Abstract
As proteomic data sets increase in size and complexity, the necessity for database-centric software systems able to organize, compare, and visualize all the proteomic experiments in a lab grows. We recently developed an integrated platform called high-throughput autonomous proteomic pipeline (HTAPP) for the automated acquisition and processing of quantitative proteomic data, and integration of proteomic results with existing external protein information resources within a lab-based relational database called PeptideDepot. Here, we introduce the peptide validation software component of this system, which combines relational database-integrated electronic manual spectral annotation in Java with a new software tool in the R programming language for the generation of logistic regression spectral models from user-supplied validated data sets and flexible application of these user-generated models in automated proteomic workflows. This logistic regression spectral model uses both variables computed directly from SEQUEST output in addition to deterministic variables based on expert manual validation criteria of spectral quality. In the case of linear quadrupole ion trap (LTQ) or LTQ-FTICR LC/MS data, our logistic spectral model outperformed both XCorr (242% more peptides identified on average) and the X!Tandem E-value (87% more peptides identified on average) at a 1% false discovery rate estimated by decoy database approach.
Keywords: Decoy database, Logistic regression model, SEQUEST, Software, Spectral validation
1 Introduction
LC coupled with high-throughput MS has become a powerful tool in proteomics. To interpret a tandem mass spectrum, peptide identification in LC/MS experiments follows one of two general approaches: (i) comparison of the obtained tandem mass spectrum to the theoretical spectrum corresponding to a database of sequences and (ii) de novo construction of peptide sequences to match the obtained spectrum. Commonly used algorithms for the implementation of database searches include heuristic algorithms, e.g. SEQUEST [1], X!Tandem [2, 3], and Protein Prospector [4, 5]. Database search algorithms can also be based on probabilistic scoring such as MASCOT [6]. Algorithms for the implementation of de novo sequencing include those implemented in the software Lutefisk [7] and PEAKS [8]. Methods for peptide identification, which combine aspects of database searching and de novo sequencing, have also been proposed such as GutenTag [9] and InsPecT [10].
The identification of peptides via database searches typically results in a large number of candidate peptides. If a final assignment is determined by picking the “best” match from the search algorithm output, a substantial portion of the final selections may be incorrect due to the poor ionization and fragmentation efficiency of peptides, especially phosphopeptides. This necessitates the validation of thousands of spectra per experiment. A number of statistical methods have been developed to automate validation of large-scale data sets [5, 11–19]. These approaches combine key output from identification algorithms with other available information and also include statistical modeling in order to make reliable predictions of whether a “best” match is correct. A number of other recent proposals in the literature use advanced tools from discriminant and cluster analysis to improve on the performance of peptide identification algorithms and to develop statistical measures of performance [9, 11, 16, 18, 20–23]. Recent work has applied statistics to the validation of phosphorylation sites from CAD-MS/MS data using statistical multiple testing and a support vector machine analysis [11], Bayesian network scoring [24], or target-decoy approach with a probability-based phosphorylation site localization score [13, 25].
Although some researchers prefer to train models on manually validated data sets, others prefer the use of single protein digests or thresholding on spectral parameters while optimizing decoy database tests to train their models. Unfortunately, flexible training of existing algorithms to user-specified validated data sets is not a feature that is currently directly supported in existing software tools, reducing their flexibility to alternative proteomic workflows. For example, MS/MS spectra from phopshorylated peptides have different spectral characteristics such as the abundant neutral loss of phosphate when compared with unphosphorylated peptides. A model trained on phosphorylated data sets may more closely capture the characteristics unique to correctly assigned phopshopeptide spectra, compared with a model trained on data sets lacking phosphorylated peptides. Furthermore, the validation philosophy employed to generate the training data set may also impact the subsequent performance of a model when applied to a newly acquired data set.
On the other hand, statistical validation alone is not enough to reach a determinate conclusion in many cases, given that no algorithm can predict with 100% accuracy all the spectra that are correctly assigned. Assigned peptides with significant biological significance still require manual validation of both sequence and sites of covalent modifications to minimize ambiguity [26–28]. Manual spectral validation will continue to be an important part of any proteomic workflow. Tools increasing the efficiency of this arduous task are critical.
Commercial software for proteomic analysis such as Bioworks (Thermo Scientific) or MASCOT [6] provides only static representations of assigned spectra, with no capability for user-driven manual annotation. One newly developed software tool, CHOMPER, enables highlighting of fragment peaks that are associated with certain user-selected amino acids from spectra loaded manually from dta and out files [29]. CHOMPER also adds the capability for users to store decisions of overall spectral quality electronically.
An ideal electronic spectral annotation tool should meet the following requirements: (i) automatically calculate theoretical fragment ion masses including neutral losses from the precursor and fragment ions, (ii) allow users to add any annotations to an MS/MS spectrum and save them for the future reference within a relational database, and (iii) tight integration within an automated proteomic pipeline. The goal of manual validation is the exhaustive assignment of all fragment ions observed in a spectrum. Often proteomic end-users are forced to mentally calculate theoretical neutral loss fragment ion masses with existing tools. Furthermore, current software tools designed to assist in the manual annotation of spectra are not integrated within lab-based relational databases. If the same peptide is observed in another data set potentially collected by a different investigator, the user of existing software is not able to compare it with previously manually annotated spectra associated with that sequence, increasing the chances of redundant manual validation and decreasing overall laboratory efficiency.
We have incorporated the improvements and addressed the limitations of existing software for MS/MS validation by developing a comprehensive validation solution. This solution includes a program for generation of statistical models based on user-validated data sets, integration of these user created models within automated proteomic workflows, and a unique visualization and annotation tool for manual spectral validation called SpecNote. These programs are fully integrated into a high-throughput proteomic platform, named high-throughput autonomous proteomic pipeline (HTAPP).
2 Materials and methods
2.1 Software architecture
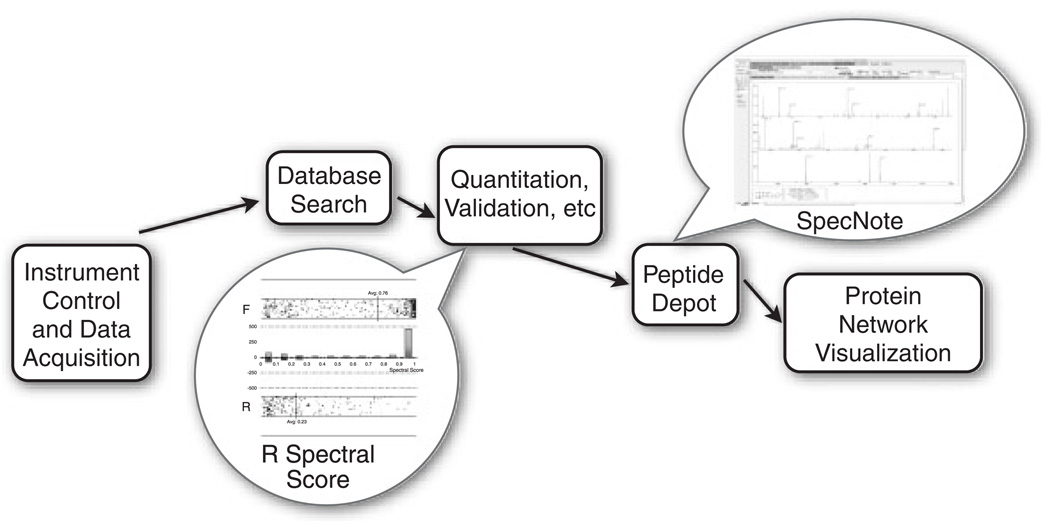
The HTAPP fully automates LC/MS data acquisition and post-acquisition analysis (Fig. 1 and HTAPP, manuscript in review). This custom-made software controls multi-dimensional LC separations of peptides (with Immobilized Metal Affinity Chromatography capability), LC/MS data acquisition, and post acquisition SEQUEST search (version 27; Thermo Scientific), peptide quantitation, decoy database analysis, spectral validation, phosphosite localization using Ascore [11], and loading data into a lab-based relational database called PeptideDepot created in FileMaker (version 9.0.3; FileMaker) with live connections to data warehoused in a MySQL database (version 5.1.16-beta-nt; MySQL). In this paper we describe the peptide validation component of HTAPP. Within this validation component, the logistic-model-based validation program is launched automatically, without user intervention. A spectral score for each peptide is calculated by an R (version 2.4.1; GNU project) program, which indicates the probability that the SEQUEST generated sequence is correctly assigned (1 = most likely, 0 = least likely). The estimation of false discovery rates (FDR) is accomplished by the decoy database approach and evaluated without user intervention. A figure illustrating the distribution of spectral score or XCorr versus decoy database search direction is dynamically generated within PeptideDepot from data stored in a custom MySQL table that represents the peptides currently viewed by the proteomic researcher. These data are represented within a custom webpage viewed through a FileMaker web portal and generated by a PHP script (version 5.2.1, http://www.php.net) hosted on Apache web server (version 2.2.4, the Apache Software Foundation), along with a table that lists the yields at a certain FDR.
Figure 1.
Schematic representation of how the statistical validation and manual validation software components fit into the HTAPP proteomic pipeline. Balloons indicate the software described in this paper.
The manual annotation software component SpecNote, which is built based on publicly available Java libraries, including interfascia.jar, pde.jar, pdf.jar, itext.jar, jogl.jar, core.jar (all from Processing distribution 0125; http://processing.org/) and mysql-connector-Java.jar (version 5.0.5; MySQL), and compiled as a Java Applet (version 1.6; Sun Microsystems) that runs in any web browser, is embedded in a web portal within PeptideDepot. A user may navigate LC/MS experiment within PeptideDeopt, select any peptide, and electronically annotate and validate the SEQUEST-assigned MS/MS spectra within SpecNote. The user annotations are transparently stored in the MySQL relational database component of PeptideDepot, accessible to other users who discover the same peptide in another proteomic experiment.
SpecNote flexibly integrates within existing user workflows that may be independent of PeptideDepot. If a user already has software for selecting a certain peptide from an LC/MS experiment, SpecNote could be utilized through a webpage. If a user’s manual validation workflow requires additional data to be represented on the spectra, a user may alter the source code of SpecNote to customize the graphical user interface or to utilize alternative data sources such as a different relational database.
2.2 Experimental data sets
Four data sets were chosen to train and evaluate the newly created logistic spectral model. The data sets represented a variety of typical proteomic data types including simple and complex mixtures of either phosphorylated or unphosphorylated peptides acquired on an linear quadrupole ion trap (LTQ) mass spectrometer (see Supporting Information for detailed protocols) [25, 26, 30–34]. The samples were (i) Mast cell phosphopeptides (MCP5), (ii) Pervanadate stimulated T-cell phosphopeptides (PVIP), (iii) 18 protein ISB standard protein mix [32] (18Mix), and (iv) BSA peptides (BSA). An additional data set, NIH3T3 phosphopeptides (3T3), was prepared to test the performance of the logistic model on an LTQ/FTICR data set.
Using the SEQUEST algorithm, tandem mass spectra were assigned to peptide sequences from species-specific NCBI non-redundant protein databases. The forward NCBI databases were reversed and appended to the forward database to estimate the FDR [35]. SEQUEST search parameters varied depending on the data set as described in Supporting Information. For all data sets, search parameters designated tryptic enzymatic cleavage. SEQUEST results were thresholded on XCorr (+1>1.5,+2>2,+3>2.5). For comparison, the same data sets were searched using X!Tandem database algorithm (version 2008.12.01.1) with the identical search parameters and protein databases used in SEQUEST searching. X!Tandem results were thresholded on E-value (≤1.0) for LTQ data, or on precursor mass error (≤20 ppm) for LTQ-FTICR data.
The MCP5 data set consisted of 1114 spectra, with 630 valid and 484 invalid spectral assignments determined by manual validation. The PVIP set consisted of 619 spectra with 193 manually validated as correct assignments; the remaining 426 were determined to be incorrect. The BSA set consisted of manually validated 605 spectra with 303 correct and 302 incorrect. The 18Mix set consisted of 25 856 spectra with 14 568 assigned correctly to the 18 proteins known to be in this sample while 11 288 were assigned as incorrect because they were not among the 18 known proteins. A subset of the 18Mix spectra was randomly selected for the model training and evaluation, including 913 valid and 687 invalid assignments.
2.3 Criteria for manual validation of spectra
Spectra were passed through intensive manual validation to ascertain whether SEQUEST-assigned sequences were consistent with MS/MS spectra for all data sets except 18Mix. Our requirements were identical to previously described manual validation metrics [36] with the additional requirements that (i) threonine and serine phosphorylated peptides should contain an abundant neutral loss of phosphate from the precursor ion (M-80/z Da or M-98/z Da), (ii) all abundant peaks should be assigned to either a b or y ion or a neutral loss of phosphate, water, or ammonia from a b, y, or precursor ion, (iii) only monoisotopic peaks are assigned, and (iv) at most two internal cleavage sites were allowed for samples digested with trypsin and peptides containing any internal cleavage sites were scrutinized more closely.
2.4 Statistical methods for spectral validation
Logistic regression was used to develop statistical models for peptide validation, with the response variable indicating whether the peptide identification is valid or invalid. Three manually validated data sets (MCP5, PVIP, BSA) along with another data set validated by matching to a known mixture of 18 proteins (18Mix) were used as training sets. For each spectrum, a number of predictor variables believed to mimic manual validation criteria were calculated and used to fit a logistic regression model. There were four groups of predictors that in total constituted 34 variables (see Supporting Information for a complete description of variables). The “SEQUEST” model was developed using only the variables in group (a); the “SEQUEST Plus” model uses variables from groups (a) and (b); and the “Spectral” model was determined by using stepwise reduction on the variables in all four groups. Unlike the SEQUEST and SEQUEST Plus models, the final variables retained in the Spectral model depended on the training data set. The stepwise reduction began with a full list of variables, sequentially removing each variable and comparing the distribution of model-predicted likelihoods to determine if the performance of the model was significantly changed (p-value <0.05). In particular, the remaining number of variables retained after stepwise model reduction was 13 for MCP5, 8 for PVIP, 9 for BSA, and 13 for 18Mix. The resulting models were applied to the validated data sets and receiver operating characteristic (ROC) analysis was used to assess their predictive performance. The ROC curves were summarized and compared via their corresponding AUC. All computations were performed using either STATA software (version 10, StataCorp LP) or software written in R (version 2.4.1; GNU project). In order to compare the effectiveness of the Spectral model at a low FDR estimated by decoy database, the number of peptide hits found in the proteomic data sets thresholded by the spectral score, Xcorr, or X!Tandem E-value to achieve an overall 1% FDR were counted and summarized into a table.
3 Results and discussion
3.1 Statistical validation
Each of three logistic regression models was applied to all four validated data sets. The AUC was computed and compared with those of the single SEQUEST variable XCorr (ΔAUC) (Table 1). All three logistic regression models, SEQUEST, SEQUEST Plus, and Spectral, performed statistically better than XCorr in most cases (ΔAUC p-value <0.05). Among our models the Spectral model performed the best compared with XCorr with a statistically significant ΔAUC for all but one case. The SEQUEST model performed the poorest with three out of 16 cases of no significant change between XCorr and SEQUEST model and one case where XCorr outperformed the SEQUEST model. The spectral model trained on MCP5 was selected for further analysis because it resulted in the highest ΔAUC of all models when cross-applied to the other data sets.
Table 1.
AUC of SEQUEST, SEQUEST Plus, and Spectral models trained on and applied to all data setsa)
| Training set | Applied to | BSA | PVIP | MCP5 | Standard mix | ||||
|---|---|---|---|---|---|---|---|---|---|
| AUC | ΔAUC p-valueb) |
AUC | ΔAUC p-valueb) |
AUC | ΔAUC p-valueb) |
AUC | ΔAUC p-valueb) |
||
| BSA | SEQUEST | 0.761 | 0.070 | 0.824 | −0.006 | 0.896 | 0.136 | 0.869 | 0.044 |
| <0.001 | 0.733 | <0.001 | <0.001 | ||||||
| SEQUEST Plus | 0.812 | 0.121 | 0.804 | −0.026 | 0.921 | 0.161 | 0.885 | 0.060 | |
| <0.001 | 0.241 | <0.001 | <0.001 | ||||||
| Spectral | 0.903 | 0.212 | 0.837 | 0.007 | 0.908 | 0.148 | 0.874 | 0.049 | |
| <0.001 | 0.739 | <0.001 | <0.001 | ||||||
| PVIP | SEQUEST | 0.693 | 0.002 | 0.900 | 0.07 | 0.858 | 0.098 | 0.867 | 0.042 |
| 0.841 | <0.001 | <0.001 | <0.001 | ||||||
| SEQUEST Plus | 0.716 | 0.025 | 0.936 | 0.106 | 0.896 | 0.136 | 0.862 | 0.037 | |
| 0.012 | <0.001 | <0.001 | <0.001 | ||||||
| Spectral | 0.852 | 0.161 | 0.947 | 0.117 | 0.920 | 0.160 | 0.875 | 0.050 | |
| <0.001 | <0.001 | <0.001 | <0.001 | ||||||
| MCP5 | SEQUEST | 0.762 | 0.071 | 0.862 | 0.032 | 0.920 | 0.160 | 0.875 | 0.050 |
| <0.001 | 0.029 | <0.001 | <0.001 | ||||||
| SEQUEST Plus | 0.782 | 0.091 | 0.890 | 0.060 | 0.961 | 0.201 | 0.890 | 0.065 | |
| <0.001 | <0.001 | <0.001 | <0.001 | ||||||
| Spectral | 0.849 | 0.158 | 0.897 | 0.067 | 0.970 | 0.210 | 0.892 | 0.067 | |
| <0.001 | <0.001 | <0.001 | <0.001 | ||||||
| Standard mix | SEQUEST | 0.662 | −0.029 | 0.744 | −0.086 | 0.800 | 0.004 | 0.891 | 0.066 |
| 0.192 | <0.001 | 0.006 | <0.001 | ||||||
| SEQUEST Plus | 0.745 | 0.054 | 0.875 | 0.045 | 0.932 | 0.172 | 0.909 | 0.084 | |
| 0.013 | 0.023 | <0.001 | <0.001 | ||||||
| Spectral | 0.796 | 0.105 | 0.883 | 0.053 | 0.944 | 0.184 | 0.914 | 0.089 | |
| <0.001 | 0.011 | <0.001 | <0.001 | ||||||
| XCorr | 0.691 | 0.830 | 0.760 | 0.825 | |||||
As a comparison, the difference in AUC from XCorr (ΔAUC) is shown with the p-value.
Gray shading indicates a model trained and applied to the same data set. The SEQUEST, SEQUEST Plus, and Spectral models outperform XCorr in all but four cases. The Spectral model performs well across all training and application data sets and has comparatively larger ΔAUC values among all models, with only one case that the Spectral model is not significantly better than XCorr (p-values <0.05 adjusted for multiple comparisons).
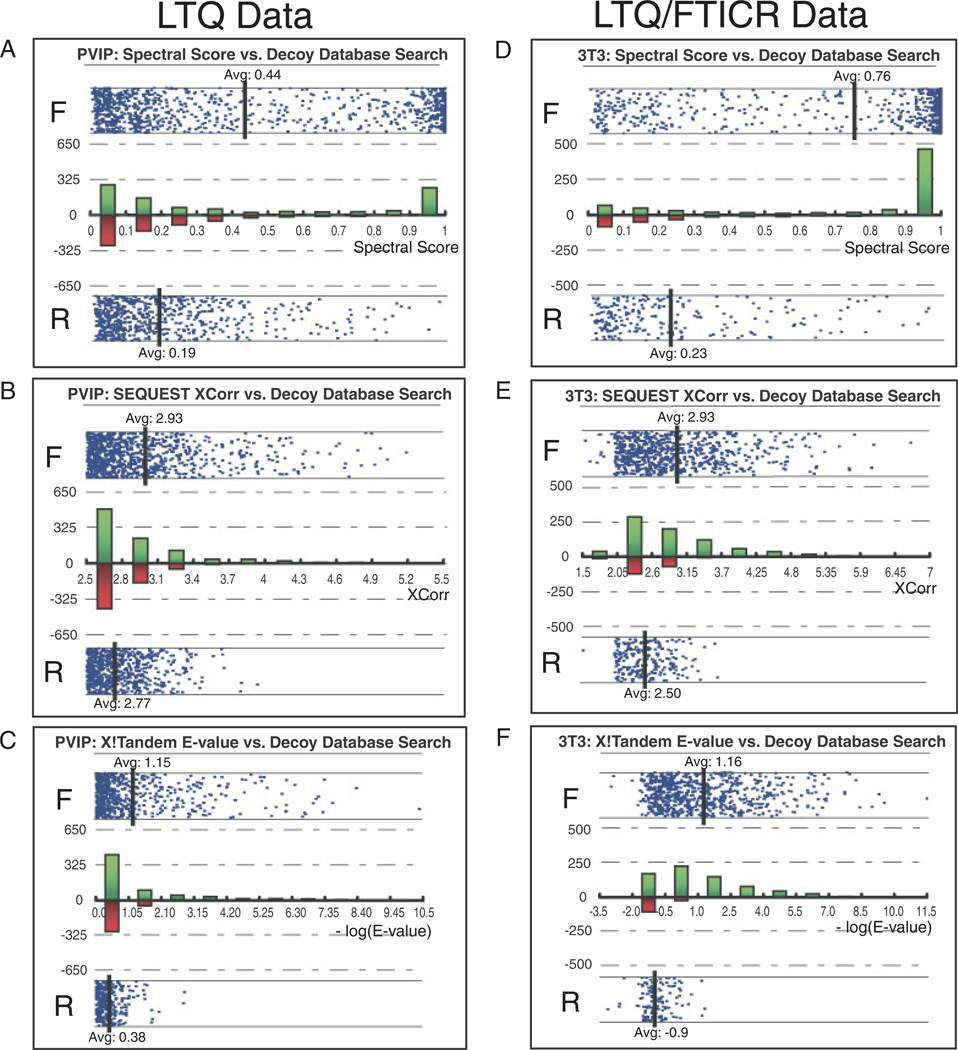
To compare the performance of the Spectral model to XCorr, we also examined the effect of the spectral score upon the distribution of forward and reversed database hits. The use of decoy-estimated FDR provides a universal metric that allows comparison of the performance of user-generated logistic models with other validation approaches. The distribution between the spectral score or XCorr and decoy database direction was examined for both LTQ and LTQ/FTICR data (Fig. 2 and Table 2). This view is useful for selection of spectral score thresholds for user-preferred FDR and is incorporated into our automated proteomic workflow using a dynamic PHP script (Figs. 3D and E). With both LTQ and LTQ/FTICR data, the forward and reversed populations using the spectral score were significantly separated when compared with XCorr or XTandem E-value (Fig. 2). This increased separation has the impact of increasing peptide yield at a user-selected FDR. For instance, to reduce the FDR of 3T3 data set to 1%, thresholding on spectral score retains 455 peptides out of a total of 959 peptides, compared with 122 peptides by XCorr and 300 peptides by E-value. For the data sets mentioned in this paper, our logistic spectral model outperformed both XCorr (242% more peptides identified on average) and the X!Tandem E-value (87% more peptides identified on average) at a 1% FDR estimated by decoy database approach.
Figure 2.
Performance of Spectral Model and XCorr evaluated with Decoy Database Approach. Scores are plotted against the protein database types, Forward (labeled as “F” in figures) and Reversed (labeled as “R” in figures) to demonstrate the distribution. Each data point in the figure represents an assigned peptide. A histogram of peptide counts was overlaid on the same figure with green bars representing forward hits and red bars representing reversed database hits. Spectral score is computed using the Spectral model trained on MCP5. Forward and reverse distribution for the PVIP data set acquired on an LTQ mass spectrometer versus (A) spectral score, (B) Xcorr, and (C) E-value calculated by X!Tandem. Forward and reverse distribution for the 3T3 data set acquired on an LTQ/FTICR mass spectrometer with a 20ppm mass error cutoff versus (D) spectral score, (E) XCorr and (F) E-value calculated by X!Tandem.
Table 2.
The Spectral model provides a substantial yield increase of confident peptide assignments when applied to a range of different proteomic data sets
| Data sets |
Average spectral score | Average XCorr | Average –log(E-value) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Forward hits |
Reversed hits |
No. of hits at 1% FDR |
Forward hits |
Reversed hits |
No. of hits at 1% FDR |
Forward hits |
Reversed hits |
No. of hits at 1% FDR |
|
| BSA | 0.81 | 0.47 | 58 | 3.2 | 2.7 | 41 | 1.42 | 0.36 | 82 |
| PVIP | 0.44 | 0.19 | 212 | 2.9 | 2.8 | 48 | 1.15 | 0.38 | 80 |
| MCP5 | 0.47 | 0.19 | 637 | 3.0 | 2.7 | 154 | 0.91 | 0.36 | 246 |
| 3T3 | 0.76 | 0.23 | 455 | 2.9 | 2.5 | 122 | 1.16 | −0.9 | 300 |
Spectral score is computed using the Spectral model trained on MCP5 data set. E-value is computed by X!Tandem and represented as −log(E-value). For each data set, thresholded by the spectral score, Xcorr, or X!Tandem E-value, number of peptide hits at 1% FDR estimated by decoy database search is calculated. The Spectral model outperformed both SEQUEST XCorr (242% more peptides identified on average) and X!Tandem (87% more peptides identified on average).
Figure 3.
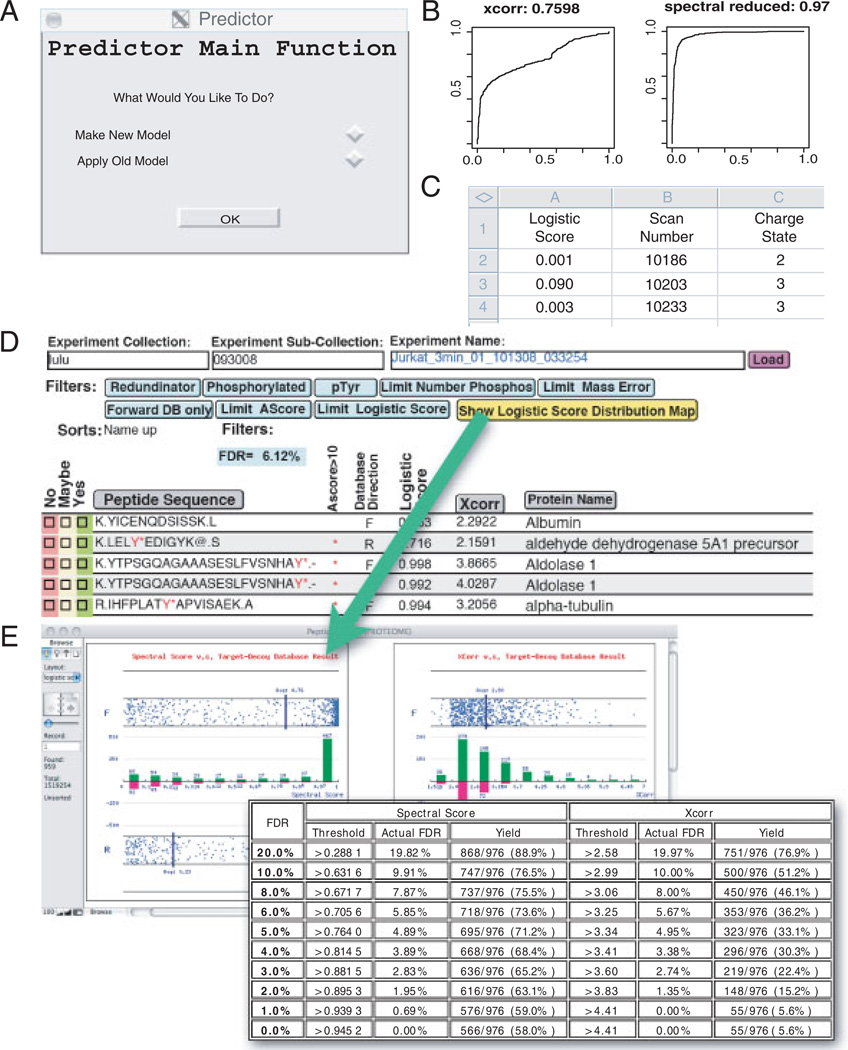
Open-source R software for training logistic models and its application. In the manual mode (A) the user selects whether to train a new model or apply an existing model to a new data set. (B) The software automatically trains logistic models based on user-provided validated data sets. User validation is provided as a boolean value using any user-preferred validation metric, such as expert manual spectral validation or simple contrived proteomic mixtures. The software calculates an ROC curve and AUC for each newly trained model. (C) When applying a model to a new data set, the software outputs a validation score for each spectra identified. In the automatic mode (D) an R program trained on MCP5 using the Spectral model is implemented in HTAPP to perform statistical validation automatically. Results are imported into a FileMaker database and the FDR is calculated. The user can specify any logistic score threshold to adjust the FDR. Clicking on the yellow button brings a live figure showing the logistic score or XCorr versus Decoy database distribution (E).
To assist proteomics researchers with high-throughput statistical analysis and generation of new statistical models, we have integrated model training and application of user-driven models within our automated data pipeline (Fig. 3). In the analyses presented in this paper, training of our new logistic model was based upon the four sets of validated data mentioned above. A user may input any validated data set from any type of mass spectrometer using any validation metrics to recompute model variables, tailoring the prediction to the user’s needs and expectations through a flexible user interface. To facilitate new model building, a freely downloadable, open-source software in the R programming language was developed to create new logistic models based either on user-supplied validated training sets or data sets described in this paper (Fig. 3A). When creating new models, this software also calculates ROC curves and the corresponding AUC for all models (Fig. 3B). Any user-created logistic model may be applied to newly collected user data manually resulting in the output of logistic scores into a flat file for every peptide (Fig. 3C). These user-generated models can also be integrated within our automated proteomic pipeline (HTAPP) that performs statistical analysis without user intervention after a data set is newly acquired. In the automated mode, the newly calculated logistic score is deposited and viewable within our proteomic relational database PeptideDepot (Fig. 3D). Within the database, a user may apply thresholds to the data, calculate FDR by the decoy database approach, and view plots of XCorr and spectral score versus decoy database direction for any filtered subset of experimental data (Figs. 3D and E). These plots assist in the selection of appropriate logistic score thresholds for any user-preferred FDR.
3.2 SpecNote for manual validation and annotation
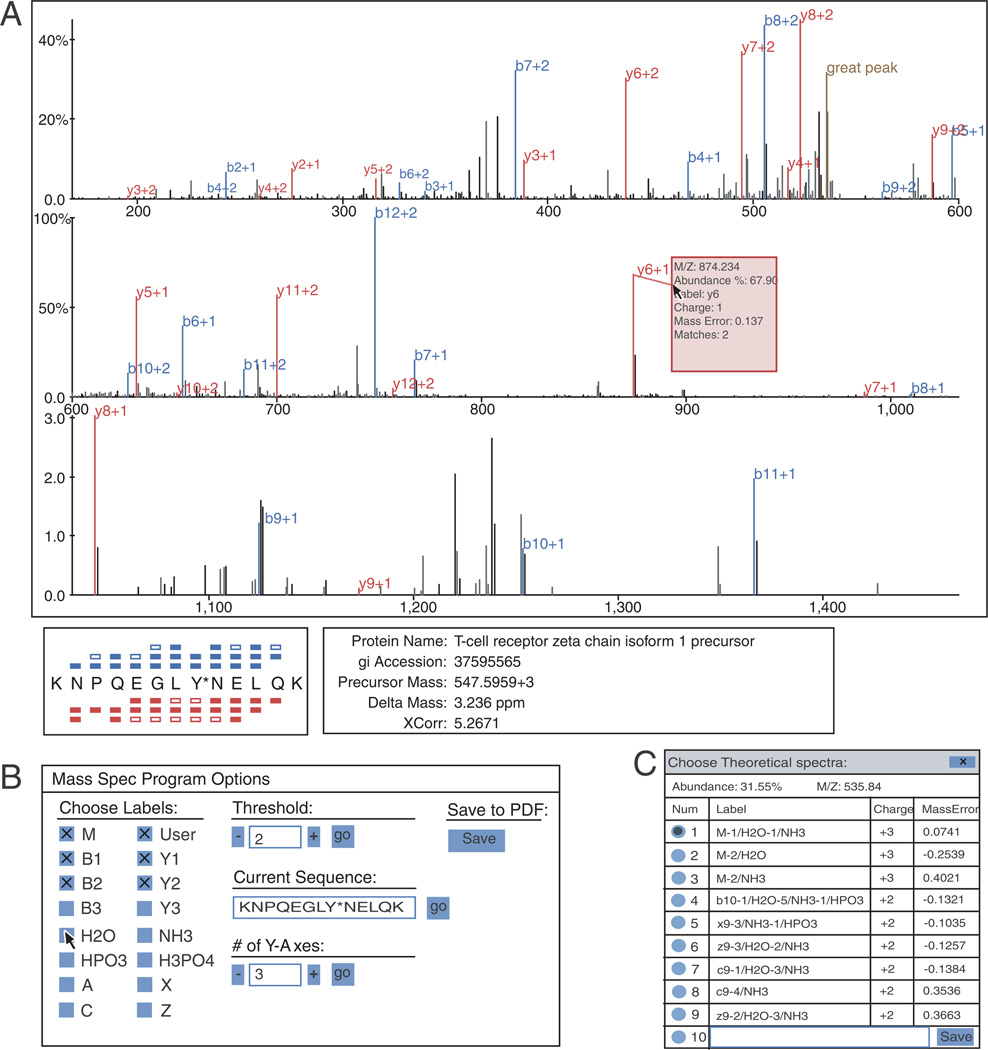
When manual validation of any peptide within the PeptideDepot database is necessary, the spectral annotation tool SpecNote is available. Manual validation involves the verification of the assigned peptide sequence and validation of any post-translational modification positions within the sequence. For both tasks, sequence coverage and spectral coverage (assigned ion current) are important parameters for successful analysis. SpecNote can provide critical information to accelerate this process. The graphical interface of SpecNote is shown in Fig. 4A. The sequence coverage of the matched peptide is indicated in the lower left corner of the display area by the peptide sequence with colored bars above or below their respective amino acids representing the matching of theoretical b and y ions to observed peaks. When the mouse pointer is hovered over amino acid letters within the peptide sequence, fragment peaks corresponding to the selected amino acid are highlighted in the spectrum, allowing the user to locate specific peaks quickly. SEQUEST-assigned phosphorylation site positions also need to be manually validated. SpecNote enables the user, by clicking on the modified amino acid and using the arrow key on the keyboard, to make a quick comparison of different repositioned post-translational modifications in both peak assignment and sequence coverage. A preference panel (Fig. 4B), hidden in the normal view, can be displayed by pressing the space bar. By default, only b/y ions and user annotations are labeled in the spectrum. Using this preference panel, other ion types, such as c, z, a, and x, as well as neutral loss of water, ammonia, and phosphate can also be labeled. In addition, a user may adjust the labeling threshold, the divisions of x-axis, and alter the sequence or modification site of the peptide.
Figure 4.
User interface of SpecNote. (A) Main display area showing essential peptide metadata and a spectrum with peaks auto-assigned to peptide fragments. Sequence coverage is illustrated with b-series ions on top and y-series ions at bottom. Detailed information such as protein name, XCorr, delta mass are provided and linked to NCBI website. (B) Preference panel allows a user to determine which ions to show by selecting ion types and intensity threshold. Assigned sequence can be changed by resubmitting a modified sequence. (C) For each peak in the spectrum, a user can reassign it to other candidate fragments or input a customized annotation. All user annotations are stored automatically within the MySQL proteomic database.
SpecNote also incorporates novel features not present in other similar tools such as a snap-to-peak function. A normal MS/MS spectrum contains many peak assignments, making it impossible to display all detailed information at the same time. With the snap-to-peak feature, the program senses the current mouse position and, if it falls within a predefined distance from an MS/MS fragment peak, the mouse pointer snaps to that peak. A window then pops up displaying details about that peak, such as m/z, relative abundance, and suggested theoretical ion assignment with the associated mass error. This feature removes the need for the proteomic end-user to zoom in and out to retrieve that same information, maintaining user orientation.
SpecNote accelerates the manual validation process by performing numerous calculations for the user and integrating calculated results within the assigned spectra. Traditionally, the user would manually calculate all possibilities to match unidentified fragments. Such procedures reduce manual validation throughput. SpecNote takes less than a second to assign all peaks to theoretical fragments by using an automatic peak assignment function to calculate mass differences (Δmass) between the observed masses and any potential theoretical fragment masses, including b/y/c/z/a/x ions. Since neutral losses of precursor or fragment ions are widely observed in CID spectra, the algorithm also searches for neutral losses of water, ammonia, and phosphate (in the case of phosphorylated peptides) from all applicable fragments and the precursor ion. All possible charge states are considered. After calculation of all possible assignments, the fragment is automatically assigned using the following hierarchy: (i) b and y ions are preferred for CID spectra by default, (ii) Δmass is minimized, and (iii) the number of neutral losses is minimized. Clicking the peak allows the user to compare among all calculated theoretical assignments for a given peak including mass errors (Fig. 4C) and select one of them or even enter a manual annotation if the user disagrees with computer-suggested assignments, increasing accuracy and efficiency. On average, SpecNote can save at least 5min per spectrum compared with printing the spectra and labeling the peaks manually.
SpecNote allows the user to export PDF reports for selected spectra along with all the assignments and annotations by incorporating iText (http://www.lowagie.com/iText/), a free Java-PDF library. iText generates PDF in vector mode, so the file size is only around 8 kilobytes per spectrum, which is convenient for distributing data between labs. (See 4897 annotated MS/MS spectra in Supporting Information 4 for an example of this capability.)
4 Concluding remarks
Recent innovations in multi-dimensional LC/MS proteomic methods have led to a deluge of data generation in proteomics. The ability to efficiently discern the true assignment of MS/MS spectra to peptide sequence is essential in this context.
Proteomic researchers sharply differ in the appropriateness of certain methods of data validation. Many investigators perform simple thresholding on SEQUEST parameters while approximating FDRs with decoy database search [37]. Other researchers prefer the development of statistical models based on simple proteinmixture digests with the assumption that true positive hits only result from the known proteins in the mixture with hits to any other protein defined as false positives. The popular PeptideProphet algorithm was trained with this type of approach [12]. Other researchers, weary of the possibility of unexpected contaminating proteins present in these contrived mixtures, prefer a manual interpretation of MS/MS spectra. Our logistic spectral score is adaptable to any user-preferred validation philosophy with model training implemented as a fully supported software feature.
An illustration of the power of model training in creating optimal models for certain proteomic workflows is the analysis of phosphoprotoemic data sets. Although the development of entirely new statistical approaches can optimize the yield of phosphopeptides at a user-selected FDR [11], we show here that our logistic spectral model trained with validated phosphoproteomic data sets (MCP5 and PVIP) outperforms logistic spectral models trained with the BSA unphosphorylated data set. For example, the AUC for cross application of PVIP and MCP5 trained models was 0.920 and 0.897 compared with 0.908 and 0.837 for application of the unphosphorylated BSA trained model onto PVIP and MCP5.
One criticism of user-driven model training is the difficulty of comparison of the predictions from multiple user-created models from different labs with each other. To address this criticism, we have integrated the standardized estimation of FDR by the decoy database approach as a central component of our fully automated data analysis pipeline. By providing a graphical representation of the distribution of spectral scores relative to forward and reversed database hits, a user may compare model performance from different user-created models and select a user-preferred FDR. Along with spectral score thresholds, a user may supply these estimated FDR from decoy database approach to provide a universal, unbiased assessment of the quality of proteomic data submitted for publication to scientific journals. Furthermore, the statistical models may be easily distributed as Supporting Information when data are submitted for publication in the form of a single binary R data file.
Overall, the combination of user-driven logistic spectral models, full automation of statistical analysis within high-throughput proteomic workflows, and accelerated manual spectral annotation within the PeptideDepot proteomic relational database increases the efficiency of proteomic workflows along with increased yield of confident peptide assignments.
Currently, the software described here is designed around a SEQUEST workflow. To adapt the software to a new search engine such as X!Tandem or MASCOT, the R software and the SpecNote annotation software would require updated parsing of the database search results. Additionally, the logistic spectral model would need to be trained on the output variables of the new search algorithm. In the future, logistic spectral models may also be trained with variables from multiple search algorithms to collate database search scores such as XCorr, E-value, and MOWSE score into a unified logistic spectral score. These small modifications could easily be implemented within the source code of our software.
Acknowledgments
We thank Sam Ulin for help in the preparation of this manuscript. This work was supported by National Institutes of Health Grant 2P20RR015578 and by a Beckman Young Investigator Award.
Abbreviations
- 3T3
NIH3T3 phosphopeptides
- 18Mix
18 protein ISB standard protein mix
- BSA
BSA peptides
- FDR
false discovery rate
- HTAPP
high-throughput autonomous proteomic pipeline
- LTQ
linear quadrupole ion trap
- MCP5
mast cell phosphopeptides
- PVIP
pervanadate stimulated T-cell phosphopeptides
- ROC
receiver operating characteristic
Footnotes
The authors have declared no conflict of interest.
References
- 1.Eng JK, McCormack AL, Yates JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 2.Craig R, Beavis RC. Amethod for reducing the time required to match protein sequences with tandem mass spectra. Rapid Commun. Mass Spectrom. 2003;17:2310–2316. doi: 10.1002/rcm.1198. [DOI] [PubMed] [Google Scholar]
- 3.Craig R, Beavis RC. TANDEM: matching proteins with tandem mass spectra. Bioinformatics. 2004;20:1466–1467. doi: 10.1093/bioinformatics/bth092. [DOI] [PubMed] [Google Scholar]
- 4.Clauser KR, Baker P, Burlingame AL. Role of accurate mass measurement (+/− 10 ppm) in protein identification strategies employing MS or MS/MS and database searching. Anal. Chem. 1999;71:2871–2882. doi: 10.1021/ac9810516. [DOI] [PubMed] [Google Scholar]
- 5.Nesvizhskii AI, Aebersold R. Analysis, statistical validation and dissemination of large-scale proteomics datasets generated by tandem MS. Drug Discov. Today. 2004;9:173–181. doi: 10.1016/S1359-6446(03)02978-7. [DOI] [PubMed] [Google Scholar]
- 6.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 7.Taylor JA, Johnson RS. Implementation and uses of automated de novo peptide sequencing by tandem mass spectrometry. Anal. Chem. 2001;73:2594–2604. doi: 10.1021/ac001196o. [DOI] [PubMed] [Google Scholar]
- 8.Ma B, Zhang K, Hendrie C, Liang C, et al. PEAKS: powerful software for peptide de novo sequencing by tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2003;17:2337–2342. doi: 10.1002/rcm.1196. [DOI] [PubMed] [Google Scholar]
- 9.Tabb DL, Saraf A, Yates JR., III GutenTag: high-throughput sequence tagging via an empirically derived fragmentation model. Anal. Chem. 2003;75:6415–6421. doi: 10.1021/ac0347462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tanner S, Shu H, Frank A, Wang LC, et al. InsPecT: identification of posttranslationally modified peptides from tandem mass spectra. Anal. Chem. 2005;77:4626–4639. doi: 10.1021/ac050102d. [DOI] [PubMed] [Google Scholar]
- 11.Lu B, Ruse C, Xu T, Park SK, Yates J., III Automatic validation of phosphopeptide identifications from tandem mass spectra. Anal. Chem. 2007;79:1301–1310. doi: 10.1021/ac061334v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 13.Beausoleil SA, Villen J, Gerber SA, Rush J, Gygi SP. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat. Biotechnol. 2006;24:1285–1292. doi: 10.1038/nbt1240. [DOI] [PubMed] [Google Scholar]
- 14.Sun S, Meyer-Arendt K, Eichelberger B, Brown R, et al. Improved validation of peptide MS/MS assignments using spectral intensity prediction. Mol. Cell. Proteomics. 2007;6:1–17. doi: 10.1074/mcp.M600320-MCP200. [DOI] [PubMed] [Google Scholar]
- 15.Anderson DC, Li W, Payan DG, Noble WS. A new algorithm for the evaluation of shotgun peptide sequencing in proteomics: support vector machine classification of peptide MS/MS spectra and SEQUEST scores. J. Proteome Res. 2003;2:137–146. doi: 10.1021/pr0255654. [DOI] [PubMed] [Google Scholar]
- 16.Higdon R, Kolker N, Picone A, van Belle G, Kolker E. LIP index for peptide classification using MS/MS and SEQUEST search via logistic regression. OMICS. 2004;8:357–369. doi: 10.1089/omi.2004.8.357. [DOI] [PubMed] [Google Scholar]
- 17.Kall L, Canterbury JD, Weston J, Noble WS, MacCoss MJ. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat. Methods. 2007;4:923–925. doi: 10.1038/nmeth1113. [DOI] [PubMed] [Google Scholar]
- 18.Razumovskaya J, Olman V, Xu D, Uberbacher EC, et al. A computational method for assessing peptide-identification reliability in tandem mass spectrometry analysis with SEQUEST. Proteomics. 2004;4:961–969. doi: 10.1002/pmic.200300656. [DOI] [PubMed] [Google Scholar]
- 19.Sun W, Li F, Wang J, Zheng D, Gao Y. AMASS: software for automatically validating the quality of MS/MS spectrum from SEQUEST results. Mol. Cell. Proteomics. 2004;3:1194–1199. doi: 10.1074/mcp.M400120-MCP200. [DOI] [PubMed] [Google Scholar]
- 20.Gentzel M, Kocher T, Ponnusamy S, Wilm M. Preprocessing of tandem mass spectrometric data to support automatic protein identification. Proteomics. 2003;3:1597–1610. doi: 10.1002/pmic.200300486. [DOI] [PubMed] [Google Scholar]
- 21.Moore RE, Young MK, Lee TD. Method for screening peptide fragment ionmass spectra prior to database searching. J. Am. Soc. Mass Spectrom. 2000;11:422–426. doi: 10.1016/S1044-0305(00)00097-0. [DOI] [PubMed] [Google Scholar]
- 22.Sadygov RG, Liu H, Yates JR. Statistical models for protein validation using tandem mass spectral data and protein amino acid sequence databases. Anal. Chem. 2004;76:1664–1671. doi: 10.1021/ac035112y. [DOI] [PubMed] [Google Scholar]
- 23.Wu FX, Gagne P, Droit A, Poirier GG. RT-PSM, a real-time program for peptide-spectrum matching with statistical significance. Rapid Commun. Mass Spectrom. 2006;20:1199–1208. doi: 10.1002/rcm.2435. [DOI] [PubMed] [Google Scholar]
- 24.Payne SH, Yau M, Smolka MB, Tanner S, et al. Phosphorylation-specific MS/MS scoring for rapid and accurate phosphoproteome analysis. J. Proteome Res. 2008;7:3373–3381. doi: 10.1021/pr800129m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Olsen JV, Blagoev B, Gnad F, Macek B, et al. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 26.Cao L, Yu K, Banh C, Nguyen V, et al. Quantitative time-resolved phosphoproteomic analysis of mast cell signaling. J. Immunol. 2007;179:5864–5876. doi: 10.4049/jimmunol.179.9.5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoffert JD, Pisitkun T, Wang G, Shen RF, Knepper MA. Quantitative phosphoproteomics of vasopressin-sensitive renal cells: regulation of aquaporin-2 phosphorylation at two sites. Proc. Natl. Acad. Sci. USA. 2006;103:7159–7164. doi: 10.1073/pnas.0600895103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lehtinen MK, Yuan Z, Boag PR, Yang Y, et al. A conserved MST-FOXO signaling pathway mediates oxidative-stress responses and extends life span. Cell. 2006;125:987–1001. doi: 10.1016/j.cell.2006.03.046. [DOI] [PubMed] [Google Scholar]
- 29.Eddes JS, Kapp EA, Frecklington DF, Connolly LM, et al. CHOMPER: a bioinformatic tool for rapid validation of tandem mass spectrometry search results associated with high-throughput proteomic strategies. Proteomics. 2002;2:1097–1103. doi: 10.1002/1615-9861(200209)2:9<1097::AID-PROT1097>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 30.Brill LM, Salomon AR, Ficarro SB, Mukherji M, et al. Robust phosphoproteomic profiling of tyrosine phosphorylation sites from human T cells using immobilized metal affinity chromatography and tandem mass spectrometry. Anal. Chem. 2004;76:2763–2772. doi: 10.1021/ac035352d. [DOI] [PubMed] [Google Scholar]
- 31.Ficarro SB, Salomon AR, Brill LM, Mason DE, et al. Automated immobilized metal affinity chromatography/nano-liquid chromatography/electrospray ionization mass spectrometry platform for profiling protein phosphorylation sites. Rapid Commun. Mass Spectrom. 2005;19:57–71. doi: 10.1002/rcm.1746. [DOI] [PubMed] [Google Scholar]
- 32.Klimek J, Eddes JS, Hohmann L, Jackson J, et al. The standard protein mix database: a diverse data set to assist in the production of improved Peptide and protein identification software tools. J. Proteome Res. 2008;7:96–103. doi: 10.1021/pr070244j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Secrist JP, Burns LA, Karnitz L, Koretzky GA, Abraham RT. Stimulatory effects of the protein tyrosine phosphatase inhibitor, pervanadate, on T-cell activation events. J. Biol. Chem. 1993;268:5886–5893. [PubMed] [Google Scholar]
- 34.Tanaka S, Ito T, Wands JR. Neoplastic transformation induced by insulin receptor substrate-1 overexpression requires an interaction with both Grb2 and Syp signaling molecules. J. Biol. Chem. 1996;271:14610–14616. doi: 10.1074/jbc.271.24.14610. [DOI] [PubMed] [Google Scholar]
- 35.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 36.Link AJ, Eng J, Schieltz DM, Carmack E, et al. Direct analysis of protein complexes using mass spectrometry. Nat. Biotechnol. 1999;17:676–682. doi: 10.1038/10890. [DOI] [PubMed] [Google Scholar]
- 37.Villen J, Beausoleil SA, Gerber SA, Gygi SP. Large-scale phosphorylation analysis of mouse liver. Proc. Natl. Acad. Sci. USA. 2007;104:1488–1493. doi: 10.1073/pnas.0609836104. [DOI] [PMC free article] [PubMed] [Google Scholar]