. Plant J. 2009;60:666–678. doi: 10.1111/j.1365-313X.2009.03992.x. [DOI] [PubMed] [Google Scholar]
- 7.Ishida T, Fujiwara S, Miura K, Stacey N, Yoshimura M, Schneider K, et al. SUMO E3 ligase HIGH PLOIDY2 regulates endocycle onset and meristem maintenance in Arabidopsis. Plant Cell. 2009;21:2284–2297. doi: 10.1105/tpc.109.068072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miura K, Rus A, Sharkhuu A, Yokoi S, Karthikeyan AS, Raghothama KG, et al. The Arabidopsis SUMO E3 ligase SIZ1 controls phosphate deficiency responses. Proc Natl Acad Sci USA. 2005;102:7760–7765. doi: 10.1073/pnas.0500778102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rodriguez MS, Dargemont C, Hay RT. SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J Biol Chem. 2001;276:12654–12659. doi: 10.1074/jbc.M009476200. [DOI] [PubMed] [Google Scholar]
- 10.Matic I, van Hagen M, Schimmel J, Macek B, Ogg SC, Tatham MH, et al. In vivo identification of human small ubiquitin-like modifier polymerization sites by high accuracy mass spectrometry and an in vitro to in vivo strategy. Mol Cell Proteomics. 2008;7:132–144. doi: 10.1074/mcp.M700173-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saracco SA, Miller MJ, Kurepa J, Vierstra RD. Genetic analysis of SUMOylation in Arabidopsis: conjugation of SUMO1 and SUMO2 to nuclear proteins is essential. Plant Physiol. 2007;145:119–134. doi: 10.1104/pp.107.102285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miura K, Lee J, Jin JB, Yoo CY, Miura T, Hasegawa PM. SUMOylation of ABI5 by the Arabidopsis SUMO E3 ligase SIZ1 negatively regulates abscisic acid signaling. Proc Natl Acad Sci USA. 2009;106:5418–5423. doi: 10.1073/pnas.0811088106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van den Burg HA, Kini RK, Schuurink RC, Takken FLW. Arabidopsis small ubiquitin-like modifier paralogs have distinct functions in development and defense. Plant Cell. 2010;22:1998–2016. doi: 10.1105/tpc.109.070961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miura K, Jin JB, Lee J, Yoo CY, Stirm V, Miura T, et al. SIZ1-mediated SUMOylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis. Plant Cell. 2007;19:1403–1414. doi: 10.1105/tpc.106.048397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Conti L, Price G, O'Donnell E, Schwessinger B, Dominy P, Sadanandom A. Small ubiquitin-like modifier proteases OVERLY TOLERANT TO SALT1 and -2 regulate salt stress responses in Arabidopsis. Plant Cell. 2008;20:2894–2908. doi: 10.1105/tpc.108.058669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Golebiowski F, Matic I, Tatham MH, Cole C, Yin Y, Nakamura A, Cox J, et al. System-wide changes to SUMO modifications in response to heat shock. Sci Signal. 2009;2:24. doi: 10.1126/scisignal.2000282. [DOI] [PubMed] [Google Scholar]
- 17.Denison C, Rudner AD, Gerber SA, Bakalarski CE, Moazed D, Gygi SP. A proteomic strategy for gaining insights into protein SUMOylation in yeast. Mol Cell Proteomics. 2005;4:246–254. doi: 10.1074/mcp.M400154-MCP200. [DOI] [PubMed] [Google Scholar]
- 18.Bossis G, Melchior F. Regulation of SUMOylation by reversible oxidation of SUMO conjugating enzymes. Mol Cell. 2006;21:349–357. doi: 10.1016/j.molcel.2005.12.019. [DOI] [PubMed] [Google Scholar]
- 19.Knipscheer P, Flotho A, Klug H, Olsen JV, van Dijk WJ, Fish A, et al. Ubc9 SUMOylation regulates SUMO target discrimination. Mol Cell. 2008;31:371–382. doi: 10.1016/j.molcel.2008.05.022. [DOI] [PubMed] [Google Scholar]
- 20.Bergink S, Jentsch S. Principles of ubiquitin and SUMO modifications in DNA repair. Nature. 2009;458:461–467. doi: 10.1038/nature07963. [DOI] [PubMed] [Google Scholar]
- 21.Xie Y, Kerscher O, Kroetz MB, McConchie HF, Sung P, Hochstrasser M. The yeast Hex3/Slx8 heterodimer is a ubiquitin ligase stimulated by substrate SUMOylation. J Biol Chem. 2007;282:34176–34184. doi: 10.1074/jbc.M706025200. [DOI] [PubMed] [Google Scholar]
- 22.Tatham MH, Geoffroy MC, Shen L, Plechanovova A, Hattersley N, Jaffray EG, et al. RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat Cell Biol. 2008;10:538–546. doi: 10.1038/ncb1716. [DOI] [PubMed] [Google Scholar]
- 23.Yoo CY, Miura K, Jin JB, Lee J, Park HC, Salt DE, et al. SIZ1 small ubiquitin-like modifier E3 ligase facilitates basal thermotolerance in Arabidopsis independent of salicylic acid. Plant Physiol. 2006;142:1548–1558. doi: 10.1104/pp.106.088831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tempe D, Piechaczyk M, Bossis G. SUMO under stress. Biochem Soc Trans. 2008;36:874–878. doi: 10.1042/BST0360874. [DOI] [PubMed] [Google Scholar]
- 25.Miura K, Lee J, Miura T, Hasegawa PM. SIZ1 controls cell growth and plant development in Arabidopsis through salicylic acid. Plant Cell Physiol. 2010;51:103–113. doi: 10.1093/pcp/pcp171. [DOI] [PubMed] [Google Scholar]
- 26.Gill G. Something about SUMO inhibits transcription. Curr Opin Genet Dev. 2005;15:536–541. doi: 10.1016/j.gde.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 27.Liu Z, Karmarkar V. Groucho/Tup1 family corepressors in plant development. Trends Plant Sci. 2008;13:137–144. doi: 10.1016/j.tplants.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 28.Ogas J, Kaufmann S, Henderson J, Somerville C. PICKLE is a CHD3 chromatin-remodeling factor that regulates the transition from embryonic to vegetative development in Arabidopsis. Proc Natl Acad Sci USA. 1999;96:13839–13844. doi: 10.1073/pnas.96.24.13839. [DOI] [PMC free article] [PubMed] [Google Scholar]


