Abstract
Recently, cytochrome P450 170A1 (CYP170A1) has been found to be a bifunctional protein, which catalyzes both monooxygenase activity and terpene synthase activity by two distinct active sites in the well established P450 protein structure. Therefore, CYP170A1 is identified clearly as a moonlighting protein. The known activities of a small number of the 13,000 members of the P450 superfamily fall into two general classes; promiscuous enzymes that are not considered as moonlighting and forms that participate in biosynthesis of endogenous compounds, such as steroids, vitamins and play different roles in different tissues, sometimes being moonlighting enzymes. Here we review examples of moonlighting P450, which add to our understanding of the large cytochrome P450 superfamily.
Introduction
The cytochrome P450 (CYP) superfamily includes more than 13,000 genes, members of which are found in all the biological kingdoms (http://drnelson.utmem.edu/CytochromeP450.html). They are protoheme containing monooxygenases, the most common activity being: RH + O2 + 2e− + 2H+ → ROH + H2O
The P450 nomenclature classifies 40% amino acid identity or greater as members of the same gene family (CYP1, CYP2, etc,). To date the 13,000 genes represent more than 400 gene families and it is well established that the vast majority of gene families contain members that metabolize multiple substrates (1). In the broadest sense, P450s are grouped into two different classes, one detoxifing drugs and chemical compounds from the environment (xenobiotics ) and the other participating in biosynthesis of endogenous compounds, steroid molecules being one example (2,3). From a different perspective P450s can be grouped into membrane bound forms (primarily eukaryotic) and soluble forms (primarily prokaryotic). An additional property of P450s is that they require electrons transferred by a reductase to carry out their monooxygenase activity (4). A majority of eukaryotic P450s are located in the endoplasmic reticulum and obtain electrons from a FAD/FMN containing membrane bound reductase (cytochrome P450 reductase). The vast majority of prokaryotic P450s are reduced by a soluble, two component reductase system consisting of a FAD-containing reductase, which transfers electrons to an iron-sulfur protein that transfers them to the heme iron of the soluble P450. In mitochondria the P450s are located in the inner mitochondrial membrane and electrons are transferred to them through a two component soluble reductase system similar to that found in prokaryotes. Electrons necessary for P450 function almost always come from NADH/NADPH.
The activities of P450s have been determined in only a small number of the 13,000 members of the superfamily. At leaset two types of moonlighting enzymes are found among these different activities. Moonlighting protein refers to a single protein having more than one function (5). Moonlighting activities can arise from the same enzyme in different cellular locations, binding of a cofactor compared to the unbound enzyme, oligomerzation of an enzyme compared to a monomer, complex formation with different proteins, different activities arising from expression of the same enzyme in different cell types or multiple enzymatic active sites within a single enzyme. Thus an additional classification of P450s can be non-moonlighting vs. moonlighting. The class of P450s that carry out xenobiotic metabolism have multiple activities, which almost always involve substrates from the environment having different chemical structures. Therefore they fall into the category of promiscuous enzymes and do not fit the moonlighting definition. The moonlighting class of P450s are involved in biosynthesis of important endogenous molecules. They include P450s that can perform different functions when expressed in different tissues or cell types, such as CYP17 and CYP7B1 and also include an enzyme possessing multiple catalytic sites, CYP170A1. To give insight as to how these examples of the large superfamily of enzymes can fit into the classification of moonlighting P450s, Table 1 shows examples of these three different types, which are then described in some detail.
Table 1.
Examples of moonlighting P450 monooxygenases
| Enzyme name | Tissues | Functions | Substrates |
|---|---|---|---|
| CYP17 | adrenal cortex | 17α-hydroxylase | pregnenolone, progesterone |
| gonads | 17–20 lyase | 17α-hydroxypregnenolone, 17α-hydroxyprogesterone | |
| CYP7B1 | brain | Steroid hormone metabolism | Pregnenolone, dehydroepiandrosterone, |
| liver | Bile salt synthesis | 25-hydroxycholesterol, 27-hydroxycholesterol | |
| Prostate | Metabolism of estrogen receptor ligands | 5α-Androstane-3β,17β-diol | |
| Immune cells | Immunoglobulin production | 25-Hydroxycholesterol | |
| CYP170A1 | Streptomyces cells | Albaflavenone synthase | Epi-isozizaene |
| Streptomyces cells | Farnesene synthase | Farnesyl pyrophosphate |
CYP17
CYP17 is known as 17α-hydroxylase/17–20 lyase cytochrome P450 (6). This microsomal P450 plays essential roles in steroid hormone biosynthesis in animals. In the adrenal gland (adrenal cortex) it is essential for cortisol (the major human glucocorticoid) biosynthesis. In the gonads CYP17 is required for the biosynthesis of androgens, the precursors for sex hormones (androgen/estrogen). In the adrenal pregnenolone is hydroxylated at the 17α position leading to the key substrate (17α-hydroxypregnenolone) for cortisol biosynthesis. In the gonads CYP17 catalyzes pregnenolone to dehydroepiandrosterone and progesterone to androstenedione (6). In the gonads these reactions involve three step reactions instead of the single hydroxylation reaction in the adrenal. The first of the three step reactions is the single hydroxylation, the same as in the adrenal cortex. The second (lyase) involves two steps (two molecules of O2 and two of reduced pyridine nucleotide) converting the 21 carbon sterols to 19 carbon androgens.
It remains uncertain as to how CYP17 can catalyze only the hydroxylase reaction in the adrenal cortex and both the hydroxylase and lyase reactions in the gonads. Clearly in the adrenal cortex the hydroxy product leaves the active site and enters the glucocorticoid biosynthetic pathway. It is not known whether the hydroxy product leaves the CYP17 active site in the gonads and then rebinds for the lyase activity to occur. There are considerable data indicating that the hemoprotein cytochrome b5 is essential for the CYP17 lyase activity (7) and perhaps in the gonads cytochrome b5 interaction with CYP17 conserves the presence of the hydroxy sterol in the CYP17 active site allowing the lyase activity to proceed. Whatever the explanation is, it is clear that CYP17 is essential for both glucocorticoid and sex hormone biosynthesis in different tissues is a moonlighting P450 (5).
CYP7B1
CYP7B1 (the first member of the 7B subfamily) can play different roles in different tissues such as brain, liver, lung, kidney, and immune cells. Russell et al recently reviewed the biochemistry, physiology, and genetics of this multi-functional enzyme (8). Briefly, CYP7B1 was initially identified as a steroid 7α-hydroxylase with activity toward pregnenolone and dehydroepiandrosterone in brain. These neurosteroids can act as allosteric modulators of neurotransmitter receptors to play regulatory, functional, and metabolic roles (9). CYP7B1 potentially could also play a role in inactivating neurosteroids in brain, however, neither of these two substrates has been observed to accumulate in CYP7B1 knock-out mice. Alternatively these steroids could be removed from the brain through CYP7B1 and/or other enzymes such as 3α-hydroxysteroid dehydrogenases. Second, CYP7B1 is also involved in the bile salt synthesis in liver. Both 25- and 27-hydroxycholesterol are substrates of CYP7B1 in hepatocytes and their amount accumulates in the sera and tissues of CYP7B1 mutant mice (10). Third, CYP7B1 plays a role in inactivation of 5α-androstane-3β,17β-diol, a 19-carbon steroid that binds to the estrogen receptor (11). Loss of CYP7B1 in males derived from one line of knock-out mice leads to smaller prostates (12) and to early ovarian failure in female mice (13). In addition, there is evidence that chronic inflammatory diseases such as rheumatoid arthritis are associated with changes in levels of this steroid. CYP7B1 catalyzes the conversion of dehydroepiandrosterone into 7α-hydroxy-dehydroepiandrosterone, which has been found in synovial biopsy tissues, synovial fluid, and serum from patients with rheumatoid arthritis. Dulos et al reported that the proinflammatory cytokine, tumor necrosis factor alpha (TNFalpha), up-regulates CYP7B1 activity in a fibroblast-like synoviocytes cell line and the enhanced CYP7B1 activity leads to higher levels of the 7α-hydroxy-dehydroepiandrosterone in synovial fluid, which may contribute to the maintenance of the chronic inflammation observed in rheumatoid arthritis (14). Of the large number of P450 enzymes CYP7B1 has much broader function than most but its substrates are all closely related sterols. In fact, not many moonlighting proteins have such a broad spectrum of functions.
CYP170A1
CYP170A1 is a truly unique bifuntional P450, which is encoded by the sco5223 gene of the Gram-positive, soil-dwelling bacterium Streptomyces coelicolor A3(2) as part of a two-gene cluster along with the sesquiterpene epi-isozizaene synthase gene (sco5222). CYP170A1 can catalyze two sequential oxidation reactions of the terpenoid epi-isozizaene through two epimers of albaflavenol to the single keto sesquiterpene product, albaflavenone (15). This metabolite has been isolated from S. albidoflavus and been shown to have antibacterial activity. Interestingly, CYP170A1 also shows terpenenoid synthase activity to generate farnesene isomers from farnesyl diphosphate (16). It is less clear how and when the organism might trigger farnesene synthesis, however, given the many signaling and defense mechanisms associated with the farnesene molecule in plants, it is a reasonable to hypothesize that farnesenes would play a similar role of chemical communication as a signaling compound or perhaps has a new function in the bacterial kingdom.
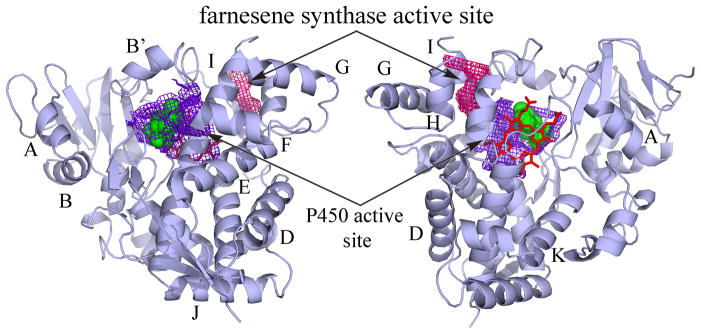
In order to explore the structural basis of the distinct P450 monooxygenase activity and the farnesene synthase activity, the crystal structures of both ligand-free CYP170A1 and in complex with epi-isozizaene were solved (16). The overall structure of CYP170A1 is the typical P450-fold consisting of α-helical and β sheet domains as seen in all other known P450 structures (Fig. 1). A conserved hydrophobic P450 catalytic active site is well defined around the heme cofactor. One epi-isozizaene molecule is situated in the proximal portion of the monooxygenase active site over the heme. The reaction carbon (C5) is within the catalytic distance ~4.8 Å of the heme iron. Since the overall CYP170A1 structure is typical for P450s, it was puzzling where the farnesene synthase active site might be located. In the P450 α-helical domain, the three-dimensional structures of C, H, I, and L helices build a common α-helical barrel, which is similar to one seen in known structures of synthases except that it contains only four α-helices rather than the typical six. The important feature is that two putative Mg2+ binding motifs in the primary sequence of CYP170A1 are located at the mouth of this α-helical barrel. These Mg2+ binding sites would anchor the diphosphate group of the farnesyl diphosphate and the α-helical barrel would accommodate the hydrophobic geranyl tail. Thus, both the sequence signature (Mg2+ binding motifs) and the helical structural domain (helical barrel) indicate the presence of a novel terpene synthase active site, which is moonlighting on the P450 structure. In addition, the mutagenesis of Mg2+ binding sites can abolish the terpene synthase activity of CYP170A1 but not the monooxygenase activity (16). The presence of two such distinct and unrelated biochemical activities in a single protein is unprecedented within the P450 superfamily. Why does the terpene synthase active site moonlight on CYP170A1? From the structure standpoint, the P450 requires more strict packing in the heme pocket for the monooxygenase function, but the remainder of the structure, such as the α-helical domain, might be less important for monooxygenase activity so that the protein could have a chance to develop a new function.
Figure 1. A view of monooxygenase active site and farnesene synthase active site in ribbon diagrams of CYP170A1.
Solvent-accessible surfaces of both P450 monooxygenase active site and farnesene synthase active site cavities of CYP170A1 represented as a mesh (PDB ID code 3EL3): the distal view (left) and the proximal view (right) were computed using the program VOIDOO (20). The active site of P450 monooxygenase is in purple blue and the active site of farnesene synthase in magenta. The overall structure of CYP170A1 is displayed as ribbons in light blue. The substrate epi-isozizaene is situated in the heme pocket on sphere mode in green. The heme is depicted as a stick model in red. The figure was generated in PyMOL (DeLano, W.L. The PyMOL Molecular Graphics System (2002), available at www.pymol.org).
The CYP170A1 structure also offers pivotal clues to explain how the two distinct catalytic activities switch between each other. The moonlighting terpene synthase active site on the surface of CYP170A1 located within the P450 α-helical domain is approximately 20 Å away from the monooxygenase catalytic active site. Probably the reason that the two conserved cation binding sites are so close together in the primary sequence compared to all other terpene synthases is due to a 4-helix active site. Furthermore, the HI loop forms a portion of the farnesene synthase active site. The disordered BC, HI, and FG loop regions in the crystal structure are conceivably consistent with the observation of two different catalytic activities and may also indicate that the protein can switch between two different functional conformations. There are limited amino acids in the single polypeptide chain in order to fully form two distinct functional active sites so that the amino acids in the three mobile loops would assume different conformations associated with P450 monooxygenase or farnesene synthase activity. Finally, it seems important to note that the different physiological states in the synthase active site have little or no affect on the monooxygenase active site. So the different active sites in this moonlighting protein seem to have quite different evolutionary histories.
Conclusions
Moonlighting proteins are a very diverse set proteins that include enzymes, transmembrane channels, chaperones, transcription factors, and are present in a large number of different types of proteins (5,17). In the cytochrome P450 superfamily, moonlighting is a new concept, even though several P450s have been known for some time to exhibit moonlighting properties including those described above. However, until the discovery of CYP170A1, this property was not recognized in P450s. We can expect to find many more cytochrome P450s having a broad substrate specificity to catalyze endogenous compounds associated with their tissue-dependent expression, implying an important role for them to regulate organ-specific functions. CYP7B1 is expressed in liver, brain, and prostate to metabolize 25-hydroxycholesterol, pregnenolone, and dehydroepiandrosterone respectively (8). All these metabolites would involve different physiological functions in the corresponding tissues. With further study of more of the 13,000 known P450 enzymes, there likely will be many more moonlighting forms found, ultimately representing several of the current descriptions of moonlighting proteins (17).
It has been challenging to explore and confirm the biological function of proteins identified from the large number of genomic sequencing studies within the last decade. However, the increasing availability of various approaches (18,19), such as evaluation of genomic context, gene knockin/knockout techniques associated with measurement of metabolic response, high throughput screening, protein structural data and sequence relatedness have provided useful clues for deciphering the biological function and activity of such proteins. Most often the second function of moonlighting proteins has no relationship with the original function. The second function usually does not have an obvious gene contact so that it is difficult to predict moonlighting activity in a protein. Therefore, not surprisingly the second function of CYP170A1 was found by luck (16). Protein expression in a new location or an organism demonstrating a novel phenotype may provide a hint on moonlighting property. Crystal structural and protein sequence information can also provide clues finding moonlighting function. For example, CYP170A1 not only provides important information regarding where the physical location of the farnesene synthase active site is and how the two distinct activities might switch from one to the other, but also gives a good example of how to identify a second activity based on known structural information combined with signature sequences (16). Since all known P450 structures have the α-helical domain and more or less have the four α-helical barrel, it may suggest some other P450s could have a similar synthase activity as well. Searches for the two conserved Mg2+ binding motifs among all known bacterial P450 primary sequences, however, revealed that only the close CYP170A1 ortholog, CYP170A2 from Streptomyces avermitilis, which is also paired with an epi-isozizaene synthase, possesses both metal binding signatures in the potential farnesene synthase active site. In spite of the difficulties in identifying moonlighting P450s, this new concept in the superfamily of monooxygenase surely provides the bases on which additional moonlighting P450 structures will be identified.
Acknowledgments
Support was provided by National Institutes of Health Grants GM69970 (M.R.W.), ES00267 (M.R.W.).
References
- 1.Nelson DR, Koymans L, Kamataki T, Stegeman JJ, Feyereisen, et al. P450 superfamily: update on new sequences, gene mapping, accession numbers and nomenclature. Pharmacogenetics. 1996;6:1–42. doi: 10.1097/00008571-199602000-00002. [DOI] [PubMed] [Google Scholar]
- 2.Porter TD, Coon MJ. Cytochrome P-450. Multiplicity of isoforms, substrates, and catalytic and regulatory mechanisms. J Biol Chem. 1991;266:13469–13472. [PubMed] [Google Scholar]
- 3.Ortiz de Montellano PR, editor. Cytochrome P450: Structure, Mechanism, and Biochemistry. 3. Kluwer Academic/Plenum Publishers; New York: 2005. pp. 1–18. [Google Scholar]
- 4.McLean KJ, Sabri M, Marshall KR, Lawson RJ, Lewis DG, et al. Biodiversity of cytochrome P450 redox systems. Biochem Soc Trans. 2005;33:796–801. doi: 10.1042/BST0330796. [DOI] [PubMed] [Google Scholar]
- 5.Jeffery CJ. Moonlighting proteins. Trends Biochem Sci. 1999;24:8–11. doi: 10.1016/s0968-0004(98)01335-8. [DOI] [PubMed] [Google Scholar]
- 6.Yanase T, Simpson ER, Waterman MR. 17 alpha-hydroxylase/17,20-lyase deficiency: from clinical investigation to molecular definition. Endocr Rev. 1991;12:91–108. doi: 10.1210/edrv-12-1-91. [DOI] [PubMed] [Google Scholar]
- 7.Katagiri M, Kagawa N, Waterman MR. The role of cytochrome b(5) in the biosynthesis of androgens by human P450c 17. Arch Biochem Biophys. 1995;317:343–347. doi: 10.1006/abbi.1995.1173. [DOI] [PubMed] [Google Scholar]
- 8.Stiles AR, McDonald JG, Bauman DR, Russell DW. CYP7B1: one cytochrome P450, two human genetic diseases, and multiple physiological functions. J Biol Chem. 2009;284:28485–28489. doi: 10.1074/jbc.R109.042168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baulieu EE, Robel P, Schumacher M. Neurosteroids: beginning of the story. Int Rev Neurobiol. 2001;46:1–32. doi: 10.1016/s0074-7742(01)46057-0. [DOI] [PubMed] [Google Scholar]
- 10.Li-Hawkins J, Lund EG, Turley SD, Russell DW. Disruption of the oxysterol 7alpha-hydroxylase gene in mice. J Biol Chem. 2000;275:16536–16542. doi: 10.1074/jbc.M001811200. [DOI] [PubMed] [Google Scholar]
- 11.Kuiper GG, Carlsson B, Grandien K, Enmark E, Haggblad J, et al. Comparison of the ligand binding specificity and transcript tissue distribution of estrogen receptors alpha and beta. Endocrinology. 1997;138:863–870. doi: 10.1210/endo.138.3.4979. [DOI] [PubMed] [Google Scholar]
- 12.Weihua Z, Lathe R, Warner M, Gustafsson JA. An endocrine pathway in the prostate, ERbeta, AR, 5alpha-androstane-3beta,17beta-diol, and CYP7B1, regulates prostate growth. Proc Natl Acad Sci USA. 2002;99:13589–13594. doi: 10.1073/pnas.162477299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Omoto Y, Lathe R, Warner M, Gustafsson JA. Early onset of puberty and early ovarian failure in CYP7B1 knockout mice. Proc Natl Acad Sci USA. 2005;102:2814–2819. doi: 10.1073/pnas.0500198102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dulos J, van der Vleuten MAJ, Kavelaars A, Heijnen CJ, Boots AM. CYP7B expression and activity in fibroblast-like synoviocytes from patients with rheumatoid arthritis: Regulation by proinflammatory cytokines. Arthritis & Rheumatism. 2005;52:770–778. doi: 10.1002/art.20950. [DOI] [PubMed] [Google Scholar]
- 15.Zhao B, Lin X, Lei L, Lamb DC, Kelly SL, et al. Biosynthesis of the sesquiterpene antibiotic albaflavenone in Streptomyces coelicolor A3(2) J Biol Chem. 2008;283:8183–8189. doi: 10.1074/jbc.M710421200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao B, Lei L, Vassylyev DG, Lin X, Cane DE, et al. Crystal structure of albaflavenone monooxygenase containing a moonlighting terpene synthase active site. J Biol Chem. 2009;284:36711–36719. doi: 10.1074/jbc.M109.064683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeffery CJ. Moonlighting proteins--an update. Mol Biosyst. 2009;5:345–350. doi: 10.1039/b900658n. [DOI] [PubMed] [Google Scholar]
- 18.Eisenberg D, Marcotte EM, Xenarios I, Yeates TO. Protein function in the post-genomic era. Nature. 2000;405:823–826. doi: 10.1038/35015694. [DOI] [PubMed] [Google Scholar]
- 19.Kinoshita K, Nakamura H. Protein informatics towards function identification. Curr Opin Struct Biol. 2003;13:396–400. doi: 10.1016/s0959-440x(03)00074-5. [DOI] [PubMed] [Google Scholar]
- 20.Kleywert GJ, Jones TA. Detection, delineation, measurement and display of cavities in macromolecular structures. Acta Crystallogr Sect D Biol Crystallogr. 1994;50:178–185. doi: 10.1107/S0907444993011333. [DOI] [PubMed] [Google Scholar]