Abstract
The lysosomal endoprotease cathepsin D (CatD) is an essential player in general protein turnover and specific peptide processing. CatD-deficiency is associated with neurodegenerative diseases, whereas elevated CatD levels correlate with tumor malignancy and cancer cell survival. Here, we show that the CatD ortholog of the budding yeast Saccharomyces cerevisiae (Pep4p) harbors a dual cytoprotective function, composed of an anti-apoptotic part, conferred by its proteolytic capacity, and an anti-necrotic part, which resides in the protein's proteolytically inactive propeptide. Thus, deletion of PEP4 resulted in both apoptotic and necrotic cell death during chronological aging. Conversely, prolonged overexpression of Pep4p extended chronological lifespan specifically through the protein's anti-necrotic function. This function, which triggered histone hypoacetylation, was dependent on polyamine biosynthesis and was exerted via enhanced intracellular levels of putrescine, spermidine and its precursor S-adenosyl-methionine. Altogether, these data discriminate two pro-survival functions of yeast CatD and provide first insight into the physiological regulation of programmed necrosis in yeast.
Keywords: chronological aging, yeast apoptosis, yeast necrosis, spermidine
Cathepsin D (CatD) is an aspartic endoprotease that is ubiquitously present in lysosomes of all mammalian cell types. Together with other members of the cathepsin family, it has a central role in general protein turnover and participates in tissue remodeling, as well as in the processing of antigens, hormones and neuropeptides.1 Mutations in CatD cause human neuronal ceroid lipofuscinoses, a group of pediatric neurodegenerative diseases collectively known as Batten Disease.1, 2
CatD seems to have a pleiotropic role with respect to programmed cell death (PCD). On the one hand, overexpression and hypersecretion of its catalytically inactive zymogene (pro-CatD) has been connected to increased tumor cell survival.3, 4, 5, 6 On the other hand, CatD has also been shown to promote cell death in response to various conditions.4, 7, 8, 9 Whether the proteolytic activity of CatD is required for its lethal function, however, remains a matter of debate.4 Thus, the differential contribution of pro-CatD and CatD to cell survival and cell death is not yet clarified and may depend on the specific cellular context.
During the last years, it has become apparent that the core machinery executing PCD is phylogenetically conserved, establishing the budding yeast Saccharomyces cerevisiae as a valuable model organism to study apoptosis and necrosis.10, 11, 12, 13 The yeast genome encodes orthologs of central mammalian apoptotic proteins, including one caspase (Yca1p), the serine protease OMI (Nma111p) or endonuclease G (Nuc1p).14, 15, 16 Yeast cell death has been linked to complex apoptotic scenarios including mitochondrial outer membrane permeabilization with cytochrome c release and histone H2B phosphorylation that also participate in PCD of mammalian cells.17, 18 Finally, chronological aging, which is tightly associated with PCD, has been instrumental in exploring phylogenetically conserved aging pathways that are applicable to postmitotic mammalian cells.19 During chronological aging, yeast cells eventually die as they exhibit biochemical and morphological markers of both apoptosis and necrosis.20, 21, 22 Importantly, here necrosis largely occurs in a coordinated manner and is not an uncontrolled process as known from ‘classical necrosis'.11 Thus, ‘programmed necrosis' represents (besides apoptosis) a further PCD subroutine present in yeast.10, 11
Driven by the aforementioned considerations, we decided to investigate the role of Pep4p (also called yeast proteinase A), the yeast CatD ortholog,23 on yeast aging and cell death. Using chronological aging as a means to physiologically induce necrotic cell death, we show that prolonged overexpression of Pep4p promotes survival by specifically reducing necrosis. We demonstrate that PEP4 deletion exacerbates both apoptosis and necrosis during chronological aging. Suppression of apoptosis by Pep4p relies on its proteolytic activity, whereas the repression of necrosis is mediated by the proteolytically inactive Pep4p propeptide, which promotes survival via a novel pathway, the stimulation of spermidine biosynthesis.
Results
Deletion of PEP4 leads to combined apoptotic and necrotic cell death during chronological aging
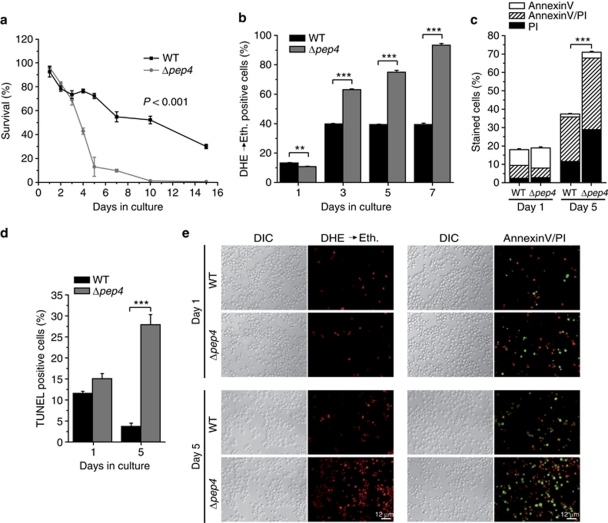
Pep4p is an essential player in the S. cerevisiae vacuolar proteolytic system, where it is important for both protein degradation and maturation of several vacuolar hydrolases.23, 24 Consistently, mutants deficient in PEP4 (Δpep4) exhibit reduced protein degradation and enhanced loss of viability under conditions of nutritional stress.24 Therefore, we first assessed the survival rate of Δpep4 during chronological aging, in which nutrients are limited. Starting at day 3 of aging, Δpep4 cells exhibited a dramatic decrease in clonogenic survival compared with the wild type (Figure 1a). This accelerated mortality was accompanied by an increased formation of reactive oxygen species (ROS; Figures 1b and e), as indicated by the enhanced superoxide-driven conversion of non-fluorescent dihydroethidium (DHE) into fluorescent ethidium (Eth). We next evaluated phosphatidylserine externalization to the outer leaflet of the plasma membrane, a typical phenotypic feature of apoptotic cells, which can be monitored by binding of fluorescein isothiocyanat (FITC)-conjugated Annexin V to the externalized phosphatidylserine. In parallel, we assessed loss of membrane integrity, a characteristic marker of necrosis, by examining membrane permeability with propidium iodide (PI). Annexin V/PI co-staining (Figures 1c and e) allowed discrimination between early apoptosis (only Annexin V stained cells), late apoptosis leading to secondary necrosis (double stained cells) and primary necrosis (only PI-stained cells).11 The Δpep4 cells showed a combined increase in apoptotic and necrotic populations (Figures 1c and e). In addition, TdT-mediated dUTP-biotin nick end labeling (TUNEL)-staining revealed enhanced apoptotic DNA fragmentation, compared with the wild-type control (Figure 1d). In conclusion, disruption of PEP4 exacerbates both apoptosis and necrosis during chronological aging.
Figure 1.
Premature death of Δpep4 cells displays markers of apoptosis and necrosis. (a) Survival determined by clonogenicity of wild type and Δpep4 cells during chronological aging. Data indicate means±S.E.M (n=4). (b) ROS production at days 1, 3, 5 and 7 of wild type and Δpep4 cells during chronological aging as determined via DHE → Eth conversion. Data in B to D represent means±S.E.M. (n=3; **P<0.01, ***P<0.001). In each experiment, 30 000 cells were evaluated using flow cytometry. (c) Phosphatidylserine externalization and loss of membrane integrity of wild type and Δpep4 cells at days 1 and 5 of chronological aging as determined using Annexin V/PI costaining. (d) DNA fragmentation of wild type and Δpep4 strains at days 1 and 5 of chronological aging as determined via TUNEL staining. (e) Fluorescence microscopic sample pictures of DHE and Annexin V/PI staining of wild type and Δpep4 cells at days 1 and 5 of the experiments shown in B and C. DIC, differential interference contrast
Overexpressed Pep4p exhibits a proteolytically independent cytoprotective function during chronological aging
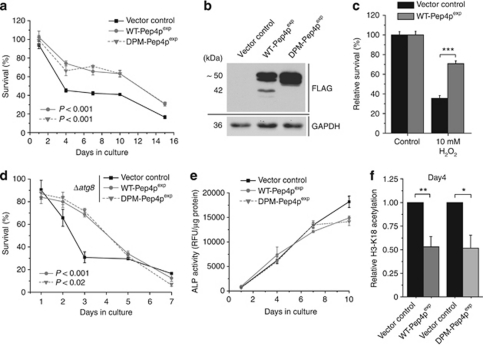
Ectopic overexpression of wild-type Pep4p (WT-Pep4p) throughout the aging experiments (Supplementary Figure S1A) significantly promoted survival, at least during the early phase of chronological aging (Figures 2a and b). In addition, overexpression of Pep4p significantly protected yeast cells from hydrogen peroxide-induced cell death (Figure 2c), whereas PEP4 deletion resulted in the opposite phenotype (Supplementary Figure S1B). This aligns the cytoprotective effect of Pep4p with the commonly observed increased resistance to oxidative stress in long-lived mutants of several model organisms, including Caenorhabditis elegans, Drosophila melanogaster, mice and yeast.25, 26
Figure 2.
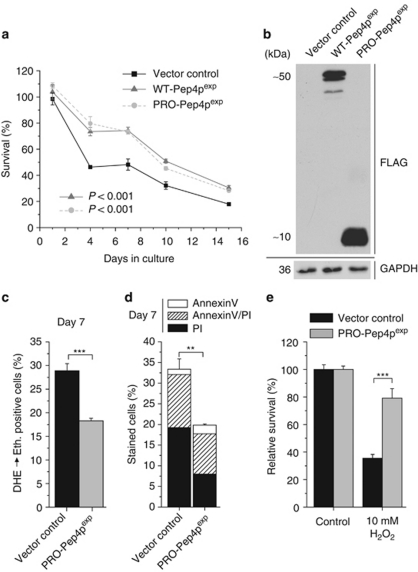
Overexpression of Pep4p or a proteolytically inactive mutant both extend chronological lifespan, which correlates with hypoacetylation of histone H3. (a) Survival determined by clonogenicity of wild-type yeast cells overexpressing WT-Pep4p or DPM-Pep4p and survival of corresponding vector control during chronological aging. Data represent means±S.E.M. (n=4). (b) Immunoblots of whole cell extracts of wild-type cells overexpressing FLAG-tagged WT-Pep4p, FLAG-tagged DPM-Pep4p and the corresponding vector control after 20 h of induction. Blots were probed with antibodies against FLAG epitope or glyceraldehyde-3-phosphate-dehydrogenase as a loading control. (c) Survival of wild-type cells overexpressing WT-Pep4p or harboring the vector control after 40 h of induction and subsequent treatment, with or without hydrogen peroxide (H2O2) for 4 h. Survival of the corresponding controls was set to 100%. Data represent means±S.E.M. (n=10 for controls, n=7 for treated cultures; ***P<0.001). (d) Survival of Δatg8 cells overexpressing WT-Pep4p or DPM-Pep4p and survival of vector control during chronological aging. Data represent means±S.E.M. (n=4). (e) Relative ALP activity indicative of autophagy during chronological aging of wild-type cells overexpressing WT-Pep4p, DPM-Pep4p or harboring the corresponding vector control. Data represent means±S.E.M. (n=4). (f) Relative acetylation of the H3 lysine residue K18 of wild-type cells overexpressing WT-Pep4p, DPM-Pep4p or harboring the vector control at day 4 of chronological aging as determined by quantification of immunoblot analyses. Data represent means±S.E.M. (n=5 for WT-Pep4p, n=3 for DPM-Pep4p; *P<0.05, **P<0.01). See also Supplementary Figure S1
Next, we generated a Pep4p-variant with two point mutations in its catalytically active aspartic residues (D109A, D294A) abrogating its proteolytic activity, as well as its ability to stabilize proteinase B (Prb1p), a key enzyme in the vacuolar degradation system.27 Surprisingly, the double-point mutant (DPM-Pep4p) was undistinguishable from WT-Pep4p, with regard to its pro-survival effect (Figures 2a and b). Of note, overexpression of Pep4p conferred a comparable rescuing effect in the deletion background of PRB1, further evidencing that the observed pro-survival effect is independent from Prb1p stabilization (Supplementary Figure S1C). In conclusion, Pep4p improves the survival of aging yeast cells independently of its proteolytic activity.
Pep4p is a crucial player in autophagy,28 a pro-survival degradative pathway central during nutrient starvation and closely associated to longevity in different organisms including yeast.29 However, overexpression of WT-Pep4p or DPM-Pep4p in a mutant lacking ATG8, which is critical for proper autophagosome formation, still retained its cytoprotective effects (Figure 2d). Autophagy deficiency accelerated cell death during chronological aging, but, at least in the early phase of chronological aging, did not prevent Pep4p-mediated survival increase (Figure 2d) or reduction of ROS accumulation (Supplementary Figure S1D). Autophagy-independency was verified using an established biochemical assay that follows the exclusively autophagy-driven vacuolar delivery and subsequent activation of a truncated form of the yeast alkaline phosphatase (ALP). As expected, overall autophagy was enhanced with ongoing aging (Figure 2e). However, overexpression of WT-Pep4p or DPM-Pep4p did not result in an increased ALP-activity, compared with the control, indicating no additional autophagy induction accompanying the pro-survival effect (Figure 2e). Thus, Pep4p overexpression protects cells from aging-induced cell death in an autophagy-independent manner.
Pep4p overexpression decreases histone H3 acetylation.
Histone deacetylation constitutes the epigenetic chromatin modification that is most consistently associated with lifespan extension and healthy aging in a wide range of organisms.20, 30 Indeed, overexpression of WT-Pep4p and DPM-Pep4p diminished acetylation of histone H3 (Figure 2f). Reduced acetylation of histone residues has been associated with lifespan extension of yeast in both replicative31 and chronological aging,20 which model the aging process in dividing cells and post-mitotic cells of higher eukaryotes, respectively. However, the mechanisms underlying hypoacetylation in each model seem to differ, as SIR2, a histone deacetylase of the sirtuin family, promotes lifespan during replicative aging, but has the opposite effects on chronological lifespan.32 Consistently, deletion of SIR2 did not affect the pro-survival effect of Pep4p during chronological aging (Supplementary Figure S1E). In conclusion, Pep4p-mediated lifespan extension during chronological aging seems to correlate with Sir2p-independent histone deacetylation.
Pep4p promotes survival by inhibiting necrosis
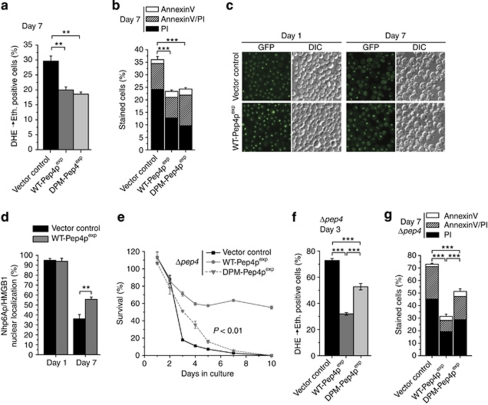
Both apoptosis and necrosis contribute to cell death with ongoing aging.20, 21, 22 The lifespan extension mediated by overexpressed WT-Pep4p or DPM-Pep4p was accompanied by diminished ROS production (Figure 3a) and a prominent reduction in primary necrotic events, yet only slight reduction in apoptosis (Figure 3b, Supplementary Figure S2A). We further evaluated the ability of prolonged Pep4p overexpression to reduce necrosis by monitoring nuclear release of Nhp6Ap, the yeast homolog of the mammalian high-mobility group box 1 protein (HMGB1). HMGB1 is a chromatin-bound non-histone protein, whose nuclear release may be associated with programmed necrosis, a controlled rather than accidental type of necrotic cell death.33 The nuclear fraction of Nhp6Ap, which was endogenously tagged with green fluorescent protein (GFP), diminished with ongoing aging as determined by fluorescence microscopy (Figures 3c and d). Pep4p overexpression had no effect on Nhp6Ap expression levels (Supplementary Figure S2B), yet stimulated the nuclear retention of Nhp6Ap (Figures 3c and d), thus confirming its capacity to inhibit necrosis.
Figure 3.
Pep4p exerts its non-proteolytic pro-survival function via reduction of necrosis. (a and b) ROS production (a), phosphatidylserine externalization (b) and loss of membrane integrity (b) at day 7 of chronological aging of wild-type cells overexpressing WT-Pep4p, DPM-Pep4p or harboring the control vector as determined via DHE → Eth conversion (a) and Annexin V/PI costaining (b). Data represent means±S.E.M. (n=3; ***P<0.001). In each experiment, 30 000 cells were evaluated using flow cytometry. (c and d) Nuclear localization of the yeast HMGB1 homolog (Nhp6Ap) via epifluorescence microscopic analysis of endogenous chimeric fusion protein Nhp6Ap-GFP upon WT-Pep4p overexpression at days 1 and 7 of chronological aging. In (c) representative sample pictures are depicted. (d) Shows manual quantification of at least 3000 cells per strain and experiment, displaying nuclear localization of Nhp6Ap-GFP. Data represent means±S.E.M. (n=3; **P<0.01). (e–g) Survival (e), ROS production (f), phosphatidylserine externalization (g) and loss of membrane integrity (g) of Δpep4 cells overexpressing WT-Pep4p or DPM-Pep4p and of the corresponding vector control at the indicated time points during chronological aging as determined via clonogenicity (e) DHE → Eth conversion (f) and Annexin V/PI costaining (g). Data represent means±S.E.M. (n=4; *P<0.05, **P<0.01, ***P<0.001). In (f) P-value was calculated for vector control and DPM-Pep4p. In each experiment for (f and g), 30 000 cells were evaluated using flow cytometry. See also Supplementary Figure S2
The proteolytic activity of Pep4p is required for its anti-apoptotic, but not for its anti-necrotic function
The drastic survival deficiency displayed by the PEP4 deletion mutant during chronological aging might involve a reduction in vacuolar proteolysis with a consequent reduction in protein recycling, degradation of oxidized proteins and/or activation of other vacuolar hydrolases. Our results, however, suggest the existence of an additional pro-survival role of Pep4p during chronological aging that is independent of proteolysis. To address this issue, we overexpressed either WT-Pep4p or proteolysis-deficient DPM-Pep4p in the PEP4 deletion mutant. DPM-Pep4p (in contrast to WT-Pep4p) could not fully complement PEP4 deficiency (Figure 3e). However, DPM-Pep4p did confer a partial, yet significant delay in the onset of clonogenic death (Figure 3e) and retarded the emergence of enhanced ROS production (Figure 3f), as compared with the empty vector control. This indicates that the requirement of the protein for healthy aging is predominantly, but not solely, mediated by its proteolytic function. Furthermore, DPM-Pep4p expressed in Δpep4 cells specifically reduced the frequency of necrotic cell death (Figure 3g), consistent with the data obtained in the wild-type background (Figure 3b). In contrast, WT-Pep4p inhibited both necrosis and apoptosis (Figure 3g, Supplementary Figure S2C). Thus, the accelerated loss of viability exhibited by Δpep4 during chronological aging is the result of the combined loss of two separable Pep4p functions, a proteolysis-dependent anti-apoptotic function and a non-proteolytic anti-necrotic function.
The non-proteolytic anti-necrotic function of Pep4p depends on polyamine biosynthesis
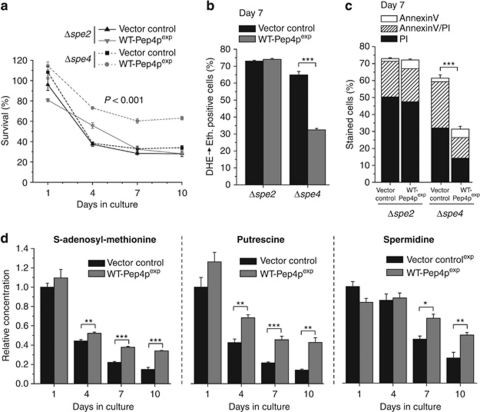
External administration of spermidine to yeast extends chronological lifespan, correlating with global histone hypoacetylation and reduced necrosis.20 To investigate a possible connection between Pep4p and polyamine biosynthesis, we overexpressed Pep4p in cells that either are unable to generate spermidine due to the lack of the S-adenosylmethionine decarboxylase SPE2, or fail to convert spermidine to spermine due to the absence of the spermine synthase SPE4. The capacity of Pep4p to prolong lifespan, reduce ROS production and inhibit necrosis was strongly diminished in Δspe2, but not in Δspe4 cells (Figures 4a–c), although all strains expressed comparable levels of Pep4p (Supplementary Figure S3A). Therefore, polyamine synthesis up to spermidine (but not spermine) generation is required for the beneficial effects of Pep4p on chronological aging.
Figure 4.
The cytoprotective role of Pep4p depends on polyamine biosynthesis. (a–c) Survival (a), ROS production (b), phosphatidylserine externalization (c) and loss of membrane integrity (c) of Δspe2 or Δspe4 cells overexpressing WT-Pep4p and of the corresponding vector controls at the indicated time points during chronological aging as determined via clonogenicity (a) DHE → Eth conversion (b) and Annexin V/PI costaining (c). Data represent means±S.E.M. (n=4; ***P<0.001). In (a) P-value corresponds to Δspe4 strains. In each experiment for (b and c), 30 000 cells were evaluated using flow cytometry. (d) Relative intracellular concentrations of SAM, putrescine and spermidine in wild-type cells overexpressing Pep4p or harboring the vector control during chronological aging as determined using LC/MS/MS, and normalized to the intracellular concentrations of the vector control at day 1. Data represent means±S.E.M. (n=4; *P<0.05, **P<0.01, ***P<0.001). See also Supplementary Figure S3
Necrotic cell death during yeast chronological aging has been previously associated with decline of internal polyamine levels.20 Indeed, the intracellular levels of spermidine and putrescine, a precursor of spermidine, declined with age in vector control cells (Figure 4d). This decline was significantly reduced in cells overexpressing WT-Pep4p (Figure 4d). Conversely, decline of spermidine levels was accelerated in the PEP4 deletion mutant, as compared with the wild type (Supplementary Figure S3B). Moreover, intracellular concentrations of S-adenosyl-methionine (SAM), a precursor to generate spermidine from putrescine, were increased upon Pep4p overexpression (Figure 4d), confirming the link between Pep4p and polyamine synthesis.
Overexpression of Pep4p was generally accompanied by an initial retardation of cell-cycle progression (data not shown). However, this arrest disappeared at day 1–2 of chronological aging, long before the pro-survival effects emerged. In addition, overexpression of Pep4p in a deletion strain that reduced its pro-survival effect (Δspe2) still displayed an initial growth delay. These observations rule out that the described effects are due to a cell cycle-mediated retardation of aging.
The propeptide of Pep4p is sufficient to mediate the anti-necrotic effect
We next generated a series of mutants with progressively shortened C-termini (not shown) and observed that the anti-necrotic effect of Pep4p was retained in all mutants, including the shortest one just encoding the N-terminal 76-amino acid propeptide of the Pep4p-zymogen, so far only known to harbor ER- and vacuolar sorting signals (Figures 5a and b). Consistently, prolonged overexpression of the Pep4p-propeptide (PRO-Pep4p) during chronological aging reduced ROS production, necrosis (Figures 5c and d) and nuclear Nhp6A-GFP release (Supplementary Figure S4A), whereas it increased the intracellular concentrations of SAM, putrescine and spermidine (Supplementary Figure S4B). PRO-Pep4p protected cells from hydrogen peroxide-induced cell death (Figure 5e) and was capable of retarding the onset of death in the Δpep4 mutant (Supplementary Figure S4C). Moreover, it extended chronological lifespan in the Δspe4 mutant, yet lost its beneficial effects in the Δspe2 mutant (Supplementary Figure S4D), although it was expressed at similar levels in both deletion strains (Supplementary Figure S3A). In addition, cytoprotection conferred by PRO-Pep4p was autophagy-independent to the same extent as shown for WT-Pep4p (Supplementary Figures S4E–G). Hence, the observed pro-survival effect of WT-Pep4p and its proteolytically inactive mutant (DPM-Pep4p) actually corresponds to a thus far unknown spermidine-dependent anti-necrotic activity of the Pep4p propeptide.
Figure 5.
The PRO-Pep4p is sufficient to mediate the anti-necrotic effect. (a) Survival determined by clonogenicity of wild-type cells overexpressing WT-Pep4p or PRO-Pep4p and survival of vector control during chronological aging. Data represent means±S.E.M. (n=4). (b) Immunoblots of whole cell extracts of wild-type cells overexpressing FLAG-tagged WT-Pep4p, FLAG-tagged PRO-Pep4p and the vector control after 20 h of induction. Blots were probed with antibodies against FLAG epitope or glyceraldehyde-3-phosphate-dehydrogenase (as a loading control). (c and d) ROS production (c), phosphatidylserine externalization (d) and loss of membrane integrity (d) of wild-type cells overexpressing PRO-Pep4p or of the vector control at day 7 of chronological aging. Data represent means±S.E.M. (n=4; **P<0.01, ***P<0.001). In each experiment, 30 000 cells were evaluated using flow cytometry. (e) Survival of wild-type cells overexpressing PRO-Pep4p or the vector control after 40 h of induction and subsequent treatment, with or without hydrogen peroxide (H2O2) for 4 h. Survival of the vector control was set to 100%. Data represent means±S.E.M. (n=10 for controls, n=7 for treated cultures; ***P<0.001). See also Supplementary Figure S4
Discussion
Combining clonogenic survival tests and the measurement of characteristic cell death markers, we show that the proteolytically inactive N-terminal propeptide of the yeast CatD ortholog (Pep4p) suppresses necrosis in a genetically regulated manner, arguing for the involvement of a cell death type known as ‘programmed necrosis'.10, 11, 33, 34 In this case, necrosis does not mainly result from strong cellular injury or physical disruption of the plasma membrane as an accidental, uncontrolled form of cell death (‘classical necrosis'), but rather follows an orchestrated program, similar to the regulated apoptotic self-execution.10, 11
The anti-necrotic effect of Pep4p seems to involve the regulation of polyamine synthesis (Figure 6), as it goes along with an increase in the intracellular levels of putrescine, spermidine and its biosynthetic precursor SAM. In addition, Pep4p loses its anti-necrotic activity when spermidine biosynthesis is blocked. Interestingly, external administration of spermidine inhibits necrosis during yeast chronological aging in a process involving histone hypoacetylation, a further correlate of the Pep4p anti-necrotic activity.20
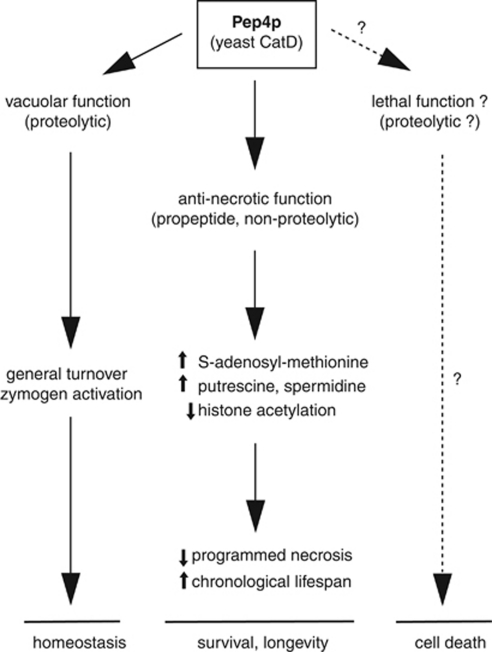
Figure 6.
Yeast CatD's functions in cell metabolism. Besides its central role in the vacuolar proteolytic system, where it is a pivotal player for general protein turnover and maturation of several vacuolar hydrolases, the yeast ortholog of CatD, Pep4p, also holds a non-proteolytic function associated with its propeptide that promotes survival via polyamine-mediated reduction of programmed necrosis. In addition, yeast CatD might be involved in the induction of PCD in specific scenarios, as it has been described for its mammalian counterpart
The pro-survival effects of spermidine have been linked to autophagy,20 which is tightly connected to lifespan extension29 and Pep4p has a central role in the autophagic process.28 However, our data reveal that autophagy is not required for the anti-necrotic effect of Pep4p, at least during the early phase of chronological aging. Of note, it has been recently reported that Pep4p overexpression confers autophagy-independent cytoprotection upon acetic acid-induced cell death.35 Though the anti-necrotic function of spermidine is largely dependent on autophagy, during early aging, autophagy-independent backup-mechanisms have been postulated to exist.20 Given the autophagy-independent yet polyamine-associated nature of the anti-necrotic effect herein described, it is probable that these backup-mechanisms might be related, at least in part, to Pep4p-driven inhibition of programmed necrosis.
Intriguingly, both CatD hyperexpression, as well as increased production of polyamines are observed in many human cancers.4, 36 The propeptide of CatD has been specifically associated to enhanced proliferation and survival of tumor cells3, 5 and polyamines have been shown to mediate tumor promotion and progression.36 It is tempting to speculate that, like Pep4p, human CatD might represent a positive regulator of polyamine biosynthesis and thus a unique target for therapeutic intervention on cellular or tissue polyamine pools. In addition, this study opens doors to investigate CatD orthologues in pathological unicellular organisms as a target for therapeutic necrosis induction.
Our results further provide evidence that Pep4p exerts dual cytoprotective activities involving an anti-necrotic and an anti-apoptotic function. Deletion of PEP4 causes premature death during chronological aging resulting from a combination of necrotic and apoptotic death. Overexpression of proteolytically inactive Pep4p in the PEP4 disruptant specifically reduces the necrotic population and significantly retards the onset of cell death. However, substantial premature demise is not prevented and the apoptotic population remains unchanged. Thus, the requirement of Pep4p for healthy aging is predominantly mediated by its proteolytic function (Figure 6), which exhibits anti-apoptotic characteristics. However, this anti-apoptotic activity is not involved in lifespan extension; wild type and enzymatically inactive Pep4p (or the sole Pep4p propeptide) confer a similar degree of improved survival through necrosis inhibition (but not apoptosis inhibition) when they are overexpressed in cells that contain baseline levels of endogenous, catalytically active Pep4p. Of note, CatD-deficient neurons exhibit, besides autophagic stress (aberrant accumulation of autophagosomes), also apoptotic markers.37, 38 However, apoptosis does not seem to be critical for neurodegeneration, suggesting alternative death mechanisms involved in neuronal death following CatD-deficiency.39 It would be interesting to analyze if a putative increase in programmed necrosis is responsible for neuron demise upon CatD deficiency.
Under the conditions tested here, Pep4p overexpression protects cells from hydrogen peroxide-induced cell death, which is in line with the enhanced resistance to oxidative stress described for long-lived mutants. Nevertheless, it is conceivable that, like its human counterpart, also yeast CatD might bear a double function in regard to PCD. Thus, it remains to be explored to which extent Pep4p may have a regulatory role as an apoptotic and/or necrotic executioner (in addition to its pro-survival roles) (Figure 6). In that case, context-dependent regulatory processes could determine whether the pro-survival or the lethal function of Pep4p prevails.
In conclusion, this study uncovers a novel pro-survival function of yeast CatD that is associated with polyamine regulation and specifically exhibits anti-necrotic characteristics, establishing that the capacity of yeast cells to undergo programmed necrosis is under tight molecular control.
Materials and Methods
Yeast strains and plasmids
Experiments were carried out in BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) and respective null mutants, obtained from Euroscarf. To monitor subcellular localization of endogenous Nhp6Ap, the GFP-tagged Nhp6Ap yeast strain (MATa leu2Δ0 met15Δ0 ura3Δ0) from the Yeast-GFP Clone Collection was used (Invitrogen, Lofer, Austria).
All strains were grown on SC medium containing 0.17% yeast nitrogen base (BD Diagnostics, Schwechat, Austria), 0.5% (NH4)2SO4, 30 mg/l of all amino acids (except 80 mg/l histidine and 200 mg/l leucine), 30 mg/l adenine and 320 mg/l uracil, with 2% glucose (SCD). For strains carrying a plasmid with a Pep4p-FLAG construct either histidine (pESC-His) or uracil (pESC-Ura) was not included and 2% galactose (SCG) was used for induction of expression.
To construct WT-Pep4pFLAG, the insert was amplified by PCR using genomic DNA as template, cut with EcoRI and NotI, and ligated into the vectors pESC-His or pESC-Ura (Agilent Technologies, Stratagene, Santa Clara, CA, USA). To construct DPM-Pep4pFLAG, the Pep4p single point mutant D294A-Pep4p was first generated by a three-step PCR: two PCR products sharing homology in the point mutation region were obtained using the correspondent WT-Pep4p primers and the corresponding mutation-primers, respectively; WT-Pep4p-pESC vector served as the template. These two PCR-products were combined and amplified using WT-Pep4p-primers. The obtained PCR product was digested with EcoRI and NotI and ligated into pESC-His or pESC-Ura (Stratagene). The same procedure was used to generate the second point mutation D109A, however, utilizing D294A-Pep4p-pESC as the template. To obtain the C-terminal Pep4p deletion mutants the different constructs were amplified by PCR using genomic DNA as template with the corresponding primers (see Supplementary Material for PRO-Pep4pFLAG primer). The resulting constructs were digested with EcoRI and NotI, and ligated into vectors pESC-His or pESC-Ura (Stratagene). Correctness of all sequences was verified (MWG Biotech, Ebersberg, Germany). All primers and restriction enzymes used are listed in Supplementary Table S1.
Survival plating and tests for cell death markers
Chronological aging experiments overexpressing Pep4p or Pep4p-mutants were performed in SCG without histidine, or without uracil, as described.16, 22 In experiments involving Δspe2 (Figures 4a–c, Supplementary Figure S4D), unable to generate spermidine, which is required for normal growth of yeast, all strains were supplemented with spermidine (S4139, Sigma, Vienna, Austria) to assure sufficient and reproducible polyamine levels during growth phase, but at concentrations (500 nM on plate to streak the strains, 10 nM in the overnight culture (ONC) and 10 nM in the inoculated culture) that allowed adequate diminution of supplemented polyamines upon entry into stationary phase, where chronological aging begins. Notably, at least three different clones were tested for any aging survival plating to rule out clonogenic variation of the observed effects. Representative aging analyses are shown with at least three independent cultures, aged at the same time. All aging analyses were performed at least twice in total, with similar outcome.
Experiments involving treatment with hydrogen peroxide were performed as previously described,14 with the following modification, in order not to challenge the overexpressor strains during the above described initial retardation of cell-cycle progression, hydrogen peroxide stress was applied after this arrest had disappeared (40 h after induction) for 4 h. As these cells were not in early exponential phase, treatment with a higher hydrogen peroxide dose was necessary to achieve similar death rates in the control strains to the ones previously described.14 To assure proper comparison to the overexpressor strains, the same modifications were applied to the knockout analyses.
DHE staining (ROS production), Annexin V/PI costaining (apoptosis/necrosis marker) and TUNEL staining (apoptosis marker) were performed and quantified by flow cytometry (BD FACSAria, BD Biosciences, Heidelberg, Germany) as previously described.14 A total of 30 000 cells per sample were evaluated using BD FACSDiva software (BD Biosciences). The same samples were analyzed by fluorescence microscopy.
As a further marker for necrosis, nuclear release of the yeast HMGB1 homolog (Nhp6Ap) was monitored by epifluorescence microscopy of endogenous chimeric fusion protein Nhp6Ap-GFP upon WT-Pep4p overexpression. For that purpose, yeast strains expressing chromosomally tagged Nhp6Ap-GFP (Invitrogen) and transformed with pESC-Ura/PEP4 were aged in SCG lacking uracil. At indicated time points, cells were washed once with PBS and directly applied to epifluorescence microscopy with the use of small-band GFP filter (Zeiss, Vienna, Austria) in order to analyze nuclear localization of Nhp6A-GFP. Quantification of cells displaying nuclear localization of Nhp6A-GFP was performed manually, counting at least 3000 cells per strain and experiment.
Microscopy
Microscopy was performed with a Zeiss Axioskop microscope using a Zeiss Plan-Neofluar objective lense with 40 × magnification and 1.30 numerical aperture in oil (using Zeiss Immersol) at room temperature. Imaging medium was AnnexinV/PI incubation buffer (10 mM HEPES, 140 mM NaCl, 5 mM CaCl2, pH 7.4) for pictures shown in Figure 1e and water for pictures shown in Figure 3c. Fluorochromes were fluorescein (used as FITC), PI and Eth (after conversion from DHE) for pictures shown in Figure 1e and GFP for pictures shown in Figure 3c. Fluorescence microscopic sample images were taken with a Diagnostic Instruments camera (Model: SPOT 9.0 Monochrome-6, Sterling Heights, MI, USA), acquired using the Metamorph software (version 6.2r4, Molecular Devices, Sunnyvale, CA, USA) and processed with IrfanView (version 3.97, Wiener Neustadt, Austria) and Adobe Photoshop (version CS2) software. Specifically, picture processing involved creation of AnnexinV and PI overlays in Figure 1e, and coloring (Figures 1e and 3c) with Metamorph and cropping of representative areas (Figure 3c) with Irfanview. In addition, for pictures in Figure 1e, contrast and brightness was adjusted with Adobe Photoshop (applying equal adjustment parameters to all pictures); for GFP-pictures in Figure 3c, equal contrast-, saturation- and gamma correction was applied for pictures of the same day with Irfanview (gamma correction: 0.35 for day1-pictures and 1.3 for day7-pictures).
Alkaline phosphatase (ALP) assay
Autophagy was monitored by ALP activity.40 Strains carrying a plasmid with either WT-Pep4p, DPM-Pep4p, PRO-Pep4p or the vector control were transformed with and selected for stable insertion of pTN9 HindIII fragment (confirmed by PCR). In order to correct for intrinsic (background) ALP activity, the corresponding strains without pTN9 were simultaneously processed and ALP activity substracted. Relative fluorescence units were determined by using a fluorescence reader (Tecan, GeniusPRO, Grödig, Austria) and applying a manual gain of 25. Retention of the pro-survival effect during chronological aging in the pTN9 transformants was confirmed by clonogenic plating assays (data not shown). For each transformed strain, two clones were tested.
Immunoblotting
Preparation of cell extracts and immunoblotting were performed as described.16 Blots were probed with murine monoclonal antibodies against FLAG-epitope (Sigma), murine monoclonal antibodies against glyceraldehyde-3-phosphate dehydrogenase (Sigma) and the respective peroxidase-conjugated affinity-purified secondary antibody (Sigma).
Quantification of histone acetylation
Trichloroacetic acid whole-cell extracts from at least three independent experiments for each strain were prepared as previously described.20 Proteins were separated on 15% SDS-PAGE for western blot analysis on PVDF membrane (Millipore, Vienna, Austria) as described,16 using CAPS buffer (10% methanol, 10% 3-(Cyclohexylamino)-1-propanesulfonic acid from Sigma) for transfer of proteins. Blots were probed with a rabbit polyclonal antibody against histone H3 (ab1791, Abcam, Cambridge, UK; 1 : 5000), which served as a loading control for total histone H3, as well as with a histone H3 modification antibody K18ac (Upstate Biotechnology, Millipore, Vienna, Austria; 1 : 10 000). Peroxidase-conjugated affinity-purified secondary antibody was obtained from Sigma.
For quantification of relative acetylation, blots were scanned using a densitometer (Molecular Dynamics, Model P.D. 300, Sunnyvale, CA, USA) and quantified with ImageQuant Version 5.1 (Molecular Dynamics). Only band densities with relative pixel intensities >500 and <7000 were considered qualitatively valid for quantification. Band densities of acetylation specific blots were normalized to the respective densities of total histone H3 blots in order to obtain specific acetylation rates for each sample. Acetylation rates of wild-type control cultures were set to 1 and the relative acetylation of each sample was calculated accordingly.
Extraction and measurements of polyamines and SAM
Extraction and measurements of polyamines were essentially performed as described in Eisenberg et al.20 For acid extraction of polyamines from yeast cells, culture equivalents of 20 OD600 were washed three times with ddH2O, resuspended in 400 μl ice-cold 5% TCA and incubated on ice for 1 h, with vortexing every 15 min. Supernatants were neutralized with 100 μl of 2 M K2HPO4 and stored at −80 °C upon polyamine measurements. Polyamines were determined using LC/MS/MS. For determination of SAM levels, acid extraction was performed as described for polyamines, and SAM-concentrations were detected using LC/MS/MS. All experiments were carried out on an Ultimate 3000 System (Dionex, LCPackings, Vienna, Austria) coupled to a Quantum TSQ Ultra AM (ThermoFinnigan, Vienna, Austria), using an APCI ion source. The system was controlled by Xcalibur Software 1.4. The stationary phase was a Sequant ZIC-HILIC column (150 × 2.1 mm, 3 μm, 100 Å). The elution solvent A was 50 mM ammonium formiate in ultra pure water and solvent B was acetonitrile. Separation was performed with 15% acetonitrile for 2 min. Thereafter, the acetonitrile content was linearly decreased to 5% over 2 min. After 1 min, acetonitrile content was increased to 15% for column equilibration. Flow rate was set to 300 μl/min.
Polyamines and SAM were detected in MRM mode using the following transitions: spermidine (m/z 146 ≥72, CE 34 eV), putrescine (m/z 89 ≥72, CE 28 eV), SAM (298 ≥136, CE 13 eV), bis(hexamethylene)-triamine as internal standard (m/z 216 ≥100, CE 36 eV). Calibration standards were prepared by spiking extraction buffer with specic concentrations of spermidine, putrescine, SAM and internal standard. Volumes of 20 μl of each sample were injected.
Statistical analyses
Error bars (±S.E.M.) are shown for independent experiments. In cases when experiments were performed in parallel, a common ONC for each strain was used and (i) fresh medium in separate flasks was inoculated directly from the corresponding ONCs (knockout studies) or (ii) common pre-cultures in glucose medium for each strain were inoculated from the corresponding ONCs and divided into separate flasks after shifting to galactose medium for expression induction (overexpression studies). The number of independent data points (n) is indicated in the figure legends of the corresponding graphs. Significances were calculated using an ANOVA and were corrected by the Bonferroni post hoc test. For aging experiments, a two-factor ANOVA with strain and time as independent factors was applied.
Acknowledgments
We thank Ulrike Potocnik, Silvia Dichtinger and Holly Stolterfoht for assistance. We are grateful to the Austrian Science Fund FWF (Austria) for grant S-9304-B05 to FM, JR and DC-G, grant ‘Molecular Enzymology' to K-UF and MB, grant ‘SFB Lipotox' to FM and DC-G, grant T414-B09 to SB and to the European Commission for project APOSYS to FM, TE. GK is supported by the Ligue Nationale contre le Cancer (Equipe labellisée), Agence Nationale pour la Recherche (ANR), European Commission (Apo-Sys, ChemoRes, ApopTrain), Fondation pour la Recherche Médicale (FRM), Institut National du Cancer (INCa) and Cancéropôle Ile-de-France.
Glossary
- ALP
alkaline phosphatase
- CatD
cathepsin D
- DHE
dihydroethidium
- DPM-Pep4p
double point mutant Pep4p
- Eth
ethidium
- GFP
green fluorescent protein
- HMGB1
high-mobility group box 1
- PCD
programmed cell death
- PI
propidium iodide
- PRO-Pep4p
Pep4p-propeptide
- ROS
reactive oxygen species
- SAM
S-adenosyl-methionine
- TUNEL
TdT-mediated dUTP-biotin nick end labeling
- WT-Pep4p
wild type Pep4p
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Cell Death and Disease website (http://www.nature.com/cddis)
Edited by G Melino
Supplementary Material
References
- Benes P, Vetvicka V, Fusek M. Cathepsin D – many functions of one aspartic protease. Crit Rev Oncol Hematol. 2008;68:12–28. doi: 10.1016/j.critrevonc.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siintola E, Partanen S, Stromme P, Haapanen A, Haltia M, Maehlen J, et al. Cathepsin D deficiency underlies congenital human neuronal ceroid-lipofuscinosis. Brain. 2006;129 (Pt 6:1438–1445. doi: 10.1093/brain/awl107. [DOI] [PubMed] [Google Scholar]
- Fusek M, Vetvicka V. Mitogenic function of human procathepsin D: the role of the propeptide. Biochem J. 1994;303 (Pt 3:775–780. doi: 10.1042/bj3030775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liaudet-Coopman E, Beaujouin M, Derocq D, Garcia M, Glondu-Lassis M, Laurent-Matha V, et al. Cathepsin D: newly discovered functions of a long-standing aspartic protease in cancer and apoptosis. Cancer Lett. 2006;237:167–179. doi: 10.1016/j.canlet.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Ohri SS, Vashishta A, Proctor M, Fusek M, Vetvicka V. The propeptide of cathepsin D increases proliferation, invasion and metastasis of breast cancer cells. Int J Oncol. 2008;32:491–498. [PubMed] [Google Scholar]
- Sagulenko V, Muth D, Sagulenko E, Paffhausen T, Schwab M, Westermann F. Cathepsin D protects human neuroblastoma cells from doxorubicin-induced cell death. Carcinogenesis. 2008;29:1869–1877. doi: 10.1093/carcin/bgn147. [DOI] [PubMed] [Google Scholar]
- Emert-Sedlak L, Shangary S, Rabinovitz A, Miranda MB, Delach SM, Johnson DE. Involvement of cathepsin D in chemotherapy-induced cytochrome c release, caspase activation, and cell death. Mol Cancer Ther. 2005;4:733–742. doi: 10.1158/1535-7163.MCT-04-0301. [DOI] [PubMed] [Google Scholar]
- Johansson AC, Steen H, Ollinger K, Roberg K. Cathepsin D mediates cytochrome c release and caspase activation in human fibroblast apoptosis induced by staurosporine. Cell Death Differ. 2003;10:1253–1259. doi: 10.1038/sj.cdd.4401290. [DOI] [PubMed] [Google Scholar]
- Kagedal K, Johansson U, Ollinger K. The lysosomal protease cathepsin D mediates apoptosis induced by oxidative stress. FASEB J. 2001;15:1592–1594. doi: 10.1096/fj.00-0708fje. [DOI] [PubMed] [Google Scholar]
- Carmona-Gutierrez D, Eisenberg T, Buttner S, Meisinger C, Kroemer G, Madeo F. Apoptosis in yeast: triggers, pathways, subroutines. Cell Death Differ. 2010;17:763–773. doi: 10.1038/cdd.2009.219. [DOI] [PubMed] [Google Scholar]
- Eisenberg T, Carmona-Gutierrez D, Buttner S, Tavernarakis N, Madeo F. Necrosis in yeast. Apoptosis. 2010;15:257–268. doi: 10.1007/s10495-009-0453-4. [DOI] [PubMed] [Google Scholar]
- Madeo F, Frohlich E, Frohlich KU. A yeast mutant showing diagnostic markers of early and late apoptosis. J Cell Biol. 1997;139:729–734. doi: 10.1083/jcb.139.3.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeo F, Frohlich E, Ligr M, Grey M, Sigrist SJ, Wolf DH, et al. Oxygen stress: a regulator of apoptosis in yeast. J Cell Biol. 1999;145:757–767. doi: 10.1083/jcb.145.4.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttner S, Eisenberg T, Carmona-Gutierrez D, Ruli D, Knauer H, Ruckenstuhl C, et al. Endonuclease G regulates budding yeast life and death. Mol Cell. 2007;25:233–246. doi: 10.1016/j.molcel.2006.12.021. [DOI] [PubMed] [Google Scholar]
- Fahrenkrog B, Sauder U, Aebi U. The S. cerevisiae HtrA-like protein Nma111p is a nuclear serine protease that mediates yeast apoptosis. J Cell Sci. 2004;117 (Pt 1:115–126. doi: 10.1242/jcs.00848. [DOI] [PubMed] [Google Scholar]
- Madeo F, Herker E, Maldener C, Wissing S, Lachelt S, Herlan M, et al. A caspase-related protease regulates apoptosis in yeast. Mol Cell. 2002;9:911–917. doi: 10.1016/s1097-2765(02)00501-4. [DOI] [PubMed] [Google Scholar]
- Ahn SH, Cheung WL, Hsu JY, Diaz RL, Smith MM, Allis CD. Sterile 20 kinase phosphorylates histone H2B at serine 10 during hydrogen peroxide-induced apoptosis in S. cerevisiae. Cell. 2005;120:25–36. doi: 10.1016/j.cell.2004.11.016. [DOI] [PubMed] [Google Scholar]
- Ludovico P, Rodrigues F, Almeida A, Silva MT, Barrientos A, Corte-Real M. Cytochrome c release and mitochondria involvement in programmed cell death induced by acetic acid in Saccharomyces cerevisiae. Mol Biol Cell. 2002;13:2598–2606. doi: 10.1091/mbc.E01-12-0161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana L, Partridge L, Longo VD. Extending healthy life span – from yeast to humans. Science. 2010;328:321–326. doi: 10.1126/science.1172539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg T, Knauer H, Schauer A, Buttner S, Ruckenstuhl C, Carmona-Gutierrez D, et al. Induction of autophagy by spermidine promotes longevity. Nat Cell Biol. 2009;11:1305–1314. doi: 10.1038/ncb1975. [DOI] [PubMed] [Google Scholar]
- Fabrizio P, Battistella L, Vardavas R, Gattazzo C, Liou LL, Diaspro A, et al. Superoxide is a mediator of an altruistic aging program in Saccharomyces cerevisiae. J Cell Biol. 2004;166:1055–1067. doi: 10.1083/jcb.200404002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herker E, Jungwirth H, Lehmann KA, Maldener C, Frohlich KU, Wissing S, et al. Chronological aging leads to apoptosis in yeast. J Cell Biol. 2004;164:501–507. doi: 10.1083/jcb.200310014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammerer G, Hunter CP, Rothman JH, Saari GC, Valls LA, Stevens TH. PEP4 gene of Saccharomyces cerevisiae encodes proteinase A, a vacuolar enzyme required for processing of vacuolar precursors. Mol Cell Biol. 1986;6:2490–2499. doi: 10.1128/mcb.6.7.2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teichert U, Mechler B, Muller H, Wolf DH. Lysosomal (vacuolar) proteinases of yeast are essential catalysts for protein degradation, differentiation, and cell survival. J Biol Chem. 1989;264:16037–16045. [PubMed] [Google Scholar]
- Fabrizio P, Pozza F, Pletcher SD, Gendron CM, Longo VD. Regulation of longevity and stress resistance by Sch9 in yeast. Science. 2001;292:288–290. doi: 10.1126/science.1059497. [DOI] [PubMed] [Google Scholar]
- Finkel T, Holbrook NJ. Oxidants, oxidative stress and the biology of ageing. Nature. 2000;408:239–247. doi: 10.1038/35041687. [DOI] [PubMed] [Google Scholar]
- Rupp S, Wolf DH. Biogenesis of the yeast vacuole (lysosome). The use of active-site mutants of proteinase yscA to determine the necessity of the enzyme for vacuolar proteinase maturation and proteinase yscB stability. Eur J Biochem. 1995;231:115–125. doi: 10.1111/j.1432-1033.1995.tb20677.x. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Cuervo AM, Seglen PO. Methods for monitoring autophagy from yeast to human. Autophagy. 2007;3:181–206. doi: 10.4161/auto.3678. [DOI] [PubMed] [Google Scholar]
- Vellai T. Autophagy genes and ageing. Cell Death Differ. 2009;16:94–102. doi: 10.1038/cdd.2008.126. [DOI] [PubMed] [Google Scholar]
- Longo VD, Kennedy BK. Sirtuins in aging and age-related disease. Cell. 2006;126:257–268. doi: 10.1016/j.cell.2006.07.002. [DOI] [PubMed] [Google Scholar]
- Dang W, Steffen KK, Perry R, Dorsey JA, Johnson FB, Shilatifard A, et al. Histone H4 lysine 16 acetylation regulates cellular lifespan. Nature. 2009;459:802–807. doi: 10.1038/nature08085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrizio P, Gattazzo C, Battistella L, Wei M, Cheng C, McGrew K, et al. Sir2 blocks extreme life-span extension. Cell. 2005;123:655–667. doi: 10.1016/j.cell.2005.08.042. [DOI] [PubMed] [Google Scholar]
- Zong WX, Thompson CB. Necrotic death as a cell fate. Genes Dev. 2006;20:1–15. doi: 10.1101/gad.1376506. [DOI] [PubMed] [Google Scholar]
- Vanlangenakker N, Vanden Berghe T, Krysko DV, Festjens N, Vandenabeele P. Molecular mechanisms and pathophysiology of necrotic cell death. Curr Mol Med. 2008;8:207–220. doi: 10.2174/156652408784221306. [DOI] [PubMed] [Google Scholar]
- Pereira C, Chaves S, Alves S, Salin B, Camougrand N, Manon S, et al. Mitochondrial degradation in acetic acid-induced yeast apoptosis: the role of Pep4 and the ADP/ATP carrier. Mol Microbiol. 2010;76:1398–1410. doi: 10.1111/j.1365-2958.2010.07122.x. [DOI] [PubMed] [Google Scholar]
- Gerner EW, Meyskens FL., Jr Polyamines and cancer: old molecules, new understanding. Nat Rev Cancer. 2004;4:781–792. doi: 10.1038/nrc1454. [DOI] [PubMed] [Google Scholar]
- Koike M, Nakanishi H, Saftig P, Ezaki J, Isahara K, Ohsawa Y, et al. Cathepsin D deficiency induces lysosomal storage with ceroid lipofuscin in mouse CNS neurons. J Neurosci. 2000;20:6898–6906. doi: 10.1523/JNEUROSCI.20-18-06898.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike M, Shibata M, Ohsawa Y, Nakanishi H, Koga T, Kametaka S, et al. Involvement of two different cell death pathways in retinal atrophy of cathepsin D-deficient mice. Mol Cell Neurosci. 2003;22:146–161. doi: 10.1016/s1044-7431(03)00035-6. [DOI] [PubMed] [Google Scholar]
- Shacka JJ, Klocke BJ, Young C, Shibata M, Olney JW, Uchiyama Y, et al. Cathepsin D deficiency induces persistent neurodegeneration in the absence of Bax-dependent apoptosis. J Neurosci. 2007;27:2081–2090. doi: 10.1523/JNEUROSCI.5577-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda T, Klionsky DJ. The quantitative Pho8Delta60 assay of nonspecific autophagy. Methods Enzymol. 2008;451:33–42. doi: 10.1016/S0076-6879(08)03203-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.