Abstract
In eukaryotic cells, roughly one-fourth of all mRNAs code for secretory and membrane proteins. This class of proteins must first be segregated to the endoplasmic reticulum, where they are either translocated into the lumen or inserted into the lipid bilayer. The study of these processes has long relied on their successful reconstitution in cell-free systems. The high manipulability of such in vitro systems has allowed the identification of key machinery, elucidation of their functional roles in translocation, and dissection of their mechanisms of action. Here, we provide the basic methodology for (i) setting up robust mammalian-based in vitro translation and translocation systems, (ii) assays for protein translocation, insertion, and topology, and (iii) methods to solubilize, fractionate, and reconstitute ER membranes. Variations of these methods should be applicable not only to forward protein translocation systems but also for dissecting other poorly understood membrane-associated processes such as retrotranslocation.
Keywords: in vitro translation, microsomes, membrane proteins, reconstitution, proteoliposomes, protease protection, protein topology
1. Introduction
The endoplasmic reticulum (ER) is the major site for the biosynthesis, maturation, quality control, and degradation of secretory and membrane proteins. Each of these basic processes employs multiple distinct pathways that operate in parallel to provide the cell considerable flexibility in handling the tremendous diversity of proteins that transit through the ER. A major goal of cell biology has long been to identify and dissect the mechanism of action of the machinery that define these pathways of secretory and membrane protein metabolism.
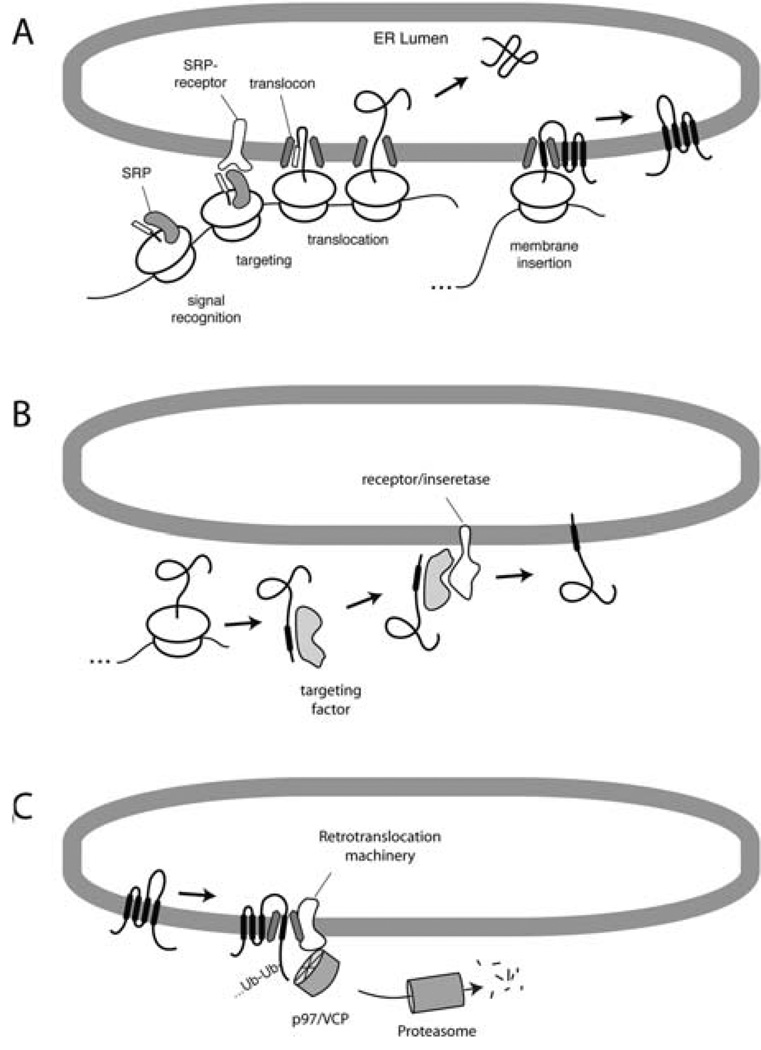
One of the principal approaches that has been applied to this problem is the reconstitution of key pathways or sub-reactions in a cell-free system. The tremendous manipulability of such systems affords a direct window into the biochemical and mechanistic dissection of any of these pathways. By understanding the basic features of the standard cell-free translation and translocation system, it can be sensibly customized in various ways to address any of a number of processes that occur at or within the ER and cytosol. A few of the major ER-associated processes that are amenable to dissection using this or similar in vitro systems are shown in Fig. 20.1.
Fig. 20.1.
Examples of ER-associated pathways amenable to in vitro reconstitution. (A) The SRP-dependent co-translational translocation pathway. (B) A post-translational translocation pathway for tail-anchored membrane protein insertion. (C) Post-translocational pathway of membrane protein metabolism involving ubiquitination, retrotranslocation, and proteasomal degradation. In each of these instances, the substrate is synthesized in vitro, making it the only protein that becomes radiolabeled. The other components of the system can be manipulated to analyze the requirements for substrate translocation, maturation, degradation, etc.
In co-translational translocation (Fig. 20.1A), secretory and membrane proteins are recognized as they are being synthesized. The signal recognition particle (SRP) binds to a hydrophobic domain (either a signal sequence or transmembrane domain) in the nascent polypeptide as it emerges from the ribosome. This complex of ribosome–nascent chain–SRP is then targeted to the membrane (via the SRP receptor) and transferred to a translocon whose central component is the Sec61 complex. Secretory proteins are translocated through the Sec61 complex, while membrane proteins are laterally released by the Sec61 complex into the membrane bilayer. Because everything occurs co-translationally, the study of these events typically depends on translating the protein of interest in a cytosolic extract in the presence of a source of ER membrane [typically rough microsomes (RM) isolated from canine pancreas (1) or other tissues/cells (2, 3)]. If the ER membranes are added to the reaction after translation is completed, translocation will not occur.
In post-translational translocation pathways (Fig. 20.1B), the protein remains competent for translocation even after it has been fully synthesized and released from the ribosome. Such pathways can be employed by a subset of secretory proteins, some small proteins, and certain types of membrane proteins. These pathways, particularly in higher eukaryotes, are not nearly as well understood as the SRP-dependent co-translational translocation pathway. For example, a novel and well-conserved pathway for tail-anchored membrane proteins was only recently discovered (4, 5) and whose full complement of machinery remains to be clarified. From a practical standpoint, these post-translational pathways are in many ways easier to study because the translation reaction can be uncoupled from the translocation reaction. Thus, proteins can be translated, after which various manipulations can be applied (e.g., removal of energy, addition of inhibitors, change in conditions) before initiating the translocation reaction by adding a source of ER membranes. This provides greater flexibility than co-translational reactions where conditions must be maintained within the narrow range that is compatible with efficient protein synthesis.
And finally, reactions after both synthesis and translocation have been completed can also be studied using these same in vitro systems (Fig. 20.1C). Examples of such processes include maturation events in the ER lumen (6), quality control of misfolded proteins, retrotranslocation, ubiquitination, and degradation (7). Again, many of these pathways are still relatively poorly understood, particularly from a mechanistic point of view. As with posttranslational translocation, these events can often be uncoupled from protein synthesis (and in some cases, even translocation), allowing experimental flexibility.
The study of all of these processes in vitro depends on three basic tools. An in vitro translation (IVT) system, clear and definitive assays for translocation and topology, and, in the case of events occurring at the ER, methods to manipulate the composition of the membrane. These basic tools can be applied in a wide range of ways. The IVT system allows one to produce in a physiologic system a protein radiolabeled with very high specific activity that can be followed. By scaling up these reactions, biochemical amounts can also be generated to identify interacting partners (4). Highly specific assays for translocation can be used to identify protein or lipid requirements (4, 8, 9) and analyze the action of small molecule inhibitors (10). Manipulation of the membrane provides access to the requirements at this usually inaccessible compartment (8, 9, 11). And the ability to isolate the membrane after insertion provides the ability to study subsequent events (such as degradation) in isolation (7).
2. Materials
2.1. Preparation of the Transcription Mix (T1)
For general advice regarding these reagents, see the following:
1 M HEPES, pH 7.6: Prepare a solution of 1 M HEPES (free acid), titrated with 0.45MNaOH. This will be the correct pH when diluted in the buffers below. Filter and store at 4°C (see Notes 1, 2, 8, and 9).
2 M MgCl2, store at RT or 4°C.
100 mM spermidine (Sigma); very hygroscopic. Dissolve 145 mg spermidine in 10 mL water. Freeze in nitrogen and store at −20°C.
1 M DTT (1,4-dithiothreitol; Roche). Dissolve 1.54 g DTT in 10 mL water. Aliquot and freeze in nitrogen. Store at −80°C. Do not freeze-thaw more than twice to prevent oxidation.
10X NTPs. 5 mM each of ATP, UTP, and CTP and 1 mM of GTP in water. Adjust to pH ~7 with NaOH as needed. Aliquot and freeze in nitrogen. Store at −80°C. Do not freeze-thaw more than five times.
10X Cap: 7-methyl diguanosine triphosphate cap structure analog (New England Biolabs). Each vial contains 25 A260 units. Add 300 µL water directly to the vial (to make ~5 mM solution), mix well to dissolve, aliquot, and freeze in nitrogen. Store at −80°C. Do not freeze-thaw more than five times. (see Note 3).
2.2. Preparation of the Translation Mix (T2)
Crude rabbit reticulocyte lysate (RRL). This can be prepared in-house (12), although very few labs currently do this due to practical limitations. Purchase from Green Hectares. They offer two products. We buy the more dilute material which they say is for ‘purification of factors.’ The other product ‘for in vitro translation’ is somewhat more concentrated (and expensive), but we have not found this to be a significant advantage. The more dilute product typically comes in rather large aliquots (~50 mL ), which should be stored at −80°C. Do not freeze-thaw more than twice. The first time you thaw a 50 mL aliquot, make 10 mL aliquots in 15 mL polypropylene tubes. To one of these 10 mL aliquots, add hemin, treat with micrococcal nuclease, and further sub-aliquot it (see Section 3.2.). The other 10 mL aliquots can be frozen directly in nitrogen and stored at −80 until needed. Crude lysate is stable for up to 10 years. Nuclease-treated RRL may be less stable, which is why we do not nuclease everything at once.
Micrococcal nuclease (Calbiochem; 15,000 units/vial). Dissolve in 1 mL 50 mM HEPES buffer pH 7.4 (i.e., 15 units per µL); aliquot and freeze in liquid nitrogen and store at −20°C.
Hemin (Sigma). Prepare 10 mL of a 100 µM stock by mixing the following in this exact order (to avoid problems with precipitation): 6.5 mg hemin, 250 µL 1 N KOH, 500 µL 200 mM Tris, pH 8.0, 8.9 mL ethylene glycol, 190 µL 1 N HCl, 50 µL water. Mix by vortexing and store at −20°C; will not freeze due to ethylene glycol.
100 mM CaCl2: Dissolve in water. Keep at 4°C.
200 mM EGTA. Prepare as follows: To 760 mg EGTA powder, add ~7 or 8 mL water and vortex to resuspend the powder (it will not go into solution). Add 950 µL 5 N NaOH (at which point the EGTA will go into solution). Adjust to 10 mL with additional water. Store at 4°C.
Calf liver tRNA (Novagen). This is typically supplied as a 10 mg/mL stock. Freeze in nitrogen and store at −80°C. Stable to multiple freeze-thaws as long as it is kept on ice when thawed and frozen in liquid nitrogen immediately after use (see Note 4).
1.2 M creatine phosphate (Roche). Dissolve in water, freeze in nitrogen, and store at −80°C.
20 mg/mL creatine kinase (Roche). Dissolve 100 mg in 5 mL of 10 mM HEPES, pH 7.5, 50% glycerol. Store at −20°C.
5 M KOAc stock (for each 500 mL add 2 mL of 12 N HCl to bring pH to ~ 7). Store at RT.
Amino acid stocks and mixes – each one is made up individually as a 20 mM stock, and these are mixed to prepare an amino acid mix of 19 amino acids (1 mM each) without methionine (which will be supplied as a 35 S-methionine to label translated proteins). Prepare stock solutions of each amino acid (purchased as powders from Sigma) at 20 mM in either 0.01 N HCl (Trp, Val, Ile, Asn, Phe, Asp, Glu, and Lys), 0.1 N HCl (Tyr), or water (the remaining ones). To make the 19 amino acid mix (1 mM each) simply mix 0.5 mL of each of the 19 with 0.5 mL of H2O. Mixes lacking different amino acids for labeling with residues other than methionine can also be made. Freeze in aliquots and store both the stock amino acids and the mixes at −80°C (see Note 5).
10X Emix: for 1 mL, mix 305 µL water, 400 µL 19 amino acid mix (1 mM each; step 10 above), 10 µL 1 M HEPES (not pH adjusted), 1.87 µL 8 N KOH, 83.3 µL 1.2 M creatine phosphate (step 7 above), 100 µL 0.1 M ATP, 100 µL 0.1MGTP. Freeze in nitrogen and store at −80°C.
CB20X: 627.5 µL water, 240 µL 1 M HEPES (not pH adjusted), 22.5 µL 8 N KOH, 100 µL 5 M KOAc pH 7 (step 9 above), 10 µL 2 M MgCl2. Freeze in nitrogen and store at −80°C (see Note 10).
2.3. Linked Transcription and Translation
Reagents for PCR amplification of the desired cDNA.
Qiagen PCR purification kit.
RNAsin (Promega). Do not freeze in nitrogen; keep at −20°C. This comes in 50% glycerol. Stable for at least 2 years.
T7 or SP6 polymerase (New England Biolabs). Do not freeze in nitrogen; keep at −20°C. This comes in 50% glycerol. Stable for at least 2 years.
35S-methionine (from PerkinElmer; 1,000 Ci/mmol, in aqueous solution). Store in aliquots of 100 µL or less at −80°C. Suitable precautions should be taken when working with radioisotopes.
Canine pancreatic rough microsomes (RM). RMs can be obtained in small amounts (50–200 µL) from commercial sources (e.g., Promega). However, for large amounts as needed for fractionation and purification of membrane proteins, purchasing RMs becomes prohibitively expensive (see Notes 6 and 11).
2.4. Assays for Translocation and Topology
Physiologic salt buffer (PSB): 100 mM KOAc, 2 mM Mg(OAc)2, 50 mM HEPES, pH 7.4. It is often convenient to prepare a 10X PSB stock, which is diluted with various other components (e.g., sucrose) as needed.
Proteinase K (PK; Roche): 10 mg/ mL dissolved in 20 mM HEPES, pH 7.4. Prepare single-use 20 µL aliquots, freeze in nitrogen, and store at −80. After thawing and use, discard remainder. We usually prepare ~100 aliquots at a time (see Note 7).
10% Triton X-100 solution. Store at 4°C.
PK-kill buffer (PKB): 1% SDS, 0.1 M Tris, pH 8.0.
2.5. Reconstitution of Membrane Proteins into Proteoliposomes
Bovine liver phosphatidylcholine (PC, supplied in organic solvent, usually chloroform or chloroform:methanol; from Avanti Polar Lipids) (see Note 12).
Bovine liver phosphatidylethanolamine (PE, supplied in organic solvent, usually chloroform or chloroform: methanol; from Avanti Polar Lipids).
Lissamine–rhodamine–dipalmotyl–PE (DPPE, supplied in organic solvent, usually chloroform or chloroform: methanol; from Avanti Polar Lipids).
Lipid hydration buffer: 50 mM HEPES, pH 7.4, 10 mM DTT (added from a freshly prepared 1 M DTT stock solution).
Bio-Beads SM2 (Bio-Rad). Two grades are sold. We buy standard grade product (less expensive) as it seems to behave identically to the more expensive ‘biotechnology’ grade Bio-Beads.
RM (see Note 6).
10% DeoxyBigChap (DBC; Calbiochem). Add ~9.5 mL water to a 1 g vial of DBC to bring the total volume to 10 mL. Dissolve by gentle agitation at room temperature. Store at 4°C for short-term use (6 months or less); otherwise freeze and store at either −20 or −80°C (see Note 13).
Pre-extraction buffer: 50 mM HEPES, pH 7.4, 250 mM sucrose, 2 mM MgCl2, 0.2% DBC.
Extraction buffer: 400 mM KOAc, 5 mM MgCl2, 50 mM HEPES, pH 7.4, 15% glycerol, 1 mM DTT (added just before use from a freshly prepared 1 M stock solution).
3. Methods
In our lab, we have simplified the transcription and translation reactions to essentially be in-house generated ‘kits’ that are easy to use. The kit components consist of a T1 mix (for transcription) and a RRL-based T2 mix (for translation). Because a single (meticulous and responsible) person can be charged with preparing and maintaining aliquots of T1 and T2, the experiments become highly reproducible and sufficiently straightforward that they are amenable to even the most inexperienced lab members. Furthermore, the use of this approach is far more economical and allows for much larger scale applications of in vitro translation reactions than is possible from commercial kits. This scalability actually makes it reasonable to purify the translated proteins [and identify interesting co-associating factors (4)], something that is typically not considered an option with mammalian in vitro systems.
The stock solutions in Steps 1 and 2 of Section 2.1 are all quite stable, and we have one primary person responsible for preparing and maintaining them. They are not used for any other purpose and are therefore kept in a separate place for use only in preparing transcription and translation reagents. This is to maintain reliability and prevent RNAse contamination. The T1 and T2 mixes are also quite stable, but less so than individual reagents. We therefore make it from the stock solutions from time to time. If you are doing lots of in vitro transcription and translation reactions, you can make 7.6 mL and 17.1 mL T1 and T2 mixes, respectively, at a time (as we usually do). Otherwise, 760 µL and 1.71 mL at a time is ample, as is described here.
3.1. Preparation of the Transcription Mix (T1)
To prepare 760 µL T1 Mix, put into a microcentrifuge tube on ice the following components, in order, with gentle mixing after each addition (to avoid problems with precipitation): 487 µL water, 40 µL 1 M HEPES, pH 7.6, 3 µL 2 M MgCl2, 20 µL 100 mM spermidine, 10 µL 1 M DTT, 100 µL 10X NTPs, 100 µL 10X Cap. Mix well (see Note 8).
Prepare aliquots (100 µL) into pre-chilled microcentrifuge tubes on ice, freeze in nitrogen, and store at −80°C. Do not freeze-thaw more than four times. Stable for at least 2 years.
3.2. Preparation of the Translation Mix (T2)
Before using the RRL, it must be supplemented with hemin [to prevent translational inhibition due to eIF2-α phosphorylation by the heme-regulated kinase (13)] and treated with micrococcal nuclease (to digest endogenous mRNAs, primary coding for globin). This is done on 10 mL at a time and sub-aliquoted for later use to make the T2. Steps 1–5 below describe how to nuclease the RRL, and steps 7–9 describe the preparation of T2.
Thaw a 10 mL aliquot of crude reticulocyte lysate quickly and put immediately on ice.
Add 400 µL of hemin solution (final concentration will be 4 µM), 100 µL of 100mMCaCl2, and 100 µL of micrococcal nuclease (15 U/µL stock). Mix gently but thoroughly (by repeated inversion).
Incubate in 25°C water bath for 12 min, making sure the entire sample is immersed in the water to ensure even warming. Mix gently by inversion after ~3 or 4 min of incubation.
Transfer to ice, immediately add 100 µL of 200 mM EGTA, and mix gently but thoroughly by repeated inversion.
Dispense 1 mL aliquots into pre-chilled 2 mL microcentrifuge tubes on ice, thereby leaving enough room to add other components to prepare the T2 mix below.
Freeze in liquid nitrogen and store at −80°C. Nucleased RRL should generally be stable for at least 1–2 years.
To prepare T2 mix, thaw quickly and put immediately on ice a 1 mL aliquot of hemin/nuclease-treated RRL from step 6 above.
Add the following, in order: 6 µL creatine kinase (20 mg/ mL), 30 µL tRNA (10 mg/mL), 224 µL water, 300 µL Emix, and 150 µL CB 20X. The total volume will be 1.71 mL. Mix gently but thoroughly (see Note 8).
Dispense 200 µL aliquots into pre-chilled tubes on ice, freeze in nitrogen, and store at −80°C. The T2 mix is stable for up to a year at −80°C and will tolerate around four or five freeze-thaws if handled properly.
3.3. Linked Transcription and Translation Reactions
The translation mixture is optimized such that the products of the transcription reaction should be used directly in the translation reaction without any purification of the transcript. This ‘linked’ system works because the composition of the translation mix is adjusted to account for the Mg+2, DTT, and spermidine contributed from the transcription. Thus, it is critical to use the transcription reaction directly (i.e., not purified transcript or mRNA) because it is providing Mg+2, DTT, and spermidine to the translation, all of which are important. If purified RNA is used, then you need to add Mg+2 to 1.2 mM, spermidine to 0.4 mM, and DTT to at least 0.5 mM (to maintain a reducing environment in the translation reaction). The protocol below is an example of a typical linked transcription–translation reaction. It can be scaled as necessary.
Design and obtain oligos to PCR amplify the coding region of interest (typically from a plasmid-containing cDNA of interest). The PCR product should contain a 5′ T7 promoter for transcription. The 3′ primer should anneal at or beyond the stop codon. The following 5′ oligo contains the T7 promoter (italics) followed by a few linker nucleotides, a Kozak’s site, the start codon (bold), and the nucleotides that should be chosen to anneal to the coding region to be amplified (indicated here by underlined Ns) 5′-TAATACGACTCACTATAGGGAGACCATG NNNNNNNNNNNNNNNNNN-3′.
Use the above oligos and desired template plasmid to PCR amplify the coding region of interest. Any of several thermostable polymerases can be used, following the manufacturer’s supplied reagents and protocol. As an example, a typical 100 µL PCR reaction for Taq polymerase (New England Biolabs) contains 1X PCR buffer (final concentration; supplied as a 10X stock with the polymerase), 20 ng template plasmid, 1 µM (final concentration) of each primer, 200 µM (final concentration) each dNTP, 5 units Taq polymerase, water to 100 µL. We usually perform 30 cycles.
Use the Qiagen PCR purification kit to purify the PCR product as described by the manufacturer. Elute the product from the spin column with 50 µL water (not TE). Check an aliquot (usually 1 or 2 µL) on an agarose gel to confirm and estimate the quantity of amplification. Typically, 50 ng/µL concentration is normal for an average-sized product (~750 bp). Anywhere from 25 to 200 ng/µL should suffice for use as template in the transcription reaction below (see Notes 14 and 15).
For each DNA template to be tested, set up on ice the following: 7.6 µL T1 mix, 0.2 µL RNAsin, 0.2 µL RNA polymerase (either SP6 or T7, depending on the promoter of your template DNA), and 2 µL template DNA (see Note 16).
Incubate for 60 min at 40°C for SP6 and at 37°C for T7, then transfer to ice (see Note 17).
To translate the products of the transcription reaction, add the following directly to the completed transcription reaction on ice: 28.5 µL T2 mix, 5 µL 35 S-Met, and water/other reagents as desired to a final reaction volume of 50 µL. If co-translational translocation is being carried out, rough microsomes (RMs), proteoliposomes, liposomes, etc. are included, typically at ~2 to 5 µL per 50 µL reaction (see Note 18).
Incubate the translation reaction at 32°C for 30 min (for the typical 25–50 kD product). Longer for larger products, shorter for smaller products (see Note 19).
Following the translation reaction, transfer the tubes to ice. Remove 1 µL to a separate tube containing 19 µL SDS-PAGE sample buffer for direct analysis, keeping the remaining 49 µL on ice for downstream analysis (see Section 3.2.). Typically, we run half of this (saving the other 10 µL in case of technical problems with the gel) on a 0.75 mm thick minigel, which is then fixed, Coomassie stained (to confirm equal loading of all lanes), dried, and applied to film (Kodak-MR single emulsion film). Such direct analysis of the total translation products is helpful in troubleshooting downstream assays (see Section 3.2.) in case they yield confusing results because you will know exactly what you started with before any additional manipulations were performed (see Note 20).
3.4. Assays for Translocation and Topology
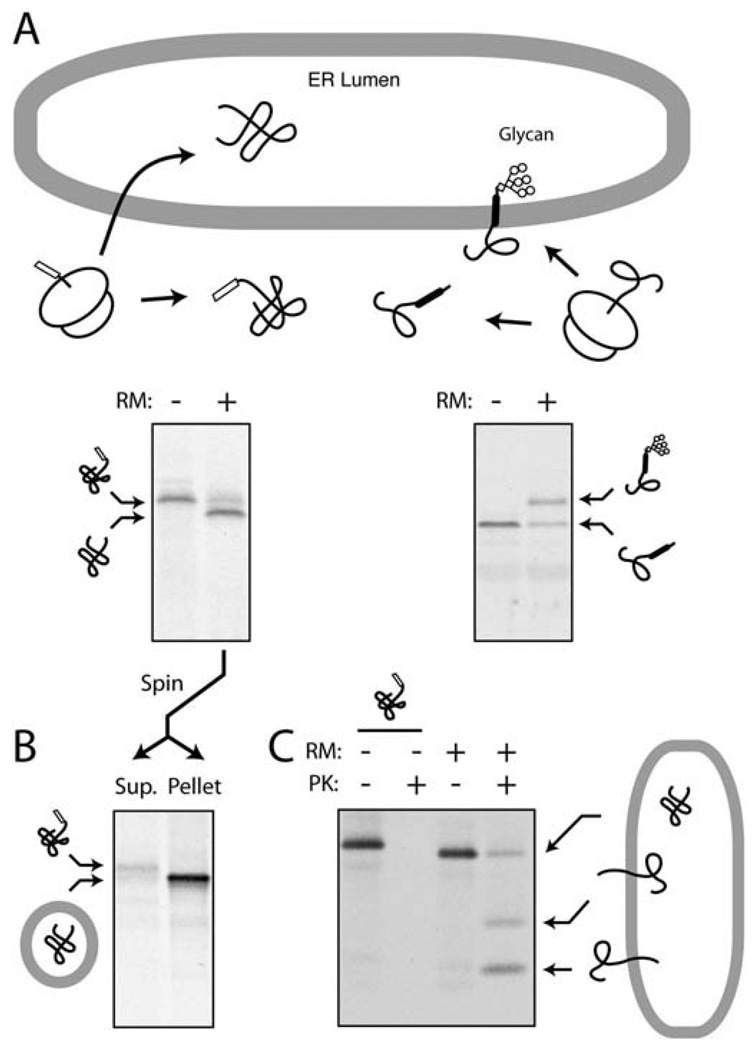
There are several ways to assay the segregation of proteins to the ER membrane. The three main ones are depicted in Fig. 20.2. Other assays have also been employed, but are not discussed further here. The first is to exploit an ER-specific modification such as glycosylation or signal sequence cleavage (Fig. 20.2A). Although this is very straightforward (typically detected as a change in migration by SDS-PAGE), it is not always applicable, may need the introduction of artificial glycosylation sites or epitopes into the substrate (8), and depends on the presence of functional and topologically restricted enzymes with very high activity and efficiency. This generally precludes its use if the ER components are fractionated. The second approach is to separate ER membranes from cytosol and determine whether the substrate cofractionates with the ER (Fig. 20.2B). Varying levels of stringency can be applied during the fractionation (e.g., high salt, pH 11.5, urea) to increase the specificity of substrate association and reduce background. However, even with these precautions, this approach cannot be used to definitively illustrate translocation (versus peripheral association) and cannot reliably provide information about protein topology. Nonetheless, such methods are very useful to re-isolate the membranes for post-translocational assays (e.g., retrotranslocation). And finally, protease protection assays can be employed to assay both translocation and topology (Fig. 20.2C). In this strategy, proteins (or portions of proteins) that are translocated or membrane inserted are shielded from proteases added to the cytosolic side of the membrane. By employing very high concentrations of an aggressive and relatively nonspecific protease, essentially all cytosolically exposed protein can be digested to leave only the specific translocated population. By combining this approach with immunoprecipitation, the background can be markedly minimized to allow detection of even very low efficiency translocation events. Below is a typical protocol for both fractionation (by both sedimentation and floatation) and protease protection assays from the 50 µL translation reaction produced in Section 3.3.
Fig. 20.2.
Assays for protein segregation to the ER. (A) ER translocation-dependent modification. In the example on the left, a precursor becomes processed by signal peptidase only upon its translocation into the lumen of rough microsomes (RM), an event that can be monitored by a change in migration on SDS-PAGE. In the example on the right, a glycosylation site becomes modified upon successful translocation (8). (B) Co-fractionation assays. A sample similar to that from panel A can be separated by centrifugation into a cytosolic supernatant and membrane pellet to assess successful translocation. (C) Protease protection assay. Upon addition of proteinase K (PK) to the products of a translocation reaction, proteins that are either fully or partially translocated into the lumen of RMs are protected. Even a protein that generates multiple topological forms (such as mammalian prion protein; see ref. 20) can be resolved by this assay. By contrast, lack of translocation leads to complete digestion upon PK treatment.
3.4.1. Isolation of Microsomes from Translation Reactions by Sedimentation
Remove 10 µL of the translation reaction and dilute with 90 µL 1X PSB on ice.
Layer this onto 100 µL of the sucrose cushion in an ultracentrifuge tube (either a tube for the TLA120.1 rotor or a micro-test tube for the TL100.3 rotor).
Spin for 5 min at 200,000 g (e.g., 70,000 rpms in the TL100.3 rotor). This spin time is suitable for traditional pancreatic RMs. If semi-permeabilized cells are used as source of ER, 5 min in a microcentrifuge may be sufficient. If proteoliposomes or smooth ER is used, longer spin times may be needed (e.g., 200,000 g for 30 min).
Remove the supernatant to a separate tube and resuspend the pellet in 1X PSB. It can be analyzed further if desired (e.g., by a protease protection assay as in Section 3.4.3.) or directly prepared for SDS-PAGE.
Analysis of the fractionation can be assessed by running equivalent amounts of the total (saved in step 8 of Section 3.3.), supernatant, and pellet fractions.
3.4.2. Isolation of Microsomes from Translation Reactions by Floatation
Remove 10 µL of the translation reaction and dilute with 90 µL of 2.2 M sucrose in 1X PSB. Considerable mixing will be required to ensure homogeneity.
Put into the bottom of a TLA120.1 tube. Layer with 100 µL of 1.8 M sucrose in 1X PSB followed by 25 µL of 1X PSB.
Centrifuge at 350,000 g for 1 h at 4°C.
Carefully remove the top 60 µL (the membranes will have floated to the top of the 1.8 M sucrose step). If the vesicles in this sample need to be recovered, the sample can be diluted in 1X PSB to 200 µL and centrifuged at 200,000 g for 20 min.
The floated vesicles can be analyzed by SDS-PAGE relative to the starting sample to assess the extent of membrane association.
3.4.3. Protease Protection Assay
Divide the translation reaction into three aliquots of 9 µL each on ice (A, B, and C).
Add 1 µL PSB to A, 0.5 µL PK and 0.5 µL PSB to B, and 0.5 µL PK and 0.5 µL Triton X-100 to C. Mix well and incubate on ice for 60 min.
To terminate the protease digest, both a protease inhibitor and rapid transfer to denaturating conditions are used. Either step alone is not fully sufficient to avoid artifacts. Approximately 10 min before the digestion reaction above is completed, start a boiling water bath.
Aliquot 100 µL of PKB into an appropriate number of empty tubes corresponding to the proteolysis reactions.
Dissolve a small amount of PMSF (2–5 mg) in DMSO to 0.25 M at room temperature.
Dip a P-2 pipette to draw up a very small amount (~0.1–0.2 µL) into the tip by capillary action and expel this into each proteolysis tube, mix, and put on ice.
After all tubes are completed, put the tubes containing the PKB into boiling water bath to pre-heat them to 100°C.
After tubes heated up (~1–2 min), transfer each proteolysis reaction directly into the boiling SDS solution and mix by rapidly pipetting up/down several times. Continue boiling sample for additional 1–2 min and remove to room temperature.
Analyze an aliquot (5 or 10 µL) of the three reactions (A–C) by SDS-PAGE and autoradiography. The remainder can be subjected to immunopreciptation if desired.
3.5. Reconstitution of Membrane Proteins into Proteoliposomes
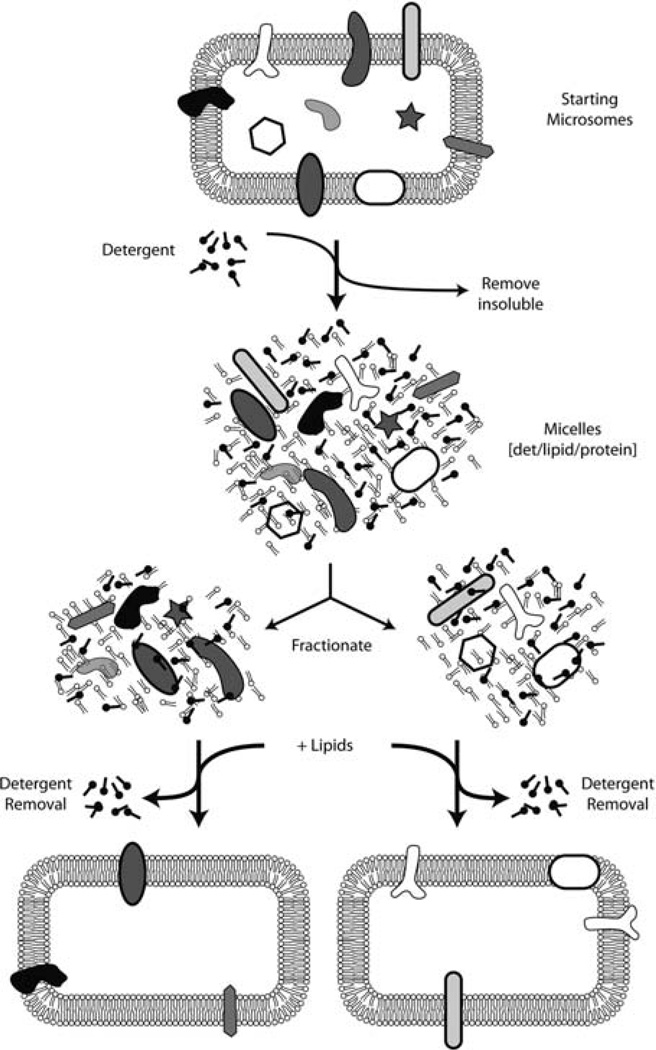
The basic steps in preparing reconstituted proteoliposomes are to prepare a mixture of membrane proteins, detergent, and lipid. The detergent is then slowly removed, during which the detergent–protein–lipid micelles will assemble into lipid vesicles containing the membrane proteins (Fig. 20.3). Removal of the detergent can be accomplished in several ways including simple dilution, dialysis, or adsorption. The method below is based on adsorption of the detergent to hydrophobic beads (Bio-Beads SM2, from Bio-Rad). The optimal conditions for reconstitution are difficult to predict and must be determined on a case-by-case basis. Furthermore, the orientation acquired by the membrane protein in the proteoliposomes is also not stochastic. Although it is often assumed that the orientation will be random, direct analysis shows this is not the case for many individual proteins.
Fig. 20.3.
Schematic depiction of membrane protein reconstitution. Crude microsomes are solubilized with detergent, fractionated, and reconstituted into proteoliposomes by removal of detergent in the presence of phospholipids. Note that not all proteins are successfully reconstituted, and the orientation achieved after reconstitution must be checked empirically.
From a practical standpoint, the protein, detergent, and lipids come from different sources and are mixed together just before reconstitution. The membrane protein(s), either a crude mixture or purified proteins, are already in detergent solutions to make them soluble. In addition, crude detergent extracts of membranes will also contain lipids. If the detergent and lipid contents need to be changed, the easiest method is to bind the membrane proteins to a chromatography resin, wash extensively with buffers containing the desired detergent, and elute in a buffer containing this new detergent. This is typically known as ‘detergent exchange’ and is often used if the detergent used for solubilization or purification is different from the one that proves to be best for reconstitution. A common example is the use of digitonin for solubilization and purification (due to its very gentle properties in maintaining membrane protein complexes), but exchange to another detergent just prior to reconstitution (because digitonin is very difficult to remove by dialysis or binding to Bio-Beads). See ref. (11) for an example.
The lipids are often provided separately, allowing the investigator to control the composition of the resulting proteoliposomes. They are usually easiest to prepare and handle as liposomes, but need to be added to the membrane protein as detergent-solubilized micelles. This way, the membrane proteins, lipid, and detergent will form mixed micelles at the start of the reconstitution.
3.5.1. Preparation of Lipids and Bio-Beads
Lipids are essential to making proteoliposomes containing solubilized membrane proteins. Although the precise composition of lipids may be largely irrelevant for reconstitution per se, they can significantly influence the activities of the reconstituted proteins. A simple mixture of phosphatidylcholine (PC) and phosphatidylethanolamine (PE) from a natural source (liver) in a 4:1 ratio is used here. However, other lipids, as well as cholesterol, can also be included if these are deemed important. Lipids are prone to oxidation and are therefore typically supplied in sealed ampules containing an inert gas (such as argon). With further manipulations, a reducing agent (e.g., DTT) is often included to prevent oxidation. In addition, lipids in organic solvents are typically not handled with plastic pipettes or put into plastic tubes. This is because the lipids often stick to the plastic, and the organic solvent can extract various contaminants from the plastic into your sample. Once lipids are hydrated in aqueous solutions (in which they form liposomes) or solubilized in detergent (in which they form micelles), they can be handled with the usual plastic pipettes and tubes.
Using glass measuring tools (e.g., glass pipettes), transfer 20 mg PC and 5 mg PE to a glass test tube or glass vial. If a tracer is desired to follow lipid recovery, include 0.5 mg rhodamine–DPPE (with accordingly less PE).
Dry the lipids under high vacuum. A conventional SpeedVac can be used. We usually leave it overnight to ensure complete removal of solvent. Alternatively, the organic solvent can be evaporated and the lipids dried to a film on the side of the tube using a stream of nitrogen (do this in the hood to avoid breathing chloroform vapors). After the bulk solvent is removed, the lipids can be lyophilized under high vacuum to remove any traces of organic solvent.
Hydrate the lipids by adding 0.4 mL lipid hydration buffer to the dried lipids and resuspend by agitation, vortexing, and/or sonication in a bath sonicator. Resist the temptation to mechanically resuspend the lipids (e.g., with a pipette) as they can be quite sticky at this stage. Be patient, and they will be fully resuspended with vortexing, at which point you will have a homogeneous cloudy/milky suspension of liposomes. Once resuspended, they can be handled with plastic pipettes and put into plastic tubes.
Transfer the lipids to an Eppendorf tube and adjust to 0.5 mL with additional lipid hydration buffer as necessary to make a 50 mg/mL suspension. This can be divided into 100 µL aliquots, frozen in liquid nitrogen, and stored at −80°C (see Note 22).
Put ~20–30 mL of dry Bio-Beads into a 50 mL polypropylene tube. Fill with MeOH, mix, and let the beads settle. Pour off the MeOH and fill with distilled water. Mix, let the beads settle (or if you are impatient, brief centrifugation), and pour off the water. Repeat this extensively (20 or more times) until all traces of MeOH are removed. Alternatively, pour the beads out into a disposable filter flask and use a vacuum to extensively wash the beads with distilled water. Put the beads back into a 50 mL polypropylene tube, fill with water, and store at 4°C. They are stable indefinitely.
3.5.2. Solubilization of ER Membranes
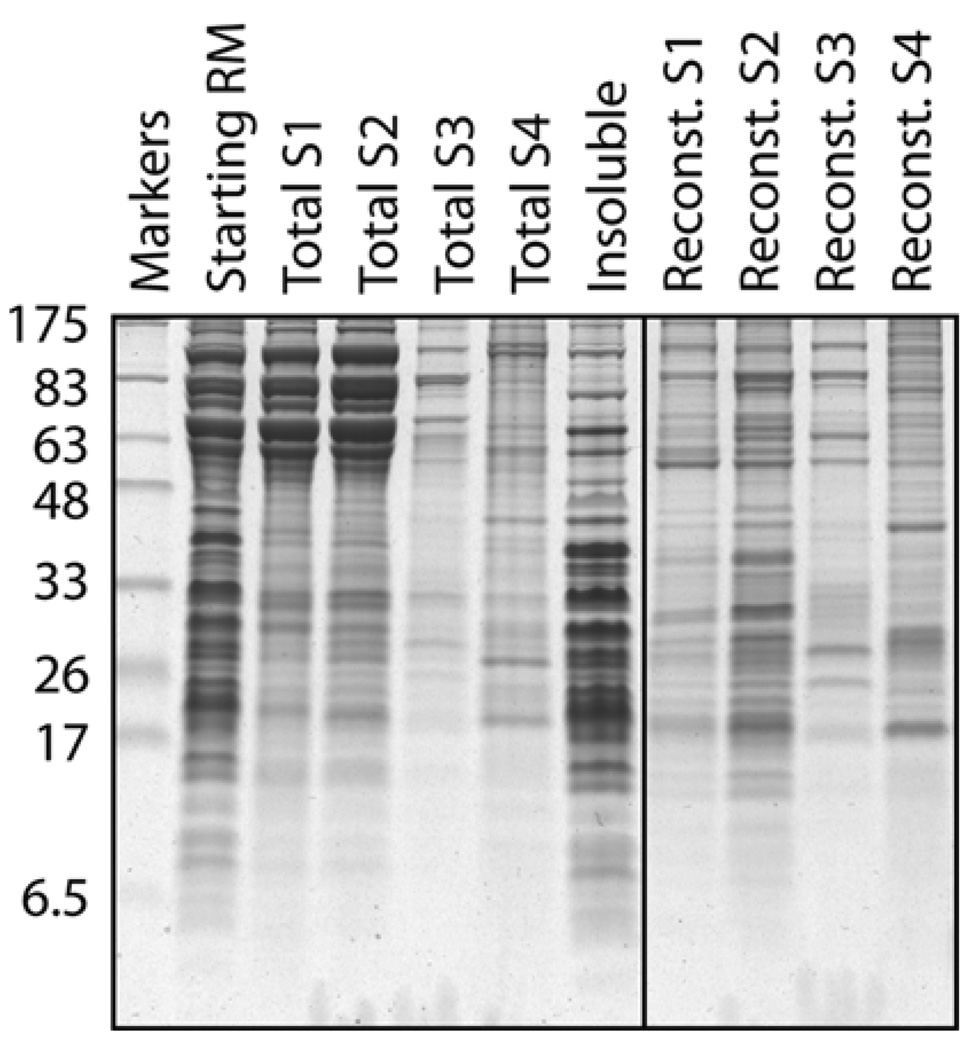
The solubilization of membrane proteins with detergent is influenced by many parameters. The most important are (i) the choice of detergent (see Note 21), (ii) the detergent concentration (more specifically, the relative ratio of detergent:protein:lipid), and (iii) the concentration and type of salt present during solubilization. The fact that different proteins are solubilized under different conditions can actually be used to effect some degree of purification via sequential selective solubilization (see Fig. 20.4). In addition to solubilization of proteins, maintaining its functionality further constrains the conditions. Hence, conditions that most efficiently solubilize a membrane protein may also result in its irreversible denaturation. While there are general guidelines, the solubilization conditions for any given protein or activity must be determined emperically. A general protein stabilizing agent such as 10–15% glycerol is often helpful. Some of the other less common parameters that influence stability of certain types of membrane proteins include divalent cations (e.g., Mg+2), nucleotides, free phospholipids, or specific co-factors. Below is a generic protocol for solubilizing and enriching for most ER membrane proteins under conditions that allow their relatively straightforward reconstitution. Although not described, the solubilized proteins can of course be fractionated (e.g., by ion exchange or lectin chromatography) prior to reconstitution of individual fractions by similar methods.
Fig. 20.4.
Example of differential solubilization and reconstitution of membrane proteins. Crude RMs (lane 1) were sequentially extracted by four buffers containing different amounts of detergent and salt to generate four supernatant fractions (S1–S4) and an insoluble pellet (primarily containing ribosomes). Each of these four fractions was then reconstituted in the presence of phospholipids by detergent removal, and the resulting proteoliposomes were also analyzed on the gel. Note that the very abundant high molecular weight proteins in S1 and S2 (primarily lumenal proteins) are not reconstituted. Note also the different protein profiles of the different proteoliposomes illustrating the utility of differential membrane protein extraction as a purification step. The detergent in this case was DeoxyBigCHAP, although similar results can be obtained with other detergents.
1 mL pancreatic rough microsomes (RM) at a concentration of 50 A260 units is put into a 3 mL thickwall polycarbonate centrifuge tube (Beckman) on ice. Add an equal volume of ice-cold pre-extraction buffer and mix well.
Centrifuge at 400,000 g in a TL100.3 rotor for 15 min at 4°C. Remove the supernatant (which will contain primarily lumenal proteins and some lipids that have been extracted by the low concentration of detergent making holes in the membrane). This can be saved for other uses if desired.
Resuspend the pellet in 0.9 mL ice-cold extraction buffer. This can be done by either gentle repeated pipetting or transfer to a small smooth glass homogenizer and manual homogenization with either a teflon or glass pestle. After resuspension, the solution should be homogeneous and turbid.
Add 100 µL of the 10% DBC on ice to solubilize the membrane. The solution should become more clear. Let sit on ice for 5–15 min to allow complete solubilization.
Centrifuge at 400,000 g in a TL100.3 rotor for 30 min at 4°C. Transfer the supernatant (the detergent extract, containing most of the membrane proteins and lipids) to a fresh tube on ice for use in the reconstitutions (see step 3 of Section 3.3.). The pellet will contain ribosomes, any tightly associated membrane proteins, and other membrane proteins that were not solubilized in step 4. If these remaining membrane proteins are desired, they can be solublized in a subsequent step using harsher conditions (e.g., a different detergent, higher salt). Otherwise, it can be discarded.
3.5.3. Reconstitution of Solubilized Membrane Proteins
The amount of Bio-Beads needed to effectively remove the detergent without significant removal of the lipids or protein, while accomplishing this slowly enough to allow formation of proteoliposomes without aggregation, needs to be determined empirically. The optimum varies depending on the detergent being used, its concentration, and the salt conditions. Below is a sample protocol for how we usually do this for any given detergent extract or sample (see Note 23).
Mix 40 µL of the liposome stock (prepared in step 1 of Section 3.3.) with 10 µL of the 10% DBC. If your samples to be reconstituted were prepared in another detergent, use that to solubilize the liposomes. Thus, the sample to be reconstituted typically contains only a single detergent.
Add the solubilized lipids from step 1 to the 1 mL of detergent extract (prepared in step 2 of Section 3.3.) on ice.
Dispense different amounts of the Bio-Beads (prepared in step 1 of Section 3.3.) into 0.5 mL microcentrifuge tubes and remove the water. A good range to start is between 30 and 200 µL of packed bead volume. An alternative is to use round-bottomed 2 mL microcentrifuge tubes.
Add 105 µL of the detergent–lipid mixture (from step 2) to each of the tubes containing the Bio-Beads.
Incubate with gentle overhead mixing for ~12–16 h at 4°C. If using the 2 mL tubes, use orbital shaking to mix (e.g., an Eppendorf Thermomixer place in the cold room, set at 700–800 rmps).
Separate the fluid from the beads. If using the 0.5 mL tubes, we briefly spin down the sample (a pulse in the microcentrifuge), cut off the lid, puncture the bottom with a 26-gauge needle, put the tube into a larger 1.5 mL microcentrifuge tube, and spin for ~1 min to recover the fluid into the larger tube (while retaining the beads in the smaller tubes). Alternatively, remove the fluid carefully with a thin long pipette tip (e.g., a gel-loading tip) and transfer to 1.5 mL tubes.
Add five volumes (500 µL) of ice-cold water (or 1X PSB). This serves to dilute any residual detergent and reduce the concentration of glycerol to reduce the density of the solution.
Centrifuge the samples in a TL100.3 rotor with micro-test tubes and adaptors at 200,000 g for 30 min.
Remove the supernatant and resuspend the pellet in 25 µL 1X PSB by careful repeated pipetting.
The efficiency of lipid recovery can be monitored by including rhodamine-DPPE in the lipid mixture (see step 1 of Section 3.3.). The amount of lipid in the starting sample (step 2 above) can then be compared to the amount in the final proteoliposomes (step 9 above) by measuring absorbance at 560 nm.
The efficiency of protein reconstitution can be assessed by comparing the starting extract and final proteoliposomes by SDS-PAGE and staining for total proteins or immunoblotting for individual membrane proteins.
Acknowledgments
Work in the Hegde lab is supported by the NICHD Intramural Research Program of the National Institutes of Health.
Footnotes
We use several precautions for handling all of the reagents for the T1 and T2 mixes. Use de-ionized, clean, RNAse-free water for all of the solutions. In general (unless otherwise indicated), we quick-freeze items by immersion in liquid nitrogen and thaw quickly using either the warmth of your hands or a room temperature water bath. The rapid freezing and thawing are important for a few reasons. First, it minimizes/avoids precipitation of various reagents that can occur upon slow freezing/thawing. Second, it minimizes oxidation. Third, proteins are better protected from damage/denaturation by ice crystals if freezing/thawing is rapid. Immediately after thawing, reagents are generally kept on ice unless otherwise noted and immediately put away after use.
The pH of solutions is important. To avoid differences in the ways different people measure pH (different brands of pH paper, pH meter, etc.), we have provided the exact amounts of acid/base to add for most solutions that require accurate buffer/pH conditions. Other items can be adjusted to approximately the indicated pH using pH paper as a rough guide. We also typically favor HEPES over Tris buffers because the former has a pKa closer to 7, and its pH does not vary with temperature.
Cap can be omitted from the transcription reaction if cost is an issue, and if an approximately twofold lower efficiency of translation can be tolerated. In this case, use 5 mM instead of 1 mM GTP in the 10X NTPs stock above.
Supplementing RRL translations with tRNA is not absolutely necessary and can be omitted if cost is an issue. It does, however, stimulate the translation of some proteins (presumably because the endogenous levels of some tRNAs in the RRL are limiting), and it is possible that some RRL preparations contain relatively low amounts of tRNA and thus would benefit from supplementation.
Endogenous RRL usually contains reasonable amounts of all of the amino acids, making supplementation optional. However, some batches of RRL may contain more or less of particular amino acids, so we always supplement with a complete mixture to avoid variability.
The most active and well-characterized source of ER-derived microsomes is from canine pancreas. Preparation of canine pancreas RMs in-house follows a well-established protocol (1). Microsomes have also been prepared from many other sources including rat liver (2) and cultured cells. In addition, semi-permeabilized cells have also been used and are a viable alternative (3). However, for the purposes of fractionation and reconstitution studies, rather large amounts of microsomes are needed and can realistically only be isolated from tissue since such large-scale cell culture is prohibitive. It is worth noting that microsomes from different sources may have different functional properties even though they are all ER-derived.
PK should be of the highest purity available commercially. Because it is purified from natural sources, contamination with other enzymes that could affect reliability or disrupt membranes (e.g., lipases) is a potential concern. Historically, preparations of PK were ‘pre-digested’ for 10 min at 25°C to proteolytically destroy any possible contaminants (PK is stable to its auto-digestion), but this has proven unnecessary in our experience.
Mix solutions gently but thoroughly. Keep in mind that many components are of considerably different density and do not always mix together with just a little tap of the tube. When mixing protein-containing solutions, avoid making bubbles or frothing the sample to minimize the possibility of denaturation.
RNAses are a potential problem because they are ubiquitous and will obviously preclude the transcription and translation reactions. We have found that simple cleanliness and care are the best strategies. The use of DEPC-treated water for the various reagents is helpful. Keeping your pipettes and tips clean and free from dust is helpful. Do not touch your tubes and tips with bare hands. The inclusion of RNAsin is helpful, but only inhibits certain RNAses, and is not really necessary. In general, we can do all of this without RNAse inhibitor by following the above precautions, and we do not have problems with degradation.
Optimal conditions for translation can vary somewhat between different transcripts. The most important parameters are Mg+2 (typically between 1.5 and 3 mM) and K+ (typically between 50 and 150 mM). Note that because a crude reticulocyte lysate is used, it contains endogenous Mg+2 and K+ (estimated to be ~1.7 and ~40 mM, respectively), probably at around one-third the levels typical for the cytosol. Hence the need to supplement these salts, especially because the added nucleotides can chelate Mg+2.
Microsomes subjected to multiple freeze-thaws will lose lumenal contents by leakage. Presumably, the integrity of the vesicles is sensitive to freeze-thawing. If the presence of these proteins in the reaction might pose a problem for your assays, it is helpful to re-sediment the microsomes and resuspend them in PSB just before use.
Cheaper alternatives to purified liver phospholipids are egg or soy-derived PC and PE. However, the acyl chain compositions can be quite different and may influence your proteins and/or activities. This needs to be tested emperically. In addition synthetic lipids of defined acyl chain composition are also available (e.g., DOPC, POPC) if you wish to precisely control lipid composition of the membrane. Other fluorescent lipids are also available to suit your requirements.
DBC is a detergent of the bile salt family. Others in the same family are Cholate, CHAPS, and BigChap. Among these, Cholate is the least expensive, but is also anionic and would therefore interfere with ion exchange fractionation. CHAPS is a good alternative that is relatively inexpensive and zwitterionic, making it more suitable for use in fractionation. Another commonly used detergent for reconstitution is octyl-glucoside because it is uncharged and is easily removed by either dialysis or Bio-Beads. A more thorough discussion of detergents is presented in ref. (14).
Although circular or linearized plasmids can be used for transcription/translation, PCR products are usually easiest and work well. In either case, you will need an SP6 or T7 promoter and a consensus Kozak’s sequence at the start codon. A poly-A tail is not necessary for efficient in vitro translation. Even if there is no promoter in the plasmid containing the coding region of interest, you can simply encode the SP6 or T7 promoter and Kozak’s sequence into the 5′ primer used for PCR (as is described in step 1 of Section 3.3.). Note that the 5′ primer can be designed to anneal internally to generate an N-terminally truncated protein. Similarly, 3′ primers containing a stop codon can be designed within the coding region to generate a C-terminally truncated protein. A DNA template lacking a stop codon will result in a translation product that remains tethered to the ribosome via the last amino-acyl tRNA (15). Such truncated translation intermediates have many applications (for example, see ref. 16–19), but are not considered further here.
If you have numerous constructs in a vector that already has an SP6 or T7 promoter (as we do), you can use the same primers to amplify them all for translation: just use a 5′ primer that anneals to the SP6 or T7 site and a 3′ primer that anneals to the vector sequence beyond the open reading frame. The circular plasmids can also be used directly in the transcription reaction. For this purpose, use DNA that is free of RNAse contamination and is at between 100 and 1000 ng/µL concentration (as is typical for standard Qiagen minipreps of high-copy plasmids).
In instances where many transcription reactions are being performed simultaneously, a master mix lacking the DNA can be prepared, aliquoted, and supplemented with the different DNAs. This can easily be scaled up or down as needed. A control reaction with water instead of DNA can be performed to determine the extent of background in the translation reactions.
Transcription reactions can be incubated for longer times (up to 2 or 3 h), but there is no significant increase in yield under these conditions (the nucleotides become limiting and the pyrophosphate that is generated inhibits the polymerase). After transcription, the sample can be kept on ice for some time (an hour or two) without any concern of degradation. If frozen in nitrogen and stored at −80°C, the transcript can be used again. However, we almost always make it fresh for each experiment and discard any leftover material (to avoid the risk of degradation with storage).
The amount of 35 S-Met can be decreased depending on the amount of labeling needed. If unlabeled translations are being performed, cold Met (at a stock concentration of 0.4 mM) should be added instead (to 40 µM final concentration).
Incubations are generally at ~30–32°C, but translation works at anywhere from 23 to 37°C. Typical translation times are 30–60 min, depending on size of expected products. In general, it takes ~5 min to complete a 100 residue protein (~10 kD) at 32°C. Thus, we generally incubate for ~10 min per 10 kD of expected size product. Longer incubations generally do not produce more product since translational activity of the lysate declines. Translation is slower at lower temperatures.
We find that for a small- to average-sized protein (~25–30 kD) containing an average proportion of methionines, a band corresponding to the translation product can be detected in as little as 5 or 10 min for optimally translating proteins. Preprolactin and prion protein are often used in our lab, and both express comparably well. Note that hemoglobin, present in the RRL at ~50 mg/mL, migrates at 14 kD. Although it is not radio-labeled, it distorts this region of the gel and can cause artifacts if your translated product is of the same size. This can be avoided by loading 10-fold less of the translation product and exposing the gel for longer times. Alternatively, the translation products can be immunoprecipitated.
There have been many instances where detergents vary sufficiently from lot to lot to affect the results of functional reconstitution (e.g., ref. 11). It is worth keeping this in mind if you have issues with reproducibility. Some detergents are more variable than others, and ones from natural sources (e.g., digitonin) are especially difficult in terms of reliability. Cost can also be an issue in detergent choice. Thus, some degree of screening can be important in initial studies. The time invested is usually very worthwhile and pays off in the long run.
Although freezing and thawing liposomes are not recommended in many protocols, this is not really relevant here (as long as oxidation is avoided), since they will be subsequently solubilized in detergent. Thus, alteration of liposome size or maintaining them as unilamellar vesicles is not an issue.
Reconstitution of purified membrane proteins in detergent solution can be accomplished using basically the same methodology. However, it may be necessary to re-optimize conditions to maximize recovery, which can be expected to be at least 50%. Generally speaking, one tries to use the least amount of lipids needed to fully incorporate the protein of interest into vesicles (to maximize density in the membrane).
References
- 1.Walter P, Blobel G. Preparation of microsomal membranes for cotranslational protein translocation. Methods Enzymol. 1983;96:84–93. doi: 10.1016/s0076-6879(83)96010-x. [DOI] [PubMed] [Google Scholar]
- 2.Adelman MR, Blobel G, Sabatini DD. An improved cell fractionation procedure for the preparation of rat liver membrane-bound ribosomes. J Cell Biol. 1973;56:191–205. doi: 10.1083/jcb.56.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wilson R, Allen AJ, Oliver J, Brookman JL, High S, Bulleid NJ. The translocation, folding, assembly and redox-dependent degradation of secretory and membrane proteins in semi-permeabilized mammalian cells. Biochem J. 1995;307:679–687. doi: 10.1042/bj3070679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stefanovic S, Hegde RS. Identification of a targeting factor for posttranslational membrane protein insertion into the ER. Cell. 2007;128:1147–1159. doi: 10.1016/j.cell.2007.01.036. [DOI] [PubMed] [Google Scholar]
- 5.Schuldiner M, Metz J, Schmid V, Denic V, Rakwalska M, Schmitt HD, Schwappach B, Weissman JS. The GET complex mediates insertion of tail-anchored proteins into the ER membrane. Cell. 2008;134:634–645. doi: 10.1016/j.cell.2008.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daniels R, Kurowski B, Johnson AE, Hebert DN. N-linked glycans direct the cotranslational folding pathway of influenza hemagglutinin. Mol Cell. 2003;11:79–90. doi: 10.1016/s1097-2765(02)00821-3. [DOI] [PubMed] [Google Scholar]
- 7.Oberdorf J, Skach WR. In vitro reconstitution of CFTR biogenesis and degradation. Methods Mol Med. 2002;70:295–310. doi: 10.1385/1-59259-187-6:295. [DOI] [PubMed] [Google Scholar]
- 8.Brambillasca S, Yabal M, Soffientini P, Stefanovic S, Makarow M, Hegde RS, Borgese N. Transmembrane topogenesis of a tail-anchored protein is modulated by membrane lipid composition. EMBO J. 2005;24:2533–2542. doi: 10.1038/sj.emboj.7600730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fons RD, Bogert BA, Hegde RS. Substrate-specific function of the translocon-associated protein complex during translocation across the ER membrane. J Cell Biol. 2003;160:529–539. doi: 10.1083/jcb.200210095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garrison JL, Kunkel EJ, Hegde RS, Taunton J. A substrate-specific inhibitor of protein translocation into the endoplasmic reticulum. Nature. 2005;436:285–289. doi: 10.1038/nature03821. [DOI] [PubMed] [Google Scholar]
- 11.Görlich D, Rapoport TA. Protein translocation into proteoliposomes reconstituted from purified components of the endoplasmic reticulum membrane. Cell. 1993;75:615–630. doi: 10.1016/0092-8674(93)90483-7. [DOI] [PubMed] [Google Scholar]
- 12.Jackson RJ, Hunt T. Preparation and use of nuclease-treated rabbit reticulocyte lysates for the translation of eukaryotic messenger RNA. Methods Enzymol. 1983;96:50–74. doi: 10.1016/s0076-6879(83)96008-1. [DOI] [PubMed] [Google Scholar]
- 13.Trachsel H, Ranu RS, London IM. Regulation of protein synthesis in rabbit reticulocyte lysates: purification and characterization of heme-reversible translational inhibitor. Proc Natl Acad Sci USA. 1978;75:3654–3658. doi: 10.1073/pnas.75.8.3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Helenius A, Simons K. Solubilization of membranes by detergents. Biochim Biophys Acta. 1975;415:29–79. doi: 10.1016/0304-4157(75)90016-7. [DOI] [PubMed] [Google Scholar]
- 15.Perara E, Rothman RE, Lingappa VR. Uncoupling translocation from translation: implications for transport of proteins across membranes. Science. 1986;232:348–352. doi: 10.1126/science.3961485. [DOI] [PubMed] [Google Scholar]
- 16.Wiedmann M, Kurzchalia TV, Hartmann E, Rapoport TA. A signal sequence receptor in the endoplasmic reticulum membrane. Nature. 1987;328:830–833. doi: 10.1038/328830a0. [DOI] [PubMed] [Google Scholar]
- 17.Görlich D, Hartmann E, Prehn S, Rapoport TA. A protein of the endoplasmic reticulum involved early in polypeptide translocation. Nature. 1992;357:47–52. doi: 10.1038/357047a0. [DOI] [PubMed] [Google Scholar]
- 18.Crowley KS, Reinhart GD, Johnson AE. The signal sequence moves through a ribosomal tunnel into a noncytoplasmic aqueous environment at the ER membrane early in translocation. Cell. 1993;73:1101–1115. doi: 10.1016/0092-8674(93)90640-c. [DOI] [PubMed] [Google Scholar]
- 19.Hegde RS, Lingappa VR. Sequence-specific alteration of the ribosome-membrane junction exposes nascent secretory proteins to the cytosol. Cell. 1996;85:217–228. doi: 10.1016/s0092-8674(00)81098-3. [DOI] [PubMed] [Google Scholar]
- 20.Hegde RS, Mastrianni JA, Scott MR, DeFea KA, Tremblay P, Torchia M, DeArmond SJ, Prusiner SB, Lingappa VR. A transmembrane form of the prion protein in neurodegenerative disease. Science. 1998;279:827–834. doi: 10.1126/science.279.5352.827. [DOI] [PubMed] [Google Scholar]