Abstract
When cells sense a decrease in oxygen availability (hypoxia), they develop adaptive responses in order to sustain this condition and survive. If hypoxia lasts too long or is too severe, the cells eventually die. Hypoxia is also known to modulate the p53 pathway, in a manner dependent or not of HIF-1 (hypoxia-inducible factor-1), the main transcription factor activated by hypoxia. The p53 protein is a transcription factor, which is rapidly stabilised by cellular stresses and which has a major role in the cell responses to these stresses. The aim of this review is to compile what has been reported until now about the interconnection between these two important pathways. Indeed, according to the cell line, the severity and the duration of hypoxia, oxygen deficiency influences very differently p53 protein level and activity. Conversely, p53 is also described to affect HIF-1α stability, one of the two subunits of HIF-1, and HIF-1 activity. The direct and indirect interactions between HIF-1α and p53 are described as well as the involvement in this complex network of their respective ubiquitin ligases von Hippel Lindau protein and murine double minute 2. Finally, the synergistic or antagonistic effects of p53 and HIF-1 on some important cellular pathways are discussed.
Keywords: apoptosis, cancer, hypoxia-inducible factor-1 (HIF-1), hypoxia, p53
In case of a decrease in oxygen availability for the cells, the hypoxic pathway is induced with the activation of the HIF-1 (hypoxia-inducible factor 1) transcription factor. HIF-1 is constituted of two subunits called HIF-1α and HIF-1β (or ARNT for aryl hydrocarbon receptor nuclear translocator). Although HIF-1β is constitutively expressed, HIF-1α is precisely regulated by hypoxia.1 On the other hand, p53 pathway is activated by various stresses. The p53 protein is coded by the TP53 gene, which is the most frequently mutated gene in human tumours.2 The p53 pathway is indeed inactivated in most tumours because it functions as the ‘guardian of the genome'.2 p53 acts as a tetrameric transcription factor that induces the transcription of hundreds of target genes, which are involved in regulating apoptosis, cell cycle and DNA repair among others.3 Furthermore, p53 has DNA-binding-independent functions. It indeed interacts with proteins of the Bcl-2 family and translocates to the mitochondria to induce cell death.4 In basal conditions, p53 is kept at very low levels by MDM2 (murine double minute 2)-mediated ubiquitination and subsequent proteasomal degradation,5, 6 while it is stabilised by various cellular stresses. HIF-1 and p53 pathways have been broadly investigated since they are both involved in the emergence and progression of various diseases, cancer being one of the most important. In this review, we discuss how hypoxia modulates the p53 pathway, the influence of p53 on the hypoxic pathway, the direct and indirect interactions that have been described between the HIF-1 and p53 transcription factors to finally conclude on how these factors modulate various cellular pathways.
Effects of Hypoxia on p53 Protein Level and Activity
Hypoxia has frequently been described to be a p53 inducer.7 However, in some cases, hypoxia, alone or in combination with other stresses, has no effect or even decreases p53 protein level.8
Hypoxia increases p53 protein level
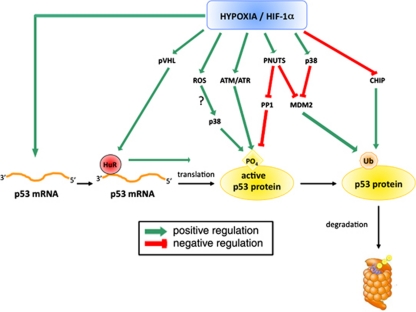
Already in 1994, Graeber et al.9 showed that cells exposed to hypoxia accumulate p53 protein. This accumulation increased with the duration of the hypoxia incubation as well as with the decrease in the pO2 level.10 Most studies showed that the increase in p53 protein level during hypoxia is due to a stabilisation that is dependent on the presence of HIF-1α.11, 12 However, HIF-1α upregulation is not sufficient for p53 induction.13, 14 Hammond et al.15 indeed showed that HIF-1α protein accumulated with similar kinetics in response to 0.02 and 2% oxygen, whereas p53 protein accumulated in response to 0.02% oxygen only. Many different hypotheses have been elaborated to explain the increase in p53 protein level during hypoxia but a complete comprehension has not been reached yet (Figure 1).
Figure 1.
Schematic representation of the different pathways that have been proposed to explain the hypoxia-induced increase in p53 mRNA abundance, translation, p53 protein stability and activity as well as the hypoxia-induced decrease in p53 proteasomal degradation. ATM, ataxia-telangiectasia mutated; ATR, ataxia-telangiectasia mutated and Rad3-related; CHIP, carboxyl terminus of HSP70-interacting protein; HIF-1α, hypoxia-inducible factor-1α; MDM2, murine double minute 2; PNUTS, protein phosphatase-1 nuclear targeting subunit; PP1, protein phosphatase-1; pVHL, von Hippel Lindau protein; ROS, reactive oxygen species; Ub, ubiquitin
Many investigators highlight a putative important role of MDM2 in this process. Chen et al.16 proposed that HIF-1α suppresses MDM2-mediated p53 ubiquitination and blocks MDM2-mediated nuclear export of p53. A hypoxia-induced decrease in MDM2 level has indeed been observed in several studies.17, 18, 19 However, the way by which hypoxia is modulating MDM2 abundance and/or activity may vary, involving p38 MAPK19 or PNUTS (protein phosphatase-1 nuclear targeting subunit)20 activation.
MDM2-independent pathways have also been suggested to explain increased p53 stability under hypoxia. Stabilisation via serine 15 phosphorylation is frequently observed under hypoxia and could be the result of the activation of ATM (ataxia-telangiectasia mutated) or ataxia-telangiectasia mutated and Rad3-related kinases by hypoxia.15, 21 The hypoxia-induced PNUTS could also increase p53 activity via the inhibition of protein phosphatase-1, a phosphatase for p53.20 Mitochondrial generation of reactive oxygen species (ROS) could also contribute to p53 induction.22 Finally, the hypoxia-induced decrease in carboxyl terminus of HSP70-interacting protein transcription, a protein involved in the induction of p53 proteasomal degradation, has been shown to be responsible for p53 accumulation in the heart after myocardial infarction.23
Although most of these reports suggested p53 protein stabilisation under hypoxia, two reports explained increased p53 protein level by either increased p53 mRNA translation without any change in mRNA level24 or by an increase in p53 mRNA level.25 Galban et al.24 indeed showed that pVHL (Von Hippel Lindau protein), which is a transcriptional target of HIF-1 and therefore induced under hypoxia, could enhance p53 translation in a way involving the RNA-binding protein HuR.
In conclusion, hypoxia was frequently shown to increase p53 protein level, with HIF-1α and MDM2 being probably involved in most cases. However, a lot of different hypotheses have been pointed out, most of them only by one group, to explain the p53 increase under hypoxia. It would therefore be interesting to investigate whether these different results could be reproduced in different settings and whether several of these complementary pathways could be activated simultaneously in one setting.
Downstream effects of the hypoxia-induced increase in p53 protein level
An increase in p53 protein level usually induces cell cycle arrest or apoptosis. p53-dependent cell cycle arrest of hypoxic cells has indeed been described.26 However, several reports observed that hypoxia-induced cell cycle arrest occurred independently of p53.9, 27 In most cases, the increase in p53 level in cells exposed to hypoxia induces apoptosis,7 although an increase in p53 protein level without subsequent apoptosis has also been observed.28 The presence of both HIF-1α and p53 seems to be essential for the hypoxia-induced cell death.
The induction of p53-dependent apoptosis by hypoxia or by DNA damage are both described to involve the mitochondrial outer membrane permeabilisation, cytochrome c release and the subsequent activation of caspase-9 and caspase-3. However, these stresses do not activate p53 through the same pathway.29, 30 Although DNA damage induces an increase in p53-target gene expression, hypoxia is mainly described to induce apoptosis via p53-dependent trans-repression, that is, via a decrease in gene expression.18, 31
In 1998, An et al.11 showed that HIF-1α overexpression under normoxia increased p53 activity. However, the responses of cells within physiological levels of HIF-1α during real hypoxia could be different. Indeed, in most cases, the hypoxia-induced p53 is not coupled with the induction of the most common p53 target genes.31, 32, 33 The first explanation that is given to explain the weak transactivation activity of hypoxia-induced p53 is that p53 is not bound to the promoter of BAX or PUMA during hypoxia.32 However, it was observed that p53 is actually bound to the promoter of most p53 target genes but that the transactivation is not induced29 because CBP (CREB-binding protein) and p300 co-activators are not recruited.31 Two main hypotheses have been given to explain the absence of co-activator recruitment. The first one is that it could be due to a competition between p53 and HIF-1 for the interaction with these co-activators.34 p300 is indeed required for the transactivation activity of HIF-1 as well as of p53 and both proteins are interacting with the same domain (CH1) of p300.35 This competition has been shown to be regulated by cockayne syndrome B (CSB) protein, which is a HIF-1 target gene.36 In CSB mutant cells exposed to hypoxia-mimicking agents, the p53 pathway is indeed massively activated, as is the recruitment of RNA polymerase II at the promoter of p53 target genes. On the contrary, for the HIF-1 target genes, despite correct HIF-1 recruitment on their promoter, downstream events such as RNA polymerase II, TFIIB and p300 recruitment do not actually occur.36 These investigators demonstrate that CSB and p300 interact with p53, these interactions being mutually exclusive. This led them to postulate that a HIF-1-dependent increase in CSB level would decrease the level of p53-bound p300 and consequently increase the level of p300 available for HIF-1.36 The second reason that is evoked for the absence of co-activators recruitment by hypoxia-induced p53 is that the posttranslational modifications of p53 occurring during hypoxia are not the ones needed for the binding of co-activators or are mainly leading to the recruitment of transrepressors.31 For example, it was observed that acetylation of p53 on K320 amino acid, probably by p300/CBP-associated factor, is reduced under hypoxia-mimicking conditions while the acetylation of this residue is known to be important for the induction of some p53 target genes.37 Hypoxia-induced p53 is also described to interact with the transcriptional corepressor mSin3A instead of the transcriptional co-activator p300 whereas p53 induced by DNA damage interacts with both proteins.18
The major activity of hypoxia-induced p53 is therefore reported to be transrepression, this transrepression activity being essential for the p53-induced apoptosis under hypoxia.31 The p53-repressed genes that are really involved in triggering apoptosis are not defined yet but HIAP/BIRC3, a protein interfering with caspase activation, could be one of them.33 The preferential activation of transrepression compared with transactivation makes sense in the context of hypoxia because the cell tends to reduce its energetic expenditure when the limited oxygen concentration reduces ATP production.
Nevertheless, p53 is described to induce the transcription of a small subset of its target genes including BNIP3L32 and FAS/CD95.33 As the promoter of these two genes also contains a hypoxia response element, their transactivation could necessitate the cooperative binding of both HIF-1 and p53 in order to be properly induced under hypoxia.32, 33 The presence of HIF-1-binding sites in the proximity of the p53-binding sites could contribute to elevate the local level of CBP.32 However, in some conditions, p53-transactivating activity has been observed with induction of PUMA or BAX mRNA during hypoxia.25, 26, 38 The induction of p21 by hypoxia is also used in some reports to suggest a p53-transactivating activity9, 39 but it is important to note that p21 has also been described to be induced by hypoxia in a p53-independent manner.40
Hypoxia-induced p53 could also have DNA-binding-independent activity. Sansome et al.41 reported that hypoxia induces p53 translocation toward mitochondria. In a rat model, it was also observed that transient global cerebral ischaemia-induced mitochondrial translocation of p53 as well as its binding to the anti-apoptotic protein BCL-XL, inducing the release of cytochrome c from the mitochondria and subsequent cell death.42 However, others observed p53 only in the nuclear compartment.31
Altogether, these data indicate that hypoxia-induced p53 is able to induce apoptosis but in a different way that when it is activated by DNA damage. Indeed, under hypoxia, p53 transactivation activity is weak probably due to the lack of co-activator recruitment. Some p53 target genes are nevertheless transcribed, mainly if p53 works in cooperation with other hypoxia-induced transcription factors. However, its transrepression and DNA-binding-independent activities are sufficient to provoke cell death.
Finally, it has to be noted that the role of HIF-2α in regulating the p53 pathway has only been recently investigated. In 2009, Bertout et al.43 showed that HIF-2α deficiency promotes p53 Ser15 phosphorylation and p53-mediated responses by disrupting cellular redox homeostasis. Similarly, Roberts et al.44 showed that accumulation of HIF-2α, in VHL−/− and HIF-1α−/− clear cell renal cell carcinoma, led to Akt-mediated activation of MDM2 and subsequent suppression of p53. According to these two reports, HIF-2α would have a negative effect on p53 suggesting an opposite role as compared with what is described for HIF-1α.
Hypoxia has no effect on p53 protein level or decreases it
Although frequently described, the induction of p53 by hypoxia is not occurring in all cases. It seems to be depending on various factors such as the severity of hypoxia, its duration as well as according to the cell line. Halterman and Federoff45 suggested that under mild conditions, hypoxia induces HIF-1-dependent transcriptional activation of adaptive genes whereas under conditions of sustained hypoxia, HIF-1α stabilisation increases cellular p53 level and promotes cell death. It was indeed shown that in RKO cells, p53 is stabilised at 0.02% of oxygen but not at 2% after 24h incubation although HIF-1α is stabilised at 2%.15 At 0% oxygen, Schmid et al.34 observed an increase in p53 protein level after 16h incubation but not before. Moreover, Cosse et al.46 showed that if hypoxia (1% oxygen during 16 h) induced an increase in p53 protein level in MCF-7 cells, it was not the case in A549 cells and the p53 level was even reduced by hypoxia in HepG2 cells. Chen et al.47 showed that hypoxia increased p53 protein level in cytotrophoblasts while it reduced it in syncytiotrophoblasts. Other reports stress the fact that hypoxia alone is not sufficient to induce p53 but that DNA damage, acidosis (a secondary effect of the Pasteur effect caused by hypoxia) or reoxygenation are needed to observe this effect.14, 48, 49 Situations in which hypoxia does not affect p53 protein level have therefore been frequently described, hypoxia inducing or not apoptosis.50, 51 Wouters et al.52 have even not observed any effect of hypoxia (<0.1% oxygen) on p53 protein level for up to 48 h. Furthermore, Amellem et al.53 showed that severe hypoxia induced apoptosis in a p53-dependent manner but without any change in p53 protein level.
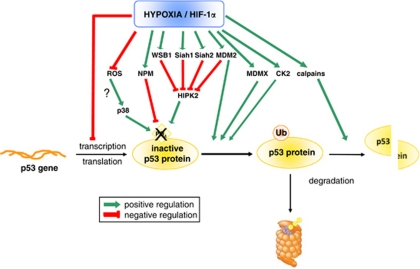
In some situations, as already evoked, hypoxia can even induce a decrease in p53 protein level.14, 47, 54, 55, 56, 57 As for the reasons explaining how hypoxia increased p53 level, several hypotheses have been made to understand it (Figure 2). It is interesting to see that some hypotheses are just the opposite that the ones described here above, suggesting that hypoxia could have totally opposite effects according to the cell type, the severity and the duration of hypoxia. Most groups suggested that decreased p53 level under hypoxia was due to p53 protein degradation. It could occur after p53 ubiquitination by MDM2, which has been described to be induced by hypoxia.58, 59 Zhang and Hill58 proposed that hypoxia induces MDM2 upregulation at the mRNA level in a p53-independent manner. An increase in MDMX expression has also been observed in hypoxic cells together with a reduced phosphorylation of p53 at Ser392.47 The involvement of hypoxia-induced increase in casein kinase 2 level and activity60 or p53 cleavage by calpains56 has also been proposed to contribute to the decrease in p53 protein level observed during hypoxia. Furthermore, hypoxia could as well decrease p53 protein stability by other means. The phosphorylation of p53 at Ser15 is inhibited by hypoxia-induced nucleophosmin.61 Rohwer et al.62 also showed that HIF-1α knock-down strongly increased the amount of p53 protein in the presence of 5-fluorouracil, suggesting that HIF-1α reduced the formation of ROS leading to decreased p53 stabilisation. Finally, hypoxia is known to induce HIPK2 (homeodomain-interacting protein kinase 2) disappearance, resulting in increased p53 ‘mutant-like' conformation and inhibition of apoptosis.63 This decrease in HIPK2 level could depend on the hypoxia-induced MDM2, on the RING family ligase seven in absentia homolog-2 (SIAH2), on SIAH1 or on WD40-repeat/SOCS box protein-1.63 At last, a transcriptional repression of the human p53 gene was shown when HIF-1α was overexpressed.64 To conclude, these numerous studies showed that mild hypoxia could decrease p53 protein level. This decrease could contribute to protect the cells against apoptosis and allow their adaptation to the hypoxic environment.
Figure 2.
Schematic representation of the different pathways that have been proposed to explain the hypoxia-induced decrease in p53 gene transcription, p53 protein stability and activity as well as the hypoxia-induced increase in p53 protein degradation. CK2, casein kinase 2; HIF-1α, hypoxia-inducible factor-1α; HIPK2, homeodomain-interacting protein kinase 2; MDM2, murine double minute 2; NPM, nucleophosmin; ROS, reactive oxygen species; SIAH, seven in absentia homolog; Ub, ubiquitin; WSB-1, WD40-repeat/SOCS box protein
Effects of the combination of hypoxia with other stresses on p53
When hypoxia is followed by a reoxygenation phase, p53 can be upregulated during this latter phase and promote apoptosis, mitigating the concomitant activation of the protective HIF-1 pathway. This has been described, for example, in ischaemia-reperfusion injury in the kidney.65 Pires et al.49 showed that, in RKO cells, after reoxygenation, there was a significant increase in p53-dependent apoptosis. p53 target gene transcription is induced during the reoxygenation phase, as it has been observed for p53 apoptosis effector related to PMP-22 in the outer medullary proximal tubular cells in post-reperfusion ischaemic kidneys.66 Hammond et al.31 showed also that reoxygenation induces phosphorylation of p53 Ser15 in a ATM-dependent manner. However, if the duration of the reoxygenation phase is long enough, p53 level returns to normal level,17 cancelling the effect of the hypoxia phase.
When hypoxia is combined with DNA damage, different cell responses have been described. Some studies showed that hypoxia does not affect the p53-dependent DNA damage response or that a small additive effect is observed.18, 67 Others showed that hypoxia inhibits the increase in p53 protein level as well as cell death in response to etoposide,68 5-fluorouracil69 or cisplatin.70
In conclusion, according to the severity and duration of hypoxia, as well as to the cell line, the effects of the oxygen deficiency on p53 can be different. When p53 level is increased, it seems to be dependent on HIF-1α and p53 has mainly trans-repressional activity. Most of the time, the hypoxia-induced increase in p53 protein level leads to cell death.
Hypoxia Selects for Cells with Mutant p53
In tumours, hypoxic zones often correlate with overexpression of mutant p53 protein. It was shown that the study of p53 and HIF-1α protein level in tumours in vivo can be used to discriminate different prognostic subgroups. Indeed, several studies showed that the immunohistological detection of a functionally inactive p53 and the presence of hypoxia have no prognostic impact if analysed as single parameters, but the combination of both parameters indicates an aggressive phenotype with an adverse prognosis in various cancer types.71
It has also been shown that hypoxia contributes to the selection of cells with decreased apoptotic potential and high metastatic capability.57, 72 This could be due to the selection of cells expressing mutant p53. Indeed, in 1996, Graeber et al.73 showed that hypoxia induces p53-dependent apoptosis and therefore counterselects cells with wild-type p53. A small number of transformed cells lacking functional p53 then overtake cells expressing wild-type p53 under hypoxic conditions. This process therefore facilitates the clonal expansion of cells with compromised p53 protein function.
The hypoxia-induced selection of mutant p53 has adverse effect for patients because mutant p53 protein has a dominant-negative effect on the wild-type protein, thus preventing apoptosis. Some mutants might even present a gain of function leading to oncogenic development.2, 74 Kamat et al.75 showed indeed that, in MCF-7 cells, mutant p53 facilitates pro-angiogenic and hyperproliferative phenotype with decreased apoptosis in response to chronic hypoxia as compared with wild-type p53. These data suggest that a change in p53 status in tumour cells switches their phenotype in response to chronic hypoxia into a more aggressive phenotype.
Effects of p53 on HIF-1α Protein Level and HIF-1 Activity
Although hypoxia and HIF-1 are described to have various effects on p53 protein level and activity, the reverse has also been demonstrated.
p53 decreases HIF-1α protein level
In 2000, Ravi et al.76 observed higher HIF-1α protein level in p53−/− cells than in p53+/+ cells. They showed that p53 promotes ubiquitination and proteasomal degradation of HIF-1α. Others confirmed this observation by showing higher basal HIF-1α level in response to hypoxia in p53-mutant cells than in p53-wild-type cells.75 Both studies suggest that the binding of MDM2 to p53 is needed for this effect. However, these observations are contradictory to others. Choy et al.77 showed indeed that prolonged desferrioxamine incubation of rat cardiac myoblasts H9c2 induces p53 activation and HIF-1α proteasomal degradation but this degradation occurred independently of MDM2. Furthermore, it was shown that the interaction of MDM2 with HIF-1α increases HIF-1 activity,78, 79 while others did not observe any effect of MDM2 loss on HIF-1 target genes expression.38 Although a negative effect of p53 on HIF-1α protein level has frequently been observed, it is therefore difficult to conclude about the involvement of MDM2. It is possible that MDM2 implication in mediating this effect could be cell-type dependent and may vary according to the severity of the hypoxia. The involvement of MIF (macrophage migration inhibitory factor) in this effect has also been suggested. MIF, which is induced in a HIF-1-dependent manner, increases HIF-1α protein level under hypoxic conditions by binding p53 and sequestrating it from HIF-1α.80 Finally, it was also shown that the decrease in HIF-1α protein level induced by p53 overexpression occurs via a pVHL- and p300-independent mechanism34 but that it can be inhibited by AKT activation.77
These observations were not only reported in vitro but also in vivo. p53 KO mice displayed higher HIF-1α level than WT mice following chronic hypoxia exposure.81 Sustained pressure overload, because of severe transverse aorta constriction, induces p53 accumulation that decreases HIF-1α level and thereby impairs cardiac angiogenesis and systolic function.82 In the kidney, p53 activation after ischaemia-reperfusion injury mitigates the activation of the protective HIF-1 pathway by decreasing HIF-1α protein level.65 Several immunohistological studies also suggest that p53 modulates HIF-1α amount and activity: indeed, expression of HIF-1α protein is positively correlated with aberrant p53 accumulation, which means accumulation of mutated p53, as well as with low p53-dependent transcription.83
Besides targeting HIF-1α, it was recently shown that p53 could also decrease HIF-1β protein level.84, 85 This effect is mediated by the transcriptional upregulation of miR-107 that targets HIF-1β 3′UTR. This leads to a decrease in HIF-1 activity.84
p53 has no effect on HIF-1α protein level
On the other hand, several reports showed that hypoxia-induced p53 has no effect on HIF-1α protein level. Indeed Achison and Hupp69 found comparable HIF-1α protein level in HCT116 p53+/+ or p53−/− cells under mild hypoxia. Others also observed that p53 has an effect on HIF-1α abundance only if artificially overexpressed34 or if concomitantly activated by DNA damage.48 This was confirmed by Yang et al.86 who showed that induction of p53 alone is insufficient to block HIF-1α induced by hypoxia but that p53 activation by the small molecule RITA (reactivation of p53 induction of tumour cell apoptosis) inhibits HIF-1α and VEGF (vascular endothelial growth factor) expression in vivo. This effect was probably mediated by the activation of a DNA damage response by RITA, inducing a p53-dependent increase in phosphorylation of the alpha subunit of eukaryotic initiation factor 2, which leads to the subsequent downregulation of HIF-1α protein synthesis.86 It is therefore likely that the induction of HIF-1α protein degradation by p53 occurs only in cases of severe stress.
p53 inhibits HIF-1 activity
A number of studies also showed that p53 can inhibit HIF-1 activity without affecting HIF-1α protein level.34 This effect seems to be independent of p53 DNA-binding activity60, 87 but depends on a competition between p53 and HIF-1 for the binding of the transcriptional co-activator p300.34, 87 In vivo also it was shown by immunohistochemistry in early-stage invasive breast cancers that the most unfavourable situation for HIF-1α to initiate downstream gene expression would be the absence or low expression of p300 and the presence of wild-type p53.88 However, Liu et al.89 did not find that the HIF-1-mediated gene expression in wild-type p53 cells was lower than in cells with mutated p53 but that some forms of mutant p53, when constitutively accumulated, can decrease the HIF-1-dependent transcription of genes. Finally, others observed that, although p53 null astrocytes harbour a markedly increased HIF-1α-dependent target expression compared with control cells, this altered expression was the result of increased cell density in p53 null cultures and of the accompanying acidosis but not of the loss of p53 protein by itself.38 These apparently opposite results could be explained as follows: if p300 abundance is limited, a strong increase in p53 protein level would decrease HIF-1 activity by sequestering p300. However, if the basal p53 level is low, no effect of its suppression would be observed on HIF-1 activity because the abundance of free p300 is sufficient to sustain HIF-1 activity.
In conclusion, p53 is frequently described to have negative effects on HIF-1. In case of severe stress, p53 could induce HIF-1α protein degradation although in milder conditions, p53 can inhibit HIF-1 activity without affecting HIF-1α protein level.
Direct and Indirect Interactions between p53 and HIF-1α
As previously described, hypoxia has been shown to decrease or increase p53 protein level according to the duration and severity of hypoxia and/or the cell type, while p53 is described to decrease HIF-1 activity and/or HIF-1α protein level in case of severe stress. A part of these observations have been explained by direct interaction between these two proteins while other hypotheses imply the formation of larger complexes containing both proteins. Several reports have indeed shown that, in some conditions, HIF-1α and p53 co-immunoprecipitate in cells incubated in vitro under hypoxia as well as in vivo after ischaemia.11, 60, 82
Direct interaction between HIF-1α and p53 has been demonstrated. Although phosphorylated HIF-1α binds HIF-1β, the dephosphorylated form of HIF-1α seems to be the major form that binds to p53, both forms being induced by hypoxia.90 Using binding affinity experiments, it was shown that p53 DNA-binding domain interacts with two sequence motifs of HIF-1α, leading to the hypothesis that each sequence motif in HIF-1α binds to a different subunit of the p53 tetramer.91 These binding data support the hypothesis that p53 provides a route for the degradation, in hypoxic cells, of HIF-1α that is not hydroxylated at the two proline residues.91 Sanchez-Puig et al.92 also showed that the natively unfolded HIF-1α ODD (oxygen-dependent degradation domain) binds to full-length p53 to give a complex containing one HIF-1α ODD and a p53 dimer. This binding probably abolishes the ability of p53 to bind DNA and thus hampers gene transactivation and p53-mediated apoptosis. However, it is not sure whether this kind of interaction can really occur in vivo.93
Trimeric complexes containing p53, MDM2 and HIF-1α have also been proposed with the hypothesis that it could promote HIF-1α degradation.76 This ternary complex could possibly form with a direct interaction between p53 and HIF-1α as described before.90, 92 However, according to Chen et al.,16 MDM2 is required to bridge the two proteins, thereby facilitating HIF-1α degradation and the activation of p53-mediated transcription. HIF-1α has indeed been shown to interact directly with MDM2, even in the absence of p53.78 LaRusch et al.79 confirmed the direct interaction of HIF-1α with MDM2 but their results do not support the trimeric model with p53 because they could not co-immunoprecipitate the three proteins together. They showed indeed that HIF-1α binds to the p53-binding domain of MDM2, leading then to the formation of two distinct complexes: MDM2-HIF-1α and MDM2-p53. In these conditions, MDM2 does not promote HIF-1α protein degradation but, on the contrary, increases its abundance as well as HIF-1 activity in hypoxic cells.78, 79 A trimeric complex containing MDM2, HIF-1α and p300 may be involved in the induction of HIF-1 target genes79 because the binding of MDM2 to p300 has been evidenced.94 Finally, a trimeric complex with the stoichiometric binding of p53 to a HIF-1/p300 complex has also been suggested.87 The existence of the trimeric complex p53/MDM2/HIF-1α is therefore still controversial.
Finally, an important role for JAB1 (jun activating binding protein 1, also called CSN5) in regulating the HIF-1/p53 interactions has been proposed.95 JAB1 and p53 bind competitively to the same domain of HIF-1α resulting in either stabilisation or degradation of HIF-1α respectively.96 Moreover, p53 is stabilised by its binding to HIF-1α, whereas its interaction with JAB1 induces p53 degradation.97 This suggests that the ratio between p53 and JAB1 determines whether hypoxia-induced accumulation of HIF-1α results in apoptosis through p53 stabilisation or adaptation through p53 degradation, with JAB1 being the factor promoting adaptation.95 The study of JAB1 expression and regulation at different oxygen levels and in various cell types would be of great interest. Indeed, it could help to reconcile the different, and frequently opposite, models cited here about the reciprocal effects between p53 and HIF-1α.
Interplay between the HIF-1α/pVHL Couple and the p53/MDM2 Couple
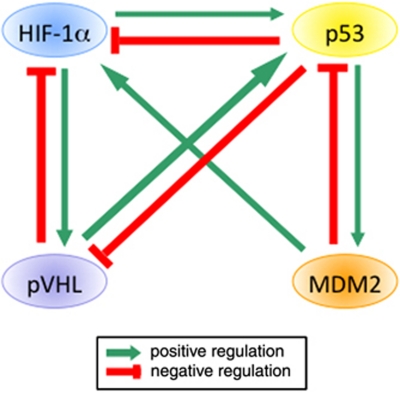
The main proteins known to regulate HIF-1α and p53 levels are their ubiquitin ligases: pVHL and MDM2, respectively. It is thus interesting to understand how these four proteins regulate each other (Figure 3). As previously described, pVHL has a negative role on HIF-1α98 and MDM2 on p53.5, 6 However HIF-1α has a positive role on pVHL99 and p53 on MDM2100 because these regulators are target genes of the transcription factor they regulate. These two couples are tightly interconnected and there obviously exists a functional Yin-Yang relationship between MDM2-p53 and pVHL-HIF-1α pairs.101
Figure 3.
Schematic representation of the reciprocal modulation of HIF-1α, p53, pVHL and MDM2
As described previously, p53 has negative effects on HIF-1α while HIF-1α seems to have mainly positive effects on p53. MDM2 is also described to have mainly positive effects on HIF-1α via a direct interaction between both proteins.78, 79 Furthermore, p53 has negative effects on pVHL by inducing dramatic changes in its expression and intracellular localisation in the kidney after ischaemia-reperfusion injury.65 On the other hand, pVHL has a positive effect on p53. Indeed, Galban et al.24 showed that pVHL elevates p53 expression by enhancing p53 translation while Roe et al.102 showed that pVHL directly associates with and stabilises p53 by suppressing MDM2-mediated ubiquitination and p53 nuclear export. After genotoxic stress, pVHL blocks MDM2-mediated degradation of p53 by recruiting ATM and mediating ATM-dependent Ser15 p53 phosphorylation. pVHL also increases the interaction between p53 and p300 and the p53 acetylation, which ultimately leads to an increase in p53 transcriptional activity and hence in p53-mediated cell cycle arrest and apoptosis. Therefore, they suggest that pVHL serves as a cofactor for strengthening the formation of ATM-p53-p300 complex when cells sense DNA damage.102 These studies suggest again a potential reciprocal regulatory effect for the pVHL/p53 couple. Until now, nothing has been reported about a hypothetical reciprocal effect of MDM2 and pVHL on each other but Roe and Youn101 suggest that pVHL could negatively regulate the function of MDM2. The conclusion is that each of these four proteins (HIF-1α, pVHL, p53 and MDM2) can affect the three others leading to a very complex network.
Contradictory or Synergistic Effects of p53 and HIF-1 in Regulating the Same Pathways
p53 and HIF-1α are proteins that have a lot of common points. They both function as transcription factors and both proteins are present in very low amounts in basal conditions because of their degradation mediated by ubiquitin ligase, degradation that is hampered during stresses. In addition, most pathways regulated by p53 and by HIF-1 are the same. Both transcription factors are indeed involved in regulating apoptosis, survival, cell cycle, glycolysis, respiration and angiogenesis among others. However, they sometimes act synergistically and sometimes in a contradictory manner.
Apoptosis and survival
Several p53 target genes have a pro-apoptotic function and p53 also induces apoptosis by repressing the expression of pro-survival genes or even by acting directly at the mitochondrial level.103 Paradoxically, it seems that in basal conditions, the low level of p53 induces anti-apoptotic and anti-oxidant genes as well as genes involved in DNA repair, therefore promoting cell survival.104 p53 would induce apoptosis only when the cell is severely damaged.
Similarly to p53, hypoxia also has pro- or anti-apoptotic effects according to the intensity of the hypoxic stress.105 Exposure to severe hypoxia leads to p53 accumulation and to the induction of apoptosis.7, 105 Moreover, some HIF-1 target genes code for pro-apoptotic proteins such as BNIP3 or NIX.106 However, hypoxia can also induce the expression of pro-survival genes such as Mcl-1 or IAP-2 and inhibits apoptosis.68 It is currently unknown if the HIF-1 target genes that are expressed under mild or severe hypoxia are the same but we suggest that they could be different because HIF-1 possesses two transcriptional activation domains that are activated at different pO2.107
Cell cycle arrest
When a cell is slightly damaged, p53 induces cell cycle arrest in order for the cell to repair this damage. The G1-arrest is mainly mediated via induction of the cyclin-dependent kinase inhibitor p21 and the control of the G2 checkpoint by p53 depends on induction of 14-3-3δ. These cycle arrest proteins also possess anti-apoptotic activities.104 The cessation of growth is also a classical cellular response to hypoxia especially via the induction of the cyclin-dependent kinase inhibitors p21 and p27 as well as via the hypophosphorylation of retinoblastoma protein.27 p53 and hypoxia therefore acts synergistically to induce cell cycle arrest via p21 upregulation for example. However, in some conditions, hypoxia also induces cell proliferation via the induction of various growth factors such as insulin-like growth factor-2 and platelet-derived growth factor.1 Such a pro-proliferative effect has not been described for p53.
Glycolysis, respiration and ROS production
During tumourigenesis, one of the main functions of hypoxia is to induce a metabolic switch of glucose metabolism from respiration to glycolysis. It is now more and more accepted that the loss of p53 function also contributes to this switch.108 Under hypoxia, HIF-1 has been shown to regulate the expression of nearly all the enzymes of the glycolytic pathway, as well as to induce the expression of glucose transporters.1 Besides activating glycolysis, hypoxia also actively inhibits mitochondrial respiration by inhibiting the conversion of pyruvate into acetyl-coenzyme 1 and by modulating components involved in the electron transport chain.109
p53 also modulates the balance between the utilisation of the respiratory and glycolytic pathways but in an opposite way to hypoxia, by promoting oxidative phosphorylation and dampening glycolysis.110 p53 promotes mitochondrial respiration especially by upregulating synthesis of cytochrome c oxidase 2 and the loss of its expression in p53-mutated cells recapitulates the metabolic switch.111 The observation that protein levels of the cytochrome c oxidase II subunit as well as mitochondrial cytochrome c oxidase activity are decreased in p53−/− cells suggests also the involvement of p53 in the positive regulation of mitochondrial respiration.112 Recently, glutaminase 2, a protein known to enhance mitochondrial respiration, has also been identified as a p53 target gene.113 Furthermore, glycolysis was shown to be inhibited by p53 via the upregulation of TIGAR (TP53-induced glycolysis and apoptosis regulator) that lowers fructose-2,6-bisphosphate level.114 The expression of glucose transporters is also decreased by p53.115
These metabolic pathways are associated with ROS production. The relationship between hypoxia and the production of ROS is controversial. Depending on the studies, hypoxia was described to increase or to decrease ROS production.116 p53 has also opposite effects. It seems that, under severe stress, p53 increases ROS production and cell death whereas in the absence of stress, low levels of p53 induce a decrease in intracellular ROS level.110, 117 Basal p53 level has indeed an antioxidant function.104 For example, p53-induced upregulation of glutathione peroxidase, TIGAR, glutaminase 2 and sestrins contributes to decrease intracellular ROS levels.104, 113, 114 p53 and hypoxia could therefore act together or in an opposite way to positively or negatively regulate ROS production.
Angiogenesis
Hypoxia is a well-known inducer of angiogenesis and vascularisation of hypoxic zones is necessary for further tumour development. HIF-1α stabilisation leads to the transcription of target genes involved in growth of blood vessels such as VEGF, inducible nitric oxide synthase, fibroblast growth factor and matrix metalloproteinases.1 The notion that p53 has a role in limiting tumour vascularisation has come from a number of clinical studies showing correlations between mutated p53, greater VEGF levels and microvessel density.118 At least three mechanisms are used by p53 to inhibit angiogenesis. First, it increases the production of antiangiogenic factors118 among them, is thrombospondin-1. Second, p53 directly inhibits the HIF-1 pathway. Finally, p53 transcriptionally represses genes encoding proangiogenic factors, including VEGF and basic fibroblast growth factor.119, 120
Conclusion
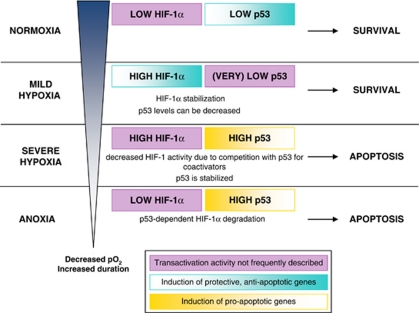
p53 and HIF-1 are two transcription factors that have been extensively studied during the last two decades since they both have major roles in numerous pathways of cells living in a hypoxic environment. One of the main common features of these proteins is that both factors have the possibility to promote cell survival and adaptation to a mild stress as well as to induce cell death when the cell is confronted to a more severe stress. Under hypoxia and depending on the conditions, it seems that they can both work in the same direction or show opposing activities. According to all what has been reported until now, we hypothesise the following model (Figure 4). Under basal normoxic conditions, the abundance of p53 and HIF-1α is very low. When the oxygen concentration decreases, HIF-1α is stabilised and HIF-1 induces the expression of its target genes that are involved in survival. Hypoxia could even reduce p53 level in order to protect the cells from apoptosis. If the oxygen concentration decreases more and/or if hypoxia lasts too long, HIF-1 induces p53 stabilisation, leading to apoptosis. A competition between these two proteins then occurs for co-activators resulting in a decrease in HIF-1-transactivating activity. Finally, under strongly anoxic conditions, p53 eventually leads to HIF-1α degradation. All the factors responsible for the transition between the pro-survival state and the pro-death state are not completely understood yet but it certainly depends on the duration and severity of the hypoxia. Moreover, the exact conditions responsible for the transition from one state to the other seem to be different according to the cell type. The study of the expression of proteins such as JAB1 or CSB in different conditions will help to better understand how p53 and HIF-1α expression and activity are modulated. Other additive stresses such as reoxygenation, acidosis or DNA damage also modulate this balance. Among the questions that have not been assessed yet, it is interesting to know if the HIF-1 target genes expressed under mild hypoxia are the same that the ones expressed under severe hypoxia. It is indeed not clear whether, after p53 hypoxic stabilisation, HIF-1 and p53 are both working for inducing apoptosis or whether both factors promote opposite cell behaviour. The regulation of survival and death mediated by p53 and HIF-1 under hypoxia is therefore extremely complex. It is, however, very important to progress in the comprehension of the interplay between these two transcription factors in order to improve the understanding of various diseases and to design new therapeutic approaches.
Figure 4.
Model for the regulation of p53 and HIF-1α protein level as well as of p53 and HIF-1 activity depending on the severity and the duration of hypoxia
Acknowledgments
Audrey Sermeus is Research Fellow of FNRS (Fonds de la Recherche Scientifique, Belgium). This article presents results of the Belgian Program on Interuniversity Poles of Attraction initiated by the Belgian State, Prime Minister's Office, Science Policy Programming. The responsibility is assumed by its authors.
Glossary
- ARNT
aryl hydrocarbon receptor nuclear translocator
- ATM
ataxia-telangiectasia mutated
- CBP
CREB-binding protein
- CSB
cockayne syndrome B
- HIF-1
hypoxia-inducible factor-1
- HIPK2
homeodomain-interacting protein kinase 2
- JAB1
jun activating binding protein 1
- MDM2
murine double minute 2
- MIF
macrophage migration inhibitory factor
- ODD
oxygen-dependent degradation domain
- PNUTS
protein phosphatase-1 nuclear targeting subunit
- pVHL
von Hippel Lindau protein
- RITA
reactivation of p53 induction of tumour cell apoptosis
- ROS
reactive oxygen species
- SIAH
seven in absentia homolog
- TIGAR
TP53-induced glycolysis and apoptosis regulator
- VEGF
vascular endothelial growth factor
The authors declare no conflict of interest.
Footnotes
Edited by G Melino
References
- Semenza GL. Targeting HIF-1 for cancer therapy. Nat Rev Cancer. 2003;3:721–732. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- Soussi T, Beroud C. Assessing TP53 status in human tumours to evaluate clinical outcome. Nat Rev Cancer. 2001;1:233–240. doi: 10.1038/35106009. [DOI] [PubMed] [Google Scholar]
- Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol. 2008;9:402–412. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- Green DR, Kroemer G. Cytoplasmic functions of the tumour suppressor p53. Nature. 2009;458:1127–1130. doi: 10.1038/nature07986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- Hammond EM, Giaccia AJ. The role of p53 in hypoxia-induced apoptosis. Biochem Biophys Res Commun. 2005;331:718–725. doi: 10.1016/j.bbrc.2005.03.154. [DOI] [PubMed] [Google Scholar]
- Schmid T, Zhou J, Brune B. HIF-1 and p53: communication of transcription factors under hypoxia. J Cell Mol Med. 2004;8:423–431. doi: 10.1111/j.1582-4934.2004.tb00467.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graeber TG, Peterson JF, Tsai M, Monica K, Fornace AJ, Jr, Giaccia AJ. Hypoxia induces accumulation of p53 protein, but activation of a G1-phase checkpoint by low-oxygen conditions is independent of p53 status. Mol Cell Biol. 1994;14:6264–6277. doi: 10.1128/mcb.14.9.6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banasiak KJ, Haddad GG. Hypoxia-induced apoptosis: effect of hypoxic severity and role of p53 in neuronal cell death. Brain Res. 1998;797:295–304. doi: 10.1016/s0006-8993(98)00286-8. [DOI] [PubMed] [Google Scholar]
- An WG, Kanekal M, Simon MC, Maltepe E, Blagosklonny MV, Neckers LM. Stabilization of wild-type p53 by hypoxia-inducible factor 1alpha. Nature. 1998;392:405–408. doi: 10.1038/32925. [DOI] [PubMed] [Google Scholar]
- Carmeliet P, Dor Y, Herbert JM, Fukumura D, Brusselmans K, Dewerchin M, et al. Role of HIF-1alpha in hypoxia-mediated apoptosis, cell proliferation and tumour angiogenesis. Nature. 1998;394:485–490. doi: 10.1038/28867. [DOI] [PubMed] [Google Scholar]
- Wenger RH, Camenisch G, Desbaillets I, Chilov D, Gassmann M. Up-regulation of hypoxia-inducible factor-1alpha is not sufficient for hypoxic/anoxic p53 induction. Cancer Res. 1998;58:5678–5680. [PubMed] [Google Scholar]
- Pan Y, Oprysko PR, Asham AM, Koch CJ, Simon MC. p53 cannot be induced by hypoxia alone but responds to the hypoxic microenvironment. Oncogene. 2004;23:4975–4983. doi: 10.1038/sj.onc.1207657. [DOI] [PubMed] [Google Scholar]
- Hammond EM, Denko NC, Dorie MJ, Abraham RT, Giaccia AJ. Hypoxia links ATR and p53 through replication arrest. Mol Cell Biol. 2002;22:1834–1843. doi: 10.1128/MCB.22.6.1834-1843.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Li M, Luo J, Gu W. Direct interactions between HIF-1 alpha and Mdm2 modulate p53 function. J Biol Chem. 2003;278:13595–13598. doi: 10.1074/jbc.C200694200. [DOI] [PubMed] [Google Scholar]
- Alarcon R, Koumenis C, Geyer RK, Maki CG, Giaccia AJ. Hypoxia induces p53 accumulation through MDM2 down-regulation and inhibition of E6-mediated degradation. Cancer Res. 1999;59:6046–6051. [PubMed] [Google Scholar]
- Koumenis C, Alarcon R, Hammond E, Sutphin P, Hoffman W, Murphy M, et al. Regulation of p53 by hypoxia: dissociation of transcriptional repression and apoptosis from p53-dependent transactivation. Mol Cell Biol. 2001;21:1297–1310. doi: 10.1128/MCB.21.4.1297-1310.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Mao XO, Sun Y, Xia Z, Greenberg DA. p38 Mitogen-activated protein kinase mediates hypoxic regulation of Mdm2 and p53 in neurons. J Biol Chem. 2002;277:22909–22914. doi: 10.1074/jbc.M200042200. [DOI] [PubMed] [Google Scholar]
- Lee SJ, Lim CJ, Min JK, Lee JK, Kim YM, Lee JY, et al. Protein phosphatase 1 nuclear targeting subunit is a hypoxia inducible gene: its role in post-translational modification of p53 and MDM2. Cell Death Differ. 2007;14:1106–1116. doi: 10.1038/sj.cdd.4402111. [DOI] [PubMed] [Google Scholar]
- Hammond EM, Dorie MJ, Giaccia AJ. ATR/ATM targets are phosphorylated by ATR in response to hypoxia and ATM in response to reoxygenation. J Biol Chem. 2003;278:12207–12213. doi: 10.1074/jbc.M212360200. [DOI] [PubMed] [Google Scholar]
- Chandel NS, Vander Heiden MG, Thompson CB, Schumacker PT. Redox regulation of p53 during hypoxia. Oncogene. 2000;19:3840–3848. doi: 10.1038/sj.onc.1203727. [DOI] [PubMed] [Google Scholar]
- Naito AT, Okada S, Minamino T, Iwanaga K, Liu ML, Sumida T, et al. Promotion of CHIP-mediated p53 degradation protects the heart from ischemic injury. Circ Res. 2010;106:1692–1702. doi: 10.1161/CIRCRESAHA.109.214346. [DOI] [PubMed] [Google Scholar]
- Galban S, Martindale JL, Mazan-Mamczarz K, Lopez de Silanes I, Fan J, Wang W, et al. Influence of the RNA-binding protein HuR in pVHL-regulated p53 expression in renal carcinoma cells. Mol Cell Biol. 2003;23:7083–7095. doi: 10.1128/MCB.23.20.7083-7095.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Pakunlu RI, Tsao W, Pozharov V, Minko T. Bimodal effect of hypoxia in cancer: role of hypoxia inducible factor in apoptosis. Mol Pharm. 2004;1:156–165. doi: 10.1021/mp034031n. [DOI] [PubMed] [Google Scholar]
- Yu J, Wang Z, Kinzler KW, Vogelstein B, Zhang L. PUMA mediates the apoptotic response to p53 in colorectal cancer cells. Proc Natl Acad Sci USA. 2003;100:1931–1936. doi: 10.1073/pnas.2627984100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner LB, Li Q, Park MS, Flanagan WM, Semenza GL, Dang CV. Hypoxia inhibits G1/S transition through regulation of p27 expression. J Biol Chem. 2001;276:7919–7926. doi: 10.1074/jbc.M010189200. [DOI] [PubMed] [Google Scholar]
- Kilic M, Kasperczyk H, Fulda S, Debatin KM. Role of hypoxia inducible factor-1 alpha in modulation of apoptosis resistance. Oncogene. 2007;26:2027–2038. doi: 10.1038/sj.onc.1210008. [DOI] [PubMed] [Google Scholar]
- Krieg AJ, Hammond EM, Giaccia AJ. Functional analysis of p53 binding under differential stresses. Mol Cell Biol. 2006;26:7030–7045. doi: 10.1128/MCB.00322-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soengas MS, Alarcon RM, Yoshida H, Giaccia AJ, Hakem R, Mak TW, et al. Apaf-1 and caspase-9 in p53-dependent apoptosis and tumor inhibition. Science. 1999;284:156–159. doi: 10.1126/science.284.5411.156. [DOI] [PubMed] [Google Scholar]
- Hammond EM, Mandell DJ, Salim A, Krieg AJ, Johnson TM, Shirazi HA, et al. Genome-wide analysis of p53 under hypoxic conditions. Mol Cell Biol. 2006;26:3492–3504. doi: 10.1128/MCB.26.9.3492-3504.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei P, Wang W, Kim SH, Wang S, Burns TF, Sax JK, et al. Bnip3L is induced by p53 under hypoxia, and its knockdown promotes tumor growth. Cancer Cell. 2004;6:597–609. doi: 10.1016/j.ccr.2004.10.012. [DOI] [PubMed] [Google Scholar]
- Liu T, Laurell C, Selivanova G, Lundeberg J, Nilsson P, Wiman KG. Hypoxia induces p53-dependent transactivation and Fas/CD95-dependent apoptosis. Cell Death Differ. 2007;14:411–421. doi: 10.1038/sj.cdd.4402022. [DOI] [PubMed] [Google Scholar]
- Schmid T, Zhou J, Kohl R, Brune B. p300 relieves p53-evoked transcriptional repression of hypoxia-inducible factor-1 (HIF-1) Biochem J. 2004;380 (Part 1:289–295. doi: 10.1042/BJ20031299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman SJ, Sun ZY, Poy F, Kung AL, Livingston DM, Wagner G, et al. Structural basis for recruitment of CBP/p300 by hypoxia-inducible factor-1 alpha. Proc Natl Acad Sci USA. 2002;99:5367–5372. doi: 10.1073/pnas.082117899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippi S, Latini P, Frontini M, Palitti F, Egly JM, Proietti-De-Santis L. CSB protein is (a direct target of HIF-1 and) a critical mediator of the hypoxic response. Embo J. 2008;27:2545–2556. doi: 10.1038/emboj.2008.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xenaki G, Ontikatze T, Rajendran R, Stratford IJ, Dive C, Krstic-Demonacos M, et al. PCAF is an HIF-1alpha cofactor that regulates p53 transcriptional activity in hypoxia. Oncogene. 2008;27:5785–5796. doi: 10.1038/onc.2008.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rempe DA, Lelli KM, Vangeison G, Johnson RS, Federoff HJ. In cultured astrocytes, p53 and MDM2 do not alter hypoxia-inducible factor-1alpha function regardless of the presence of DNA damage. J Biol Chem. 2007;282:16187–16201. doi: 10.1074/jbc.M702203200. [DOI] [PubMed] [Google Scholar]
- Long X, Boluyt MO, Hipolito ML, Lundberg MS, Zheng JS, O'Neill L, et al. p53 and the hypoxia-induced apoptosis of cultured neonatal rat cardiac myocytes. J Clin Invest. 1997;99:2635–2643. doi: 10.1172/JCI119452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denko NC, Green SL, Edwards D, Giaccia AJ. p53 checkpoint-defective cells are sensitive to X rays, but not hypoxia. Exp Cell Res. 2000;258:82–91. doi: 10.1006/excr.2000.4928. [DOI] [PubMed] [Google Scholar]
- Sansome C, Zaika A, Marchenko ND, Moll UM. Hypoxia death stimulus induces translocation of p53 protein to mitochondria. Detection by immunofluorescence on whole cells. FEBS Lett. 2001;488:110–115. doi: 10.1016/s0014-5793(00)02368-1. [DOI] [PubMed] [Google Scholar]
- Endo H, Saito A, Chan PH. Mitochondrial translocation of p53 underlies the selective death of hippocampal CA1 neurons after global cerebral ischaemia. Biochem Soc Trans. 2006;34 (Part 6:1283–1286. doi: 10.1042/BST0341283. [DOI] [PubMed] [Google Scholar]
- Bertout JA, Majmundar AJ, Gordan JD, Lam JC, Ditsworth D, Keith B, et al. HIF2alpha inhibition promotes p53 pathway activity, tumor cell death, and radiation responses. Proc Natl Acad Sci USA. 2009;106:14391–14396. doi: 10.1073/pnas.0907357106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts AM, Watson IR, Evans AJ, Foster DA, Irwin MS, Ohh M. Suppression of hypoxia-inducible factor 2alpha restores p53 activity via Hdm2 and reverses chemoresistance of renal carcinoma cells. Cancer Res. 2009;69:9056–9064. doi: 10.1158/0008-5472.CAN-09-1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halterman MW, Federoff HJ. HIF-1alpha and p53 promote hypoxia-induced delayed neuronal death in models of CNS ischemia. Exp Neurol. 1999;159:65–72. doi: 10.1006/exnr.1999.7160. [DOI] [PubMed] [Google Scholar]
- Cosse JP, Sermeus A, Vannuvel K, Ninane N, Raes M, Michiels C, et al. Differential effects of hypoxia on etoposide-induced apoptosis according to the cancer cell lines. Mol Cancer. 2007;6:61. doi: 10.1186/1476-4598-6-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Longtine MS, Sadovsky Y, Nelson DM. Hypoxia downregulates p53 but induces apoptosis and enhances expression of BAD in cultures of human syncytiotrophoblasts. Am J Physiol Cell Physiol. 2010;299:C968–C976. doi: 10.1152/ajpcell.00154.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaluzova M, Kaluz S, Lerman MI, Stanbridge EJ. DNA damage is a prerequisite for p53-mediated proteasomal degradation of HIF-1alpha in hypoxic cells and downregulation of the hypoxia marker carbonic anhydrase IX. Mol Cell Biol. 2004;24:5757–5766. doi: 10.1128/MCB.24.13.5757-5766.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires IM, Bencokova Z, Milani M, Folkes LK, Li JL, Stratford MR, et al. Effects of acute versus chronic hypoxia on DNA damage responses and genomic instability. Cancer Res. 2010;70:925–935. doi: 10.1158/0008-5472.CAN-09-2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Zhang ZF, Rao JY, Sato JD, Brown J, Messadi DV, et al. Treatment with siRNA and antisense oligonucleotides targeted to HIF-1alpha induced apoptosis in human tongue squamous cell carcinomas. Int J Cancer. 2004;111:849–857. doi: 10.1002/ijc.20334. [DOI] [PubMed] [Google Scholar]
- Brunelle JK, Santore MT, Budinger GR, Tang Y, Barrett TA, Zong WX, et al. c-Myc sensitization to oxygen deprivation-induced cell death is dependent on Bax/Bak, but is independent of p53 and hypoxia-inducible factor-1. J Biol Chem. 2004;279:4305–4312. doi: 10.1074/jbc.M312241200. [DOI] [PubMed] [Google Scholar]
- Wouters A, Pauwels B, Lambrechts HA, Pattyn GG, Ides J, Baay M, et al. Chemoradiation interactions under reduced oxygen conditions: Cellular characteristics of an in vitro model. Cancer Lett. 2009;286:180–188. doi: 10.1016/j.canlet.2009.05.026. [DOI] [PubMed] [Google Scholar]
- Amellem O, Stokke T, Sandvik JA, Smedshammer L, Pettersen EO. Hypoxia-induced apoptosis in human cells with normal p53 status and function, without any alteration in the nuclear protein level. Exp Cell Res. 1997;232:361–370. doi: 10.1006/excr.1997.3497. [DOI] [PubMed] [Google Scholar]
- Cosse JP, Ronvaux M, Ninane N, Raes MJ, Michiels C. Hypoxia-induced decrease in p53 protein level and increase in c-jun DNA binding activity results in cancer cell resistance to etoposide. Neoplasia. 2009;11:976–986. doi: 10.1593/neo.09632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sendoel A, Kohler I, Fellmann C, Lowe SW, Hengartner MO. HIF-1 antagonizes p53-mediated apoptosis through a secreted neuronal tyrosinase. Nature. 2010;465:577–583. doi: 10.1038/nature09141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MJ, Oh SJ, Park SH, Kang HJ, Won MH, Kang TC, et al. Hypoxia-induced cell death of HepG2 cells involves a necrotic cell death mediated by calpain. Apoptosis. 2007;12:707–718. doi: 10.1007/s10495-006-0002-3. [DOI] [PubMed] [Google Scholar]
- Koshikawa N, Maejima C, Miyazaki K, Nakagawara A, Takenaga K. Hypoxia selects for high-metastatic Lewis lung carcinoma cells overexpressing Mcl-1 and exhibiting reduced apoptotic potential in solid tumors. Oncogene. 2006;25:917–928. doi: 10.1038/sj.onc.1209128. [DOI] [PubMed] [Google Scholar]
- Zhang L, Hill RP. Hypoxia enhances metastatic efficiency by up-regulating Mdm2 in KHT cells and increasing resistance to apoptosis. Cancer Res. 2004;64:4180–4189. doi: 10.1158/0008-5472.CAN-03-3038. [DOI] [PubMed] [Google Scholar]
- Nardinocchi L, Puca R, Sacchi A, Rechavi G, Givol D, D'Orazi G. Targeting hypoxia in cancer cells by restoring homeodomain interacting protein-kinase 2 and p53 activity and suppressing HIF-1alpha. PLoS One. 2009;4:e6819. doi: 10.1371/journal.pone.0006819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubert A, Paris S, Piret JP, Ninane N, Raes M, Michiels C. Casein kinase 2 inhibition decreases hypoxia-inducible factor-1 activity under hypoxia through elevated p53 protein level. J Cell Sci. 2006;119 (Part 16:3351–3362. doi: 10.1242/jcs.03069. [DOI] [PubMed] [Google Scholar]
- Li J, Zhang X, Sejas DP, Bagby GC, Pang Q. Hypoxia-induced nucleophosmin protects cell death through inhibition of p53. J Biol Chem. 2004;279:41275–41279. doi: 10.1074/jbc.C400297200. [DOI] [PubMed] [Google Scholar]
- Rohwer N, Dame C, Haugstetter A, Wiedenmann B, Detjen K, Schmitt CA, et al. Hypoxia-inducible factor 1alpha determines gastric cancer chemosensitivity via modulation of p53 and NF-kappaB. PLoS ONE. 2010;5:e12038. doi: 10.1371/journal.pone.0012038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puca R, Nardinocchi L, Givol D, D'Orazi G. Regulation of p53 activity by HIPK2: molecular mechanisms and therapeutical implications in human cancer cells. Oncogene. 2010;29:4378–4387. doi: 10.1038/onc.2010.183. [DOI] [PubMed] [Google Scholar]
- Lee SG, Lee H, Rho HM. Transcriptional repression of the human p53 gene by cobalt chloride mimicking hypoxia. FEBS Lett. 2001;507:259–263. doi: 10.1016/s0014-5793(01)02989-1. [DOI] [PubMed] [Google Scholar]
- Sutton TA, Wilkinson J, Mang HE, Knipe NL, Plotkin Z, Hosein M, et al. p53 regulates renal expression of HIF-1{alpha} and pVHL under physiological conditions and after ischemia-reperfusion injury. Am J Physiol Renal Physiol. 2008;295:F1666–F1677. doi: 10.1152/ajprenal.90304.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singaravelu K, Devalaraja-Narashimha K, Lastovica B, Padanilam BJ. PERP, a p53 proapoptotic target, mediates apoptotic cell death in renal ischemia. Am J Physiol Renal Physiol. 2009;296:F847–F858. doi: 10.1152/ajprenal.90438.2008. [DOI] [PubMed] [Google Scholar]
- Moeller BJ, Dreher MR, Rabbani ZN, Schroeder T, Cao Y, Li CY, et al. Pleiotropic effects of HIF-1 blockade on tumor radiosensitivity. Cancer Cell. 2005;8:99–110. doi: 10.1016/j.ccr.2005.06.016. [DOI] [PubMed] [Google Scholar]
- Sermeus A, Cosse JP, Crespin M, Mainfroid V, de Longueville F, Ninane N, et al. Hypoxia induces protection against etoposide-induced apoptosis: molecular profiling of changes in gene expression and transcription factor activity. Mol Cancer. 2008;7:27. doi: 10.1186/1476-4598-7-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achison M, Hupp TR. Hypoxia attenuates the p53 response to cellular damage. Oncogene. 2003;22:3431–3440. doi: 10.1038/sj.onc.1206434. [DOI] [PubMed] [Google Scholar]
- Wang J, Biju MP, Wang MH, Haase VH, Dong Z. Cytoprotective effects of hypoxia against cisplatin-induced tubular cell apoptosis: involvement of mitochondrial inhibition and p53 suppression. J Am Soc Nephrol. 2006;17:1875–1885. doi: 10.1681/ASN.2005121371. [DOI] [PubMed] [Google Scholar]
- Sumiyoshi Y, Kakeji Y, Egashira A, Mizokami K, Orita H, Maehara Y. Overexpression of hypoxia-inducible factor 1alpha and p53 is a marker for an unfavorable prognosis in gastric cancer. Clin Cancer Res. 2006;12:5112–5117. doi: 10.1158/1078-0432.CCR-05-2382. [DOI] [PubMed] [Google Scholar]
- Kim CY, Tsai MH, Osmanian C, Graeber TG, Lee JE, Giffard RG, et al. Selection of human cervical epithelial cells that possess reduced apoptotic potential to low-oxygen conditions. Cancer Res. 1997;57:4200–4204. [PubMed] [Google Scholar]
- Graeber TG, Osmanian C, Jacks T, Housman DE, Koch CJ, Lowe SW, et al. Hypoxia-mediated selection of cells with diminished apoptotic potential in solid tumours. Nature. 1996;379:88–91. doi: 10.1038/379088a0. [DOI] [PubMed] [Google Scholar]
- Hainaut P, Hollstein M. p53 and human cancer: the first ten thousand mutations. Adv Cancer Res. 2000;77:81–137. doi: 10.1016/s0065-230x(08)60785-x. [DOI] [PubMed] [Google Scholar]
- Kamat CD, Green DE, Warnke L, Thorpe JE, Ceriello A, Ihnat MA. Mutant p53 facilitates pro-angiogenic, hyperproliferative phenotype in response to chronic relative hypoxia. Cancer Lett. 2007;249:209–219. doi: 10.1016/j.canlet.2006.08.017. [DOI] [PubMed] [Google Scholar]
- Ravi R, Mookerjee B, Bhujwalla ZM, Sutter CH, Artemov D, Zeng Q, et al. Regulation of tumor angiogenesis by p53-induced degradation of hypoxia-inducible factor 1alpha. Genes Dev. 2000;14:34–44. [PMC free article] [PubMed] [Google Scholar]
- Choy MK, Movassagh M, Bennett MR, Foo RS. PKB/Akt activation inhibits p53-mediated HIF1A degradation that is independent of MDM2. J Cell Physiol. 2010;222:635–639. doi: 10.1002/jcp.21980. [DOI] [PubMed] [Google Scholar]
- Nieminen AL, Qanungo S, Schneider EA, Jiang BH, Agani FH. Mdm2 and HIF-1alpha interaction in tumor cells during hypoxia. J Cell Physiol. 2005;204:364–369. doi: 10.1002/jcp.20406. [DOI] [PubMed] [Google Scholar]
- LaRusch GA, Jackson MW, Dunbar JD, Warren RS, Donner DB, Mayo LD. Nutlin3 blocks vascular endothelial growth factor induction by preventing the interaction between hypoxia inducible factor 1alpha and Hdm2. Cancer Res. 2007;67:450–454. doi: 10.1158/0008-5472.CAN-06-2710. [DOI] [PubMed] [Google Scholar]
- Oda S, Oda T, Nishi K, Takabuchi S, Wakamatsu T, Tanaka T, et al. Macrophage migration inhibitory factor activates hypoxia-inducible factor in a p53-dependent manner. PLoS One. 2008;3:e2215. doi: 10.1371/journal.pone.0002215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno S, Bogaard HJ, Kraskauskas D, Alhussaini A, Gomez-Arroyo J, Voelkel NF, et al. p53 gene deficiency promotes hypoxia-induced pulmonary hypertension and vascular remodeling in mice. Am J Physiol Lung Cell Mol Physiol. 2011;300:L753–L761. doi: 10.1152/ajplung.00286.2010. [DOI] [PubMed] [Google Scholar]
- Sano M, Minamino T, Toko H, Miyauchi H, Orimo M, Qin Y, et al. p53-induced inhibition of Hif-1 causes cardiac dysfunction during pressure overload. Nature. 2007;446:444–448. doi: 10.1038/nature05602. [DOI] [PubMed] [Google Scholar]
- Salnikow K, Costa M, Figg WD, Blagosklonny MV. Hyperinducibility of hypoxia-responsive genes without p53/p21-dependent checkpoint in aggressive prostate cancer. Cancer Res. 2000;60:5630–5634. [PubMed] [Google Scholar]
- Yamakuchi M, Lotterman CD, Bao C, Hruban RH, Karim B, Mendell JT, et al. P53-induced microRNA-107 inhibits HIF-1 and tumor angiogenesis. Proc Natl Acad Sci USA. 2010;107:6334–6339. doi: 10.1073/pnas.0911082107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka Y, Shimizu S, Ito T, Taniguchi M, Nomura M, Nishida T, et al. p53 inhibits vascular endothelial growth factor expression in solid tumor J Surg Res 2011. e-pub ahead of print 15 January 2011. [DOI] [PubMed]
- Yang J, Ahmed A, Poon E, Perusinghe N, de Haven Brandon A, Box G, et al. Small-molecule activation of p53 blocks hypoxia-inducible factor 1alpha and vascular endothelial growth factor expression in vivo and leads to tumor cell apoptosis in normoxia and hypoxia. Mol Cell Biol. 2009;29:2243–2253. doi: 10.1128/MCB.00959-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagosklonny MV, An WG, Romanova LY, Trepel J, Fojo T, Neckers L. p53 inhibits hypoxia-inducible factor-stimulated transcription. J Biol Chem. 1998;273:11995–11998. doi: 10.1074/jbc.273.20.11995. [DOI] [PubMed] [Google Scholar]
- Vleugel MM, Shvarts D, van der Wall E, van Diest PJ. p300 and p53 levels determine activation of HIF-1 downstream targets in invasive breast cancer. Hum Pathol. 2006;37:1085–1092. doi: 10.1016/j.humpath.2006.03.015. [DOI] [PubMed] [Google Scholar]
- Liu J, Shibata T, Qu R, Ogura M, Hiraoka M. Influences of the p53 status on hypoxia-induced gene expression. J Radiat Res (Tokyo) 2004;45:333–339. doi: 10.1269/jrr.45.333. [DOI] [PubMed] [Google Scholar]
- Suzuki H, Tomida A, Tsuruo T. Dephosphorylated hypoxia-inducible factor 1alpha as a mediator of p53-dependent apoptosis during hypoxia. Oncogene. 2001;20:5779–5788. doi: 10.1038/sj.onc.1204742. [DOI] [PubMed] [Google Scholar]
- Hansson LO, Friedler A, Freund S, Rudiger S, Fersht AR. Two sequence motifs from HIF-1alpha bind to the DNA-binding site of p53. Proc Natl Acad Sci USA. 2002;99:10305–10309. doi: 10.1073/pnas.122347199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Puig N, Veprintsev DB, Fersht AR. Binding of natively unfolded HIF-1alpha ODD domain to p53. Mol Cell. 2005;17:11–21. doi: 10.1016/j.molcel.2004.11.019. [DOI] [PubMed] [Google Scholar]
- Fels DR, Koumenis C. HIF-1alpha and p53: the ODD couple. Trends Biochem Sci. 2005;30:426–429. doi: 10.1016/j.tibs.2005.06.009. [DOI] [PubMed] [Google Scholar]
- Grossman SR, Perez M, Kung AL, Joseph M, Mansur C, Xiao ZX, et al. p300/MDM2 complexes participate in MDM2-mediated p53 degradation. Mol Cell. 1998;2:405–415. doi: 10.1016/s1097-2765(00)80140-9. [DOI] [PubMed] [Google Scholar]
- Larsen M, Hog A, Lund EL, Kristjansen PE. Interactions between HIF-1 and Jab1: balancing apoptosis and adaptation. Outline of a working hypothesis. Adv Exp Med Biol. 2005;566:203–211. doi: 10.1007/0-387-26206-7_28. [DOI] [PubMed] [Google Scholar]
- Bae MK, Ahn MY, Jeong JW, Bae MH, Lee YM, Bae SK, et al. Jab1 interacts directly with HIF-1alpha and regulates its stability. J Biol Chem. 2002;277:9–12. doi: 10.1074/jbc.C100442200. [DOI] [PubMed] [Google Scholar]
- Bech-Otschir D, Kraft R, Huang X, Henklein P, Kapelari B, Pollmann C, et al. COP9 signalosome-specific phosphorylation targets p53 to degradation by the ubiquitin system. Embo J. 2001;20:1630–1639. doi: 10.1093/emboj/20.7.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell PH, Wiesener MS, Chang GW, Clifford SC, Vaux EC, Cockman ME, et al. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399:271–275. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- Karhausen J, Kong T, Narravula S, Colgan SP. Induction of the von Hippel-Lindau tumor suppressor gene by late hypoxia limits HIF-1 expression. J Cell Biochem. 2005;95:1264–1275. doi: 10.1002/jcb.20489. [DOI] [PubMed] [Google Scholar]
- Wu X, Bayle JH, Olson D, Levine AJ. The p53-mdm-2 autoregulatory feedback loop. Genes Dev. 1993;7:1126–1132. doi: 10.1101/gad.7.7a.1126. [DOI] [PubMed] [Google Scholar]
- Roe JS, Youn HD. The positive regulation of p53 by the tumor suppressor VHL. Cell Cycle. 2006;5:2054–2056. doi: 10.4161/cc.5.18.3247. [DOI] [PubMed] [Google Scholar]
- Roe JS, Kim H, Lee SM, Kim ST, Cho EJ, Youn HD. p53 stabilization and transactivation by a von Hippel-Lindau protein. Mol Cell. 2006;22:395–405. doi: 10.1016/j.molcel.2006.04.006. [DOI] [PubMed] [Google Scholar]
- Vousden KH, Lu X. Live or let die: the cell's response to p53. Nat Rev Cancer. 2002;2:594–604. doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- Janicke RU, Sohn D, Schulze-Osthoff K. The dark side of a tumor suppressor: anti-apoptotic p53. Cell Death Differ. 2008;15:959–976. doi: 10.1038/cdd.2008.33. [DOI] [PubMed] [Google Scholar]
- Piret JP, Mottet D, Raes M, Michiels C. Is HIF-1alpha a pro- or an anti-apoptotic protein. Biochem Pharmacol. 2002;64:889–892. doi: 10.1016/s0006-2952(02)01155-3. [DOI] [PubMed] [Google Scholar]
- Greijer AE, van der Wall E. The role of hypoxia inducible factor 1 (HIF-1) in hypoxia induced apoptosis. J Clin Pathol. 2004;57:1009–1014. doi: 10.1136/jcp.2003.015032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouyssegur J, Dayan F, Mazure NM. Hypoxia signalling in cancer and approaches to enforce tumour regression. Nature. 2006;441:437–443. doi: 10.1038/nature04871. [DOI] [PubMed] [Google Scholar]
- Yeung SJ, Pan J, Lee MH. Roles of p53, MYC and HIF-1 in regulating glycolysis – the seventh hallmark of cancer. Cell Mol Life Sci. 2008;65:3981–3999. doi: 10.1007/s00018-008-8224-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenza GL. Oxygen-dependent regulation of mitochondrial respiration by hypoxia-inducible factor 1. Biochem J. 2007;405:1–9. doi: 10.1042/BJ20070389. [DOI] [PubMed] [Google Scholar]
- Vousden KH, Ryan KM. p53 and metabolism. Nat Rev Cancer. 2009;9:691–700. doi: 10.1038/nrc2715. [DOI] [PubMed] [Google Scholar]
- Matoba S, Kang JG, Patino WD, Wragg A, Boehm M, Gavrilova O, et al. p53 regulates mitochondrial respiration. Science. 2006;312:1650–1653. doi: 10.1126/science.1126863. [DOI] [PubMed] [Google Scholar]
- Zhou S, Kachhap S, Singh KK. Mitochondrial impairment in p53-deficient human cancer cells. Mutagenesis. 2003;18:287–292. doi: 10.1093/mutage/18.3.287. [DOI] [PubMed] [Google Scholar]
- Hu W, Zhang C, Wu R, Sun Y, Levine A, Feng Z. Glutaminase 2, a novel p53 target gene regulating energy metabolism and antioxidant function. Proc Natl Acad Sci USA. 2010;107:7455–7460. doi: 10.1073/pnas.1001006107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensaad K, Tsuruta A, Selak MA, Vidal MN, Nakano K, Bartrons R, et al. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126:107–120. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- Schwartzenberg-Bar-Yoseph F, Armoni M, Karnieli E. The tumor suppressor p53 down-regulates glucose transporters GLUT1 and GLUT4 gene expression. Cancer Res. 2004;64:2627–2633. doi: 10.1158/0008-5472.can-03-0846. [DOI] [PubMed] [Google Scholar]
- Chandel NS, Budinger GR. The cellular basis for diverse responses to oxygen. Free Radic Biol Med. 2007;42:165–174. doi: 10.1016/j.freeradbiomed.2006.10.048. [DOI] [PubMed] [Google Scholar]
- Ladelfa MF, Toledo MF, Laiseca JE, Monte M.Interaction of p53 with tumor suppressive and oncogenic signaling pathways to control cellular ROS production Antioxid Redox Signal 2011. e-pub ahead of print 14 January 2011. [DOI] [PubMed]
- Teodoro JG, Evans SK, Green MR. Inhibition of tumor angiogenesis by p53: a new role for the guardian of the genome. J Mol Med. 2007;85:1175–1186. doi: 10.1007/s00109-007-0221-2. [DOI] [PubMed] [Google Scholar]
- Pal S, Datta K, Mukhopadhyay D. Central role of p53 on regulation of vascular permeability factor/vascular endothelial growth factor (VPF/VEGF) expression in mammary carcinoma. Cancer Res. 2001;61:6952–6957. [PubMed] [Google Scholar]
- Zhang L, Yu D, Hu M, Xiong S, Lang A, Ellis LM, et al. Wild-type p53 suppresses angiogenesis in human leiomyosarcoma and synovial sarcoma by transcriptional suppression of vascular endothelial growth factor expression. Cancer Res. 2000;60:3655–3661. [PubMed] [Google Scholar]