Abstract
Background Between 5 and 25 April 2009, pandemic (H1N1) 2009 caused a substantial, severe outbreak in Mexico, and subsequently developed into the first global pandemic in 41 years. We determined the reproduction number of pandemic (H1N1) 2009 by analyzing the dynamics of the complete case series in Mexico City during this early period.
Methods We analyzed three mutually exclusive datasets from Mexico City Distrito Federal which constituted all suspect cases from 15 March to 25 April: confirmed pandemic (H1N1) 2009 infections, non‐pandemic influenza A infections and patients who tested negative for influenza. We estimated the initial reproduction number from 497 suspect cases identified prior to 20 April, using a novel contact network methodology incorporating dates of symptom onset and hospitalization, variation in contact rates, extrinsic sociological factors, and uncertainties in underreporting and disease progression. We tested the robustness of this estimate using both the subset of laboratory‐confirmed pandemic (H1N1) 2009 infections and an extended case series through 25 April, adjusted for suspected ascertainment bias.
Results The initial reproduction number (95% confidence interval range) for this novel virus is 1·51 (1·32–1·71) based on suspected cases and 1·43 (1·29–1·57) based on confirmed cases before 20 April. The longer time series (through 25 April) yielded a higher estimate of 2·04 (1·84–2·25), which reduced to 1·44 (1·38–1·51) after correction for ascertainment bias.
Conclusions The estimated transmission characteristics of pandemic (H1N1) 2009 suggest that pharmaceutical and non‐pharmaceutical mitigation measures may appreciably limit its spread prior the development of an effective vaccine.
Keywords: Epidemiologic methods, infectious disease outbreak, influenza, initial reproduction number, pandemic
Introduction
Influenza A of the H1N1 subtype caused the 1918 pandemic, was replaced in 1957 by the H2N2 subtype, re‐emerged in 1976/1977, and has since variously contributed to influenza illness (alongside the dominant H3N2 subtype that emerged in 1968). 1 The A/H1N1 subtype was present in swine populations by 1930, and separate North American and Eurasian lineages are now considered endemic in these animals. 2 During the fourth week of April 2009, pandemic (H1N1) 2009 virus consisting of North American and Eurasian components was identified as the cause of sporadic but mild human illness in California and Texas and a substantial and severe outbreak in Mexico.
This novel influenza variant is believed to be the result of a recent reassortment event among three distinct swine influenza virus lineages, resulting in a novel hybrid H1N1 virus including North American swine hemagglutinin (H1), Eurasian neuraminidase (N1) and matrix proteins, and contribution of remaining segments from the classic triple reassortant swine virus. 3 The human index case of pandemic (H1N1) 2009 appears to have occurred in the town of La Gloria in the state of Veracruz, a region containing large‐scale industrial pig farms. Spread to Mexico City occurred by 15 March 2009. 4 Between 15 March and 19 April, sporadic cases in Mexico City increased erratically, with person‐to‐person transmission becoming sustained and amplified after mass population return to the city following the Holy Week holiday (5–19 April). A public health emergency was decreed in Mexico on 23 April. After laboratory confirmation of pandemic (H1N1) 2009 infection on 23 April, Direccion General de Epidemiologia, Secretaria de Salud, Mexico (DGE) developed case definitions. A suspected case was defined as severe respiratory illness with fever, cough and difficulty breathing. A probable case was defined as a suspected case in a patient from whom a specimen had been collected and tested positive for influenza A. A confirmed case was defined as a probable case that tested positive for pandemic (H1N1) 2009 by real‐time reverse‐transcription polymerase chain reaction. Health‐care officials were contacted and asked to provide retrospective and ongoing data for persons having illness consistent with these case definitions and seeking care on or after 1 March. As of 15 June, 6241 cases including 108 deaths have been reported as laboratory‐confirmed because of pandemic (H1N1) 2009 in Mexico. Returning travelers have also seeded cases in 74 other countries, including 35 928 confirmed cases globally as of 15 June with 55 deaths reported outside of Mexico to date. 5 On 11 June, the World Health Organization (WHO) declared the outbreak to be the first global influenza pandemic in 41 years. 6
The initial community outbreak in Mexico City, a metropolitan urban area with about 20 million inhabitants, provides a critical glimpse into the further epidemic potential of this virus. Early transmission in Mexico City was punctuated by two pivotal sociological events. First, from 5 to 19 April, schools were closed and approximately 10% of the population left the city for the Holy Week holiday. 7 This may have temporarily reduced transmission within the city while increasing the risk of spread to other communities. Second, on 23 April, the government of Mexico City declared a public health emergency that may have increased hospital visits for milder illness that might otherwise have gone unreported. Mexico as a whole experienced a mild and delayed 2008–2009 influenza season, with other human subtypes of influenza viruses; the extent to which human influenza and other respiratory viruses contributed to illness reports during that period is unknown.
Using the time series of the first 497 suspect cases, we have estimated the initial rate of spread – the reproduction number R– of pandemic (H1N1) 2009, using methods of contact network epidemiology that incorporates extrinsic sociological drivers and uncertainties in disease progression and underreporting. 8 , 9 , 10 To date, it is unknown whether prior infection with human A/H1N1 subtypes provides cross immunity to the novel variant; if not, our estimate reflects its basic reproduction number R 0.
Methods
We analyzed four data sets provided by public health officials in Mexico City. The first time series consists of early reports of suspect influenza infection from all hospitals in the Mexico City Metropolitan Area between 15 March and 30 April (hereafter S) (Figure 1). This data included dates of self‐reported onset of symptoms and hospitalization for approximately 3798 suspect cases of pandemic (H1N1) 2009 variant. While it is likely that these cases ranged from mild to fatal and that treatment ranged from home‐based self‐care to hospitalization, such individual‐level clinical information was not available. However, prior to laboratory testing, it was unknown which of these infections were pandemic (H1N1) 2009 rather than typical influenza A or other respiratory infectious agents. After laboratory results from suspect cases were released on 4 June 2009, we received three additional epidemiological time series provided by the Mexico City Ministry of Health (Secretaria de Salud del Distrito Federal) from Mexico City (Distrito Federal) from 1 March to 30 May. These mutually exclusive data sets include symptom onset dates for cases where laboratory tests were negative for influenza A virus (hereafter N), cases where laboratory tests were positive for any influenza A strain, excluding pandemic (H1N1) 2009 (hereafter A), and cases with laboratory confirmation for pandemic (H1N1) 2009 (hereafter C) (Figure 1).
Figure 1.
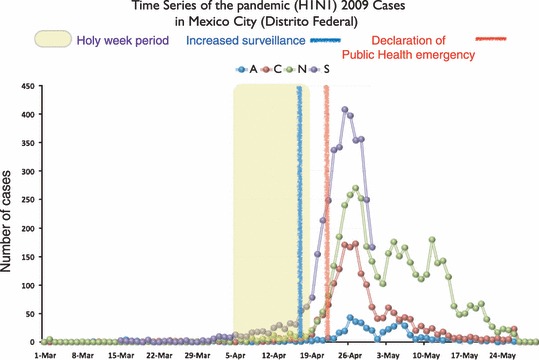
Four time series collected during the March–April 2009 pandemic (H1N1) 2009 outbreak in Mexico City. The first series (S) was provided by the Secretaria de Salud del Distrito Federal (SS‐DF) in early May during the first stage of our investigation. This series corresponds to individuals who met the case definition for a suspected case (please see the text for more information). The three remaining time series were provided in early June, also by SS‐DF. These three series correspond to the number of pandemic (H1N1) 2009 confirmed cases (C), non‐pandemic (H1N1) 2009 influenza A cases (A) and non‐influenza cases (N).
Using a network‐based statistical approach, we estimated the initial reproduction number (R) of the novel North American A/H1N1 variant during this period of spread. 11 This method initially estimates the time series of infection dates and the rates of removal through recovery, death or hospitalization, and then incorporates these values into a stochastic, network‐based model to estimate the reproduction number. The estimate is refined with every additional day of time series data. For typical unmitigated influenza epidemics, values from this method typically converge to the best estimate of R within a few days, before the acceleration of the outbreak, and remain stable throughout the epidemic period. Therefore, any deviations from the converged value likely reflect external influences including social events and/or intervention [please refer to the supplementary material for more details].
The estimation method requires information about disease progression, specifically the duration of the incubation (t l), asymptomatic infectious (t a) and symptomatic infectious (t s) periods, as well as the contact patterns underlying disease transmission (specifically the expected number of contacts for each new case). At the time of this study, little was known about these parameters for the pandemic (H1N1) 2009 outbreak in Mexico City. Empirical data from contact investigations of confirmed cases of pandemic (H1N1) 2009 in the Canadian province of Ontario suggest that the median interval between contact with a confirmed symptomatic case and development of symptoms is 6 days (95% CI 5–7 days). As such, we used a 5‐day latent period and 2‐day asymptomatic infectious period as a plausible upper bound for time from infection until development of symptomatic disease. Thus, we considered a wide range of possible values estimated for other strains of influenza and large urban centers, respectively (Table 1). The infectious period of a case may be curtailed by hospitalization or self‐isolation prior to recovery or death. We therefore directly estimated the distribution of removal rates via either natural causes or intervention by calculating the numbers of days between the onset of symptoms and hospitalization (type II and III triage protocol) or self‐isolation (type I triage protocol) for all reported cases. We assumed that unreported asymptomatic or mild cases 12 remained infectious until recovery.
Table 1.
Baseline values and ranges of epidemiological and social contact parameters that were used in the sensitivity analysis.
| Parameter symbol | Definition | Estimated values Baseline (range) | Reference |
|---|---|---|---|
| t l | Latency period (days) | 3 (1–5) | (23) |
| t a | Asymptomatic infectious period (days) | 1 (0–2) | (17, 18) |
| t s | Symptomatic infectious period (days) | 7 (4–10) | (23) |
| Z | Excess degree (normal) (please see SOM for definition) | 30 (20–40) | (24, 25, 26) |
| Excess degree (Holy Week) (please see SOM for definition) | 15 (5–20) | (24, 25, 26) |
Results
The early time series of suspect cases in Mexico City (S) revealed a 1‐month period in which daily case counts remained low (never surpassing twenty) followed by a fairly sharp climb and descent (Figure 1). The large pulse, from 78 to 407 cases occurred from 19 April – the last day of the Holy Week period – to 26 April, shortly after initial pronouncements by Mexico’s Public Health Secretary. The number of laboratory confirmed cases of pandemic (H1N1) 2009 (epidemic curve C) was significantly lower than the initial curve of suspected patients (S). Many of the original suspect cases were not infected with influenza A at all (N), and a number were infected by influenza A strains other than pandemic (H1N1) 2009 (A).
Based on suspect cases prior to 20 April, we estimate a reproduction number of 1·51 (95% CI 1·32–1·71); for confirmed cases, it is 1·43 (95% CI 1·29–1·57). (Table 2) To assess the reliability of estimates made from early epidemic data, we estimated the reproduction number of pandemic (H1N1) 2009 using both early data (Figure 1, curve A) and later data (Figure 1, other curves), and compared the results of these two analyses. For each day, we estimate R using the full time series preceding and including that day. The estimates stabilize approximately one week before the end of Holy Week. This value likely represents the intrinsic rate of transmission that would ultimately drive the exponential growth of epidemic (the so‐called ‘epidemic curve’).
Table 2.
Estimates of the initial reproduction number, R, for the four time series S, C, A and N (introduced in the text) before and after public health alerts (19 April and 25 April, respectively). The mean and the 95% confidence interval range reported in this table are derived from Figure 4.
| Time Series | Mean R (95% CI range) (estimated on 19 April 2009) | Mean R (95% CI range) (estimated on 25 April 2009) |
|---|---|---|
| Suspect (early time series) | 1·51 (1·32–1·71) | 2·04 (1·84–2·25) |
| Confirmed | 1·43 (1·29–1·57) | 2·26 (2·01–2·49) |
| N_adjusted confirmed (inflated) | 1·43 (1·29–1·57) | 1·32 (1·20–1·43) |
| N_adjusted confirmed (deflated) | 1·43 (1·29–1·57) | 1·32 (1·20–1·43) |
| A_adjusted confirmed (inflated) | 1·43 (1·29–1·57) | 1·44 (1·38–1·51) |
| A_adjusted confirmed (deflated) | 1·43 (1·29–1·57) | 1·44 (1·38–1·51) |
The analysis of seasonal influenza incidence in Mexico from 1990 to 2005 suggests that the bulk of seasonal influenza transmission occurs between November and February, declining in March and subsiding in April. 13 Therefore, the apparent peak (Figure 1, curve A) between 19 April and 10 May likely does not correspond to a seasonal influenza epidemic peak. The surprisingly synchronous changes in non‐influenza A cases (curve N) suggests that some of the fluctuations may be caused by extrinsic factors rather than underlying transmission dynamics. Assuming that there were no other respiratory epidemics in the same period, it is plausible that the marked increase in reported cases after 19 April is attributable to a surge in notification spurred by the end of Holy Week and the public health decrees.
Assuming that the improved surveillance and public health alerts led to increased case ascertainment of both A and C in a similar manner, one can use the rates of A to ‘deflate’ the number of confirmed cases with date of symptoms onset later than 17 April to estimate the underlying transmission‐driven incidence curve (Figure 2A,B). This adjusted time series should represent the number of cases identified if the public health measures described had not been implemented. Of note, the peak of the adjusted epidemic curve shifts from 27 April to 23 April, the date the public alert took place and when the social distancing interventions were implemented. This suggests that the public health interventions may have significantly mitigated the epidemic. Adjustment using both the patients without influenza (curve N) and those with non‐pandemic influenza A (curve A) has similar impacts (Figure 2). Looking at the time period immediately before 23 April, one may also use either curve A or N to ‘inflate’ the number of confirmed cases to reflect the hypothetical scenario of early heightened surveillance and public awareness (from the start of the epidemic) (Figure 3A,B). This approach increases confirmed cases closer to the ‘true’ prevalence. Estimates of R based on the four adjusted curves are also given in Table 2. Unlike the estimates based on the raw data, these estimates remain relatively stable throughout the period of heightened awareness and surveillance.
Figure 2.
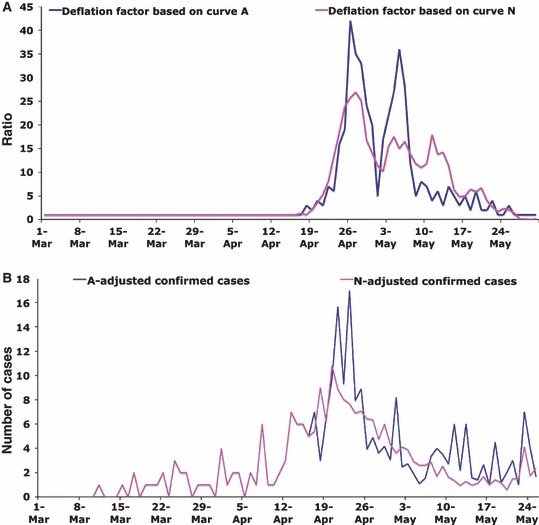
Adjusted number of confirmed cases (C): ‘deflated time series’. The ratios of the number of non‐pandemic (H1N1) 2009 influenza A cases on 18 April (from time series A in Figure 1) to the number of non‐pandemic (H1N1) 2009 influenza A cases in each subsequent day were calculated [blue curve (A)]. These ratios were used to create a ‘deflated’ time series for confirmed cases [blue curve (B)] (please see the text for more details). The procedure was repeated replacing the number of non‐pandemic (H1N1) 2009 influenza A cases with non‐influenza A cases (time series N in Figure 1) the results of which are shown in magenta curves in (A) and (B). The large visible trough in the blue curve in (A) corresponds to fewer confirmed cases reported during the International Workers’ Day (May 1) long weekend in Mexico City.
Figure 3.
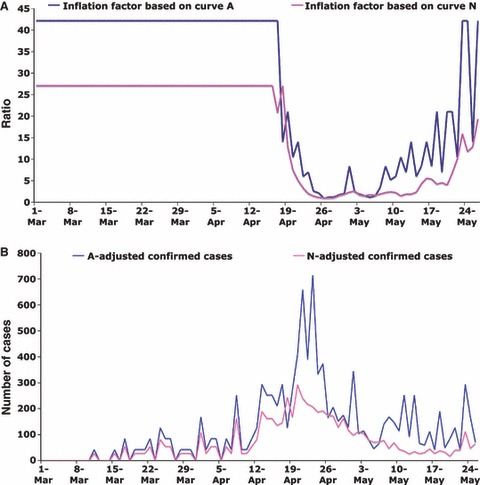
Adjusted number of confirmed cases (C): ‘inflated time series’. The same data as in Figure 2, but this time the inverse of the ratios calculated in Figure 2A were used to create an ‘inflated’ time series. For more detail, please see the Figure 2 caption and the main text.
To test the sensitivity of the estimates to underlying assumptions, we repeatedly re‐estimated R using random choices of parameters from the ranges given in Table 1. Figure 4 shows the distribution of R for case series S, C, and both A‐ and N‐deflated C, with each series run with data from before and after the surge (to 19 April and to 25 April, respectively). Each panel summarizes the estimates of R using 388 080 combinations of parameter values. The parameter with greatest impact on estimated reproduction number over the range assessed is the latency period (data not shown). The results suggest that the estimates of R for the second segment of the adjusted time series (after public health alerts) are in agreement with the first segments of both time series S and C. Figure 4 also illustrates that if the time series representing confirmed cases is not adjusted for extrinsic factors, a wider confidence interval will be achieved.
Figure 4.
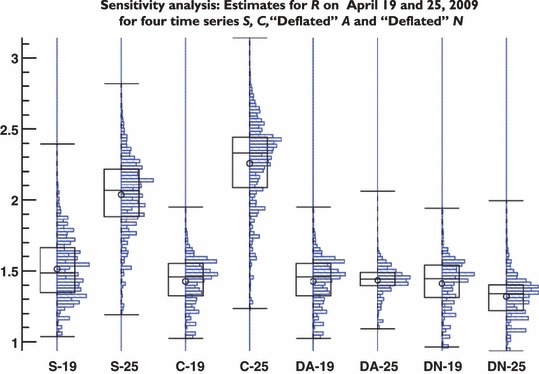
Sensitivity analysis. We simultaneously varied all parameters depicted in Table 1. This figure shows the boxplots and the distribution of outcomes estimated using the time series from 1March to 19 April and the time series from 1 March to 25 April (near the peak). While the value of R is predominantly determined by transmission dynamics, there is a pronounced impact by external social forces on the estimation of R resulting in a higher value (and wider distribution). This effect can be corrected if instead of the raw data (four left panels), the adjusted time series are used (four right panels). For each panel, we used 388 080 different combinations of parameter values from Table 1.
Discussion
With a reproduction number of approximately 1·5, the pandemic (H1N1) 2009 virus appears to exhibit community transmissibility similar to the 1957 and 1968 pandemics, or the recently emerged respiratory pathogen SARS coronavirus (SARS‐CoV) but less than the fall wave of the 1918 influenza pandemic. 10 , 14 , 15 Early global dissemination of both SARS and pandemic (H1N1) 2009 illustrates the complex inter‐connectivity of human populations globally and the epidemiological significance of individual and social behavior.
Our analysis shows that even noisy and potentially biased early epidemic outbreak data may form the basis for stable estimates of biological characteristics of emerging pathogens. Our analytical method enables us to evaluate the impact of variability in disease serial interval and contact network structure on the temporal progression of an outbreak – and the related epidemiological parameter R; additionally, we can use this method to evaluate the effect of external social drivers, while respecting the pattern observed in time series data in Mexico City (for more details, see the supplementary material).
Although most discussions of pandemic (H1N1) 2009 will likely focus on how it compares with the three 20th century influenza pandemics, we believe that comparison with SARS‐CoV will also yield critical policy implications for public health intervention. The SARS‐CoV had a relatively long 4‐ to 6‐day incubation period and a peak infectious period that was delayed until the tenth day of severe illness. 16 SARS was transmitted predominantly in the healthcare settings, with a case fatality rate exceeding 10%. These features made SARS relatively easy to detect and readily amenable to individual‐based control measures such as case isolation and contact tracing before substantial spread could occur beyond initial seeding and local hospital‐based outbreaks. Influenza viruses, in contrast, are characterized by shorter incubation (typically 1–4 days), pre‐clinical virus shedding and peak infectiousness shortly after illness onset. 17 , 18 Influenza illness comprises a spectrum including mild or asymptomatic infection with overall case fatality below 2% even during the worst pandemic on record (1918) and 10‐fold lower still during subsequent pandemics. 19 In keeping with this classic influenza profile, pandemic (H1N1) 2009 shows a larger proportion of mild infections, community‐based propagation and a lower case fatality than SARS. 20 Thus, while the reproduction numbers of the two infections are not dramatically different, they likely will require a different set of population‐based social distancing and mitigation measures. General reinforcement of voluntary self‐isolation, cough etiquette, handwashing and self‐monitoring by contacts combined with social strategies to disrupt complex contact networks and lessen virus amplification and adaptation at the community level are needed. National health authorities in North America and Europe have implemented varying school closure policies (e.g. broadly in Mexico, targeted in the UK and minimal in the US) in an attempt to contain viral transmission; as of this writing, these measures are being scaled back, but further interventions to change social contact patterns may be important as the outbreak progresses. 21
Pandemics are classically defined as the emergence of novel influenza A subtypes that have not previously been experienced by human populations, or at least not for a long time. Although pandemic (H1N1) 2009 is a new zoonotic pathogen and antigenically distinct from previous human or swine H1N1 viruses, there may be some pre‐existing immunity limiting its pandemic potential. Nevertheless, even a new variant of human influenza virus within a given subtype can significantly exacerbate seasonal morbidity and mortality (especially among persons with chronic conditions) – as was witnessed globally with the A/Sydney/05/97(H3N2)‐like virus in 1997–98 or the A/Fujian/411/2002(H3N2)‐like virus in 2003–04. Thus, delaying further spread and evolution of pandemic (H1N1) 2009 remains a worthwhile goal until a safe and effective vaccine can be developed and administered. Furthermore, global inequalities in social and economic conditions amplify the impacts of infectious diseases, including influenza. 22
As our estimates account for important sociological anomalies and are based on multiple data sources from communities rather than closed settings, they are likely to be broadly applicable. However, our optimistic prognosis relies on several critical assumptions about disease progression, virulence, and reporting rates, and it is clear that worse scenarios could evolve. Vigilant surveillance, self‐isolation, adherence to social distancing and hygiene measures, strategic school closures, and other community measures to mitigate spread, as directed by national policy, may be paramount in the months to come.
Authors’ contributions
Conception, design and implementation of mathematical and statistical models
B. Pourbohloul, B. Davoudi, R. Meza, L. A. Meyers, D. J. D. Earn, J. Dushoff, S. V. Scarpino.
Public health evaluation and interpretation of data
D. M. Skowronski, D. Fisman, W. J. Edmunds, N. Hupert, D. M. Patrick, R. C. Brunham.
Participation in drafting the manuscript
B. Pourbohloul, L. A. Meyers, D. M. Skowronski, D. Fisman, W. J. Edmunds, N. Hupert, R. C. Brunham.
Preparation of the supplementary material
B. Pourbohloul, B. Davoudi, R. Meza.
Preparation of the Mexico City data (collection, data processing, interpretation and verification for accuracy, statistical analysis) and report on public health response
A. Ahued, I. Villaseñor, F. Galván, P. Cravioto, J. Trujillo, M. Lutzow, J. Morales, A. Contreras, C. Chávez.
Supporting information
Initial Human Transmission Dynamics of the Pandemic (H1N1) 2009 Virus in North America
Please note: Wiley‐Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item
Acknowledgements
We would like to thank the generous support of the Secretaria de Salud del Distrito Federal, for providing epidemiological data and insight. This work was supported by the Canadian Institutes of Health Research (CIHR) (grants nos. PTL‐93146, PTL‐97125, PAP‐93425 and MOP‐81273), Provincial Health Services Authority of BC (PHSA), Canadian Consortium for Pandemic Preparedness Modeling (CanPan) and BC Pandemic Influenza Accelerated Vaccine Initiative (PANAVI). BP is grateful for the support of the CIHR and the Michael Smith Foundation for Health Research (MSFHR). LAM would like to acknowledge the support of the NIGMS Models of Infectious Disease Agent Study (MIDAS) (1‐U01‐GM087719‐01), the National Science Foundation (DEB‐0749097), and the James F. McDonnell Foundation.
References
- 1. Kilbourne ED. Influenza pandemics of the 20th century. Emerg Infect Dis 2006; 12:9–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Scholtissek C, Hinshaw VS, Olsen CW Influenza in pigs and their role as the intermediate host; in Nicholson KG, Webster RG, Hay AJ. (eds): Textbook of Influenza. Oxford: Blackwell Science, 1998: 137–145. [Google Scholar]
- 3. GenBank sequences from 2009 H1N1 influenza outbreak. Available at: http://www.ncbi.nlm.nih.gov/genomes/FLU/SwineFlu.html.
- 4. Centers for Disease Control and Prevention (CDC) . Outbreak of swine‐origin influenza A (H1N1) virus infection – Mexico, March–April 2009. MMWR Morb Mortal Wkly Rep. 2009; 58(Dispatch):1–3. [PubMed] [Google Scholar]
- 5. Influenza A (H1N1) – update 49, Global Alert and Response (GAR), World Health Organization. Available at: http://www.who.int/csr/don/2009_06_15/en/index.html.
- 6. Statement by WHO Director‐General, Dr Margaret Chan, 11 June 2009.
- 7. El Universal , 6 April 2009. Available at: http://www.eluniversal.com.mx/hemeroteca/edicion_impresa_20090406.html.
- 8. Bansal S, Pourbohloul B, Meyers LA. A comparative analysis of influenza vaccination programs. PLoS Med 2006; 3:e387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Newman ME. Spread of epidemic disease on networks. Phys Rev E Stat Nonlin Soft Matter Phys 2002; 66 (1 Pt 2):016128. [DOI] [PubMed] [Google Scholar]
- 10. Meyers LA, Pourbohloul B, Newman ME, Skowronski DM, Brunham RC. Network theory and SARS: predicting outbreak diversity. J Theor Biol 2005; 232:71–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Davoudi B, Pourbohloul B, Miller JC, Meza R, Meyers LA, Earn DJD Early Real‐time Estimation of Infectious Disease Reproduction Number. arXiv (arXiv:0905.0728) 2009.
- 12. Fraser C, Donnelly CA, Cauchemez S et al. Pandemic potential of a strain of influenza A (H1N1): early findings. Science 2009; 324:1557–1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kuri‐Morales P, Galvan F, Cravioto P, Zarraga Rosas LA, Tapia‐Conyer R. Mortality due to influenza and pneumonia in Mexico between 1990 and 2005. Salud Publica Mex 2006; 48:379–384. [DOI] [PubMed] [Google Scholar]
- 14. Chowell G, Fenimore PW, Castillo‐Garsow MA, Castillo‐Chavez C. SARS outbreaks in Ontario, Hong Kong and Singapore: the role of diagnosis and isolation as a control mechanism. J Theor Biol 2003; 224:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mills CE, Robins JM, Lipsitch M. Transmissibility of 1918 pandemic influenza. Nature 2004; 432:904–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Skowronski DM, Astell C, Brunham RC et al. Severe acute respiratory syndrome (SARS): a year in review. Annu Rev Med 2005; 56:357–381. [DOI] [PubMed] [Google Scholar]
- 17. Longini IM Jr, Nizam A, Xu S et al. Containing pandemic influenza at the source. Science 2005; 309:1083–1087. [DOI] [PubMed] [Google Scholar]
- 18. Ferguson NM, Cummings DA, Cauchemez S et al. Strategies for containing an emerging influenza pandemic in Southeast Asia. Nature 2005; 437:209–214. [DOI] [PubMed] [Google Scholar]
- 19. Potter CW Chronicle of influenza pandemics; in Nicholson KG, Webster RG, Hay AJ. (eds): Textbook of Influenza. Oxford: Blackwell Science, 1998: 3–18. [Google Scholar]
- 20. Centers for Disease Control and Prevention (CDC) . Swine‐origin influenza A (H1N1) virus infections in a school – New York City, April 2009. MMWR Morb Mortal Wkly Rep. 2009; 58(Dispatch):1–3. [PubMed] [Google Scholar]
- 21. Update on School (K•12) and Child Care Programs: Interim CDC Guidance in Response to Human Infections with the Novel Influenza A (H1N1) Virus, US Centers for Disease Control and Prevention. Available at: http://www.cdc.gov/h1n1flu/k12_dismissal.htm.
- 22. Murray CJ, Lopez AD, Black R et al. Global burden of disease 2005: call for collaborators. Lancet 2007; 370:109–110. [DOI] [PubMed] [Google Scholar]
- 23. WHO statement May 5th (Keiji Fukuda). Available at: http://whoterrance.vo.msecnd.net/mediacentre/audio/press_briefings/VPC_05MAY2009_Influenza_A_H1N1.mp3.
- 24. Pourbohloul B, Meyers LA, Skowronski DM, Krajden M, Patrick DM, Brunham RC. Modeling control strategies of respiratory pathogens. Emerg Infect Dis 2005; 11:1249–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Meyers LA. Predicting epidemics on directed contact networks. J Theor Biol 2006; 240:400. [DOI] [PubMed] [Google Scholar]
- 26. Miller JC. Spread of infectious disease through clustered populations. J R Soc Interface 2009. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Initial Human Transmission Dynamics of the Pandemic (H1N1) 2009 Virus in North America
Please note: Wiley‐Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item


