Abstract
The hepatocyte growth factor (HGF)/Met receptor signaling pathway is deregulated in diverse human malignancies and plays a central role in oncogenesis, tumor progression, and invasive cancer growth. Similarly, altered expression and splicing (i.e. inclusion of variant exon 5, “v5”) of the cell adhesion marker, CD44, is associated with advanced cancer phenotypes. We sought to further understand how HGF regulates CD44v5 expression. Immortalized nontumorigenic keratinocyte (HaCaT) cells abundantly express both Met receptors and CD44v5 transmembrane glycoproteins. HGF stimulated CD44v5 protein expression and HaCaT cell migration; these events required activation of the ERK1/2 MAPK module and Sam68, a protein involved in RNA processing, splicing, and v5 inclusion. Similar to HaCaT cells, highly migratory MDA-MB-231 breast cancer cells also required Sam68 expression for HGF-induced migration. However, MDA-MB-231 cell migration occurred independently of ERK1/2 and CD44v5 expression and instead required ERK5 signaling to Sam68. Phospho-mutant, but not WT-Sam68, blocked HGF-induced cell migration in both cell types; MDA-MB-435 cells behaved similarly. These results suggest that Sam68 acts as a convergence point for ERK signaling to cell migration; blockade of phospho-Sam68 may provide a new avenue for therapeutic inhibition of metastatic cancers.
Keywords: Cell Migration, ERK, Growth Factors, RNA Splicing, Signal Transduction, Sam68
Introduction
Hepatocyte growth factor (HGF),2 a pleiotropic factor initially isolated from the serum of hepatectomized rats (1, 2) and later from fibroblasts (3), is an essential regulator of cell proliferation, motility, morphogenesis, and angiogenesis (4). HGF and its specific receptor, Met tyrosine kinase, regulate both physiological and pathological processes. Lack of either HGF or Met expression leads to lethal morphogenetic defects in knock-out mice during embryogenesis (5), and in adult (transgenic) mice, deregulated HGF or Met signaling contributes to impaired tissue repair after injury (6). In wound healing models, the HGF/Met pathway promotes motility (7–9) and rapid migration (10, 11) of keratinocyte cells. Birchmeier and co-workers (6) have shown that Met is specifically expressed at the leading edge of migrating keratinocytes; skin cells expressing mutant Met receptors (loss of function) are no longer able to proliferate and migrate into the wound area relative to Met+ cells. In addition to these functions in normal processes, aberrant activation of the HGF/Met signaling pathway favors oncogenesis, tumor progression, and metastasis (12–15).
Cell migration not only requires motility but also the ability of cells to adhere to and migrate through the extracellular matrix. The CD44 family of type I transmembrane proteins mediates a variety of effects such as lymphocyte homing, cell proliferation, adhesion, and migration (16). Differential responses to extracellular stimuli are due to multiple CD44 isoforms generated by alternative splicing (17). CD44 proteins are encoded by a single highly conserved gene composed of 10 constitutively spliced exons and 10 variant exons, designated as v1-v10, that reside between constitutive exons 5 and 6. Depending on the cell type and microenvironment, CD44 primary transcripts undergo alternative splicing that occurs predominantly at the membrane proximal region of the extracellular domain and give rise to different CD44 variant (v) exon-expressing isoforms (18, 19). The smallest CD44 isoform (standard form, CD44s), lacking variant exon inclusion in the stem region, is widely expressed in most tissues. CD44 is known to interact in vitro with hyaluronic acid, a component of the extracellular matrix, thus mediating cell-extracellular matrix adhesion and promoting matrix-dependent migration (20). On the contrary, the larger CD44 isoforms (harboring multiple and diverse variant exons) are expressed in a tissue-specific manner, particularly in proliferating compartments, such as in epithelial and malignant cells (21). For example, ectopic expression of CD44 v4–v7 confers a metastatic phenotype to nonmetastatic rat pancreatic carcinoma cells (18), whereas expression of CD44v5 correlates with the invasiveness of renal cell carcinoma (22). In addition, targeting CD44v5 with small interfering RNA (siRNA) in human cervical carcinoma cells (HeLa) reduces tumor cell migration and invasion in vitro (23).
The CD44v5 isoform and the regulation of its alternative splicing in response to extracellular stimuli have been extensively studied (24). Immortalized nontumorigenic keratinocyte cells (HaCaT) represent a useful model system to study factors regulating CD44 isoform expression, as these cells synthesize high levels of CD44 isoforms containing v2–v10 (25); these isoforms localize to the leading edge of migrating cell filopodia (26).
We showed previously that breast tumor kinase (Brk/PTK6) is an important mediator of HGF-induced cell migration in both keratinocyte and breast cancer cells (27). Brk gene-silencing studies showed that Brk was important for signal transduction from activated Met receptors to ERK5. These proteins associated in HGF-treated cells. However, rescue experiments demonstrated that the kinase activity of Brk was not required for HGF-induced cell migration. ERK5 was activated by both wild-type and kinase-inactive Brk, indicating that complex formation is key to passing the HGF-initiated signal to ERK5.
Herein, we further explored the mechanisms of Met receptor-induced cell migration in HaCaT and breast cancer cells with focus on downstream (from Brk-ERK5 complexes) or distal events. We show that in HaCaT cells expressing abundant Met receptors, HGF stimulates CD44v5 up-regulation and cell migration that requires ERK1/2 and Src substrate associated in mitosis of 68 kDa (Sam68), a nuclear protein involved in RNA processing and regulated downstream of the Ras/MEK/ERK pathway. In contrast to HaCaT cells, following HGF, highly migratory MDA-MB-231 breast cancer cells migrate independently of CD44v5 and utilize the ERK5 MAPK module instead of ERK1/2 as an input to Sam68-dependent cell migration; phospho-mutant Sam68 blocks HGF-induced migration in multiple cell types. These studies highlight a key role for phospho-Sam68 in normal and cancer cell migration and suggest that although ERK1/2 induces Sam68-dependent CD44v5 splicing in keratinocytes, additional Sam68-regulated mRNA species (i.e. splice variants other than CD44 isoforms) may contribute to the mechanisms of ERK5-dependent breast cancer cell migration in response to HGF.
EXPERIMENTAL PROCEDURES
Cell Culture
HaCaT, MDA-MB-231, and MDA-MB-435 cells were cultured as described previously (27).
Gene Silencing and Transfection
Knockdown experiments were performed with 100 nm Sam68 siRNA, 50 nm ERK5 siRNA, and 25 nm CD44v5 siRNA (Dharmacon, Lafayette, CO) using Effectene transfection reagent (Qiagen, Valencia, CA) according to the manufacturer's instructions. HaCaT and MDA-MB-231 cells were plated at 2 × 105 in 60-mm dishes, and 24 h later were transiently transfected with either control or siRNA specific for the mRNA target of interest (Sam68, ERK5, or CD44v5). Cells were transfected with 2.5 μg of the pcDNA3.1 expression construct, 1–2.5 μg of Myc-tagged wild-type Sam68, or Myc-tagged phospho-mutant m1 and m4 using FuGENE 6 transfection reagent (Roche Applied Science) according to the manufacturer's instructions. 48 h post-transfection, cells were either starved and treated with either vehicle or HGF for Western blot analysis or harvested and seeded for migration assays.
Antibodies and Reagents
Antibodies to p38, ERK1/2, Sam68, ERK5, human CD44, Myc-tag and phospho-ERK1/2 (Thr-202/Tyr-204) were purchased from Cell Signaling Technology (Danvers, MA) and used at 1:1000 in 1% milk, except for Sam68 (used at 1:5000). Specific antibodies to human CD44 containing exon v5 and v6 were purchased from Bender MedSystems (Burlingame, CA), and human pan-CD44 antibody was from R&D Systems (Minneapolis, MN) and used at 1:1000 in 1% milk. HGF was obtained from Millipore (Temecula, CA); the ERK MAPK-specific inhibitor U0126 was obtained from Calbiochem; PD0325901 was from Selleck Chemicals (Houston, TX). Collagen type I was purchased from Sigma.
Migration Assay
Cell migration assays were performed essentially as described previously (27), except fewer cells (7.5 × 104 for HaCaT or 2.5 × 104 for MDA-MB-231 cells) were seeded into wells containing collagen type I (5 μg/ml), and migration assays were surveyed at 4 h. MEK inhibitors (U0126 or PD0325901) were added to both upper and lower chambers. Results represent at least three independent experiments and are presented as mean ± S.D. Each sample was seeded in quadruplicate wells, and three fields per well were counted.
Immunoprecipitation and Western Blot Analysis
Cells transfected with either siRNA or express constructs were serum-starved for 18 h and then treated with either vehicle or 50 ng/ml HGF, in the presence or absence of MAPK inhibitors, for different time periods. Whole cell lysates were collected as described previously (27) and immunoblotted with specific antibodies. For immunoprecipitation analysis, 1.5 × 106 HaCaT keratinocyte cells or MDA-MB-231 breast cancer cells were plated in 100-mm dishes and the next day were serum-starved and treated as described above. Cells were lysed as described previously (27) except for RIPA supplemented with 2 μg/ml pepstatin, 50 μm dephostatin, 10 μm cypermetrin, 50 nm calyculin (Sigma), 10 mm NaF, and 300 nm okadaic acid (Calbiochem), and 200 μg of total whole cell lysates were used for Sam68 immunoprecipitation, and the Sam68 immunoprecipitations were incubated for 2 h at 4 °C.
RESULTS
HGF Induces Keratinocyte Cell Migration via ERK1/2-dependent Up-regulation of CD44v5
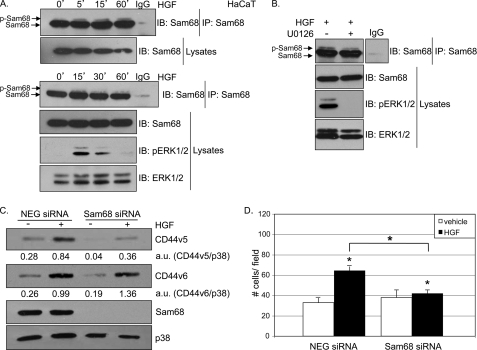
Keratinocyte cells respond to HGF with increased migration, an essential step for the healing process (7), via specific binding to Met receptors and activation of downstream ERK1/2 MAPK module (28). Immortalized nontumorigenic keratinocyte cells (HaCaT) lack the standard CD44 isoform (CD44s) but synthesize high levels of CD44 isoforms containing variant exons 4–7 (25, 29). To link these findings, we tested whether HGF-stimulated HaCaT cells migrate via MAPK-dependent signaling and regulation of CD44 isoforms. HGF activated ERK1/2 and specifically up-regulated CD44v5 (Fig. 1A). Blockade of ERK1/2 activity using the MEK1/2 inhibitor U0126 (10 μm) completely abolished HGF-induced CD44v5 expression. We then tested the ability of HGF-treated HaCaT cells to migrate in Boyden chamber assays in the absence or presence of U0126. HGF-stimulated HaCaT cell migration was entirely ERK1/2-dependent (Fig. 1B). The presence of the MEK inhibitor alone did not alter basal cell migration. The role of CD44v5 as a downstream effector of Met-induced ERK1/2 signaling to HaCaT cell migration was directly tested using siRNA knockdown (Fig. 1C). As expected, HGF induced cell migration relative to vehicle controls. However, HaCaT cells expressing CD44v5 siRNA (Fig. 1C, inset) showed blunted basal and HGF-induced migration relative to cells expressing control siRNA. Notably, CD44s was not detected in HaCaT cells (supplemental Fig. 1, A and B), and CD44v5 siRNA targeted both CD44v5 and high migrating CD44 mRNAs that also contained exon v5 (supplemental Fig. 1C), consistent with the findings of other groups (25, 29). These results demonstrate that HaCaT cells utilize HGF to induce ERK1/2-dependent cell migration that requires v5-containing CD44 isoform(s).
FIGURE 1.
ERK1/2 activation is required for HGF-induced CD44v5 up-regulation and migration in keratinocyte cells. A, HaCaT cells were serum-starved for 18 h and pretreated with 10 μm of the MEK1/2 inhibitor U0126 for 30 min. Cells were then treated with either vehicle or 50 ng/ml HGF for 4 h. CD44v5 expression and ERK1/2 activation were determined with specific antibodies. Total ERK1/2 served as a loading control. B, Boyden chamber migration assays performed with HGF-stimulated HaCaT cells in the presence or absence of 10 μm U0126. MEK inhibitor was added to both upper and lower chambers (wells). C, Boyden chamber migration assays with HaCaT cells transiently transfected with control or CD44v5 siRNA. Inset, Western blots showing CD44v5 and ERK1/2 expression upon CD44v5 knockdown. Forty eight hours post-transfection, cells were harvested and assayed for HGF-induced migration. * denotes significance (p < 0.05) relative to vehicle between indicated groups, determined by unpaired Student's t test. § denotes no statistical difference relative to vehicle.
Sam68 Is Required for HGF-induced CD44v5 Regulation and Migration
Sam68 (Src-associated in mitosis, 68 kDa), a protein involved in RNA processing, is a substrate for ERK1/2 in T-lymphoma cells stimulated with phorbol ester (30). In these studies, phosphorylation of Sam68 was detected by a partial upshift in SDS-polyacrylamide gels. We performed Western blot analyses of immunoprecipitated Sam68 from lysates of HaCaT cells treated with either vehicle or 50 ng/ml HGF. In untreated cells, Sam68 was visible as a predominantly unmodified single band at the expected size of 68 kDa. Upon HGF treatment (0–60 min), a portion of immunopurified Sam68 was slightly upshifted (Fig. 2A, top panel), indicating multisite phosphorylation (30); ERK1/2 was activated at similar time points (Fig. 2A, bottom panel). Additionally, the HGF-induced upshift in Sam68 mobility (15 min) was blunted by pretreatment of cells with the MEK1/2 inhibitor U0126 (Fig. 2B).
FIGURE 2.
Sam68 is required for HGF-induced MAPK-dependent CD44v5 up-regulation and keratinocyte cell migration. A, HaCaT cells were starved for 18 h and treated with either vehicle or 50 ng/ml HGF for 5, 15, and 60 min (top panel) or 15, 30, and 60 min (bottom panel). B, HaCaT cells were starved for 18 h, pretreated with either vehicle or 10 μm of the MEK inhibitor U0126 for 30 min, and treated with 50 ng/ml HGF for 15 min. Cellular lysates (A and B) were immunoprecipitated (IP) for Sam68 and immunoblotted (IB) using Sam68-specific antibodies. Normal rabbit IgG was used as specificity control. ERK1/2 activation was evaluated using phospho-specific antibodies; total ERK1/2 served as a loading control. C, cells transiently expressing either control or Sam68 siRNA were starved for 18 h, induced with 50 ng/ml HGF for 4 h, and Western-blotted (IB) with antibodies specific to CD44v5, CD44v6, and Sam68. Total p38 served as a loading control. Densitometry analysis was performed and indicated below the proper lane as a ratio between CD44v5 and p38 intensity or CD44v6 and p38. D, control and Sam68 siRNA-expressing cells were assayed for their ability to migrate in the presence of HGF. * denotes significance (p < 0.05) relative to vehicle and between indicated groups, as determined by unpaired Student's t test.
Sam68 has been referred to as a tumor suppressor gene (31, 32). However, other studies suggest that Sam68 is required downstream of Src kinase for cell polarization, changes in morphology, and migration (33). To determine whether Sam68 mediates HGF-induced up-regulation of CD44v5 in HaCaT cells, we knocked down Sam68 using siRNA and measured CD44v5 expression. HaCaT cells transiently expressing control or Sam68 siRNA were serum-starved, treated with either vehicle or 50 ng/ml HGF and analyzed for CD44v5 protein expression. Sam68 protein levels were efficiently knocked down by Sam68-targeted siRNA; HGF treatment (4 h) induced robust expression of CD44v5 protein in control siRNA-expressing cells but not in cells expressing Sam68-specific siRNA (Fig. 2C and supplemental Fig. 1E). HGF also induced CD44v6 in HaCat cells, but expression of this variant was unaffected by Sam68 knockdown (Fig. 2C); high migrating CD44 isoforms also remained intact upon Sam68 knockdown (supplemental Fig. 1D). To address the role of Sam68 in HGF-induced keratinocyte migration, HaCaT cells were transiently transfected with either control or Sam68 siRNA and subjected to Boyden chamber assays. Again, HGF induced increased cell migration compared with vehicle controls. Interestingly, however, loss of Sam68 expression significantly inhibited HGF-induced migration (Fig. 2D).
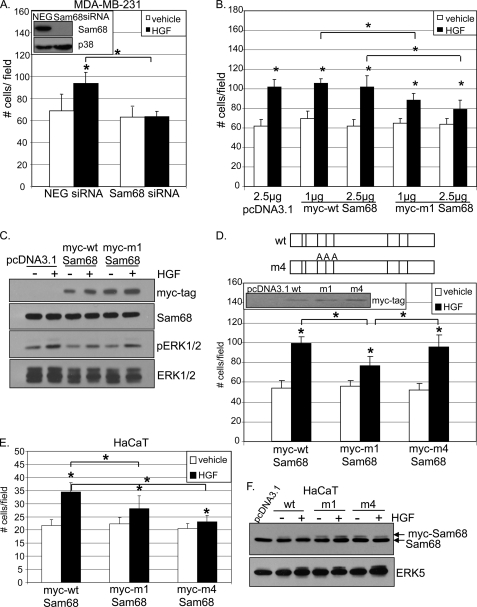
Next, we sought to test the requirement for ERK1/2-mediated Sam68 phosphorylation in HaCaT cell migration. Sam68 contains eight potential proline-directed MAPK phosphorylation sites (34). We obtained the m1-Sam68 expression construct wherein all eight Thr or Ser residues are replaced by Ala (Fig. 3A, top panel) (30). HaCaT cells were transiently transfected with increasing doses (1.0 and 2.5 μg) of vector, Myc-tagged wild-type (WT) Sam68, or Myc-tagged m1 phospho-mutant Sam68 and subjected to Boyden chamber assays in the presence of either vehicle or HGF. HGF induced migration in vector controls and cells expressing Myc-tagged WT Sam68. However, HGF-induced cell migration was significantly impaired in cells expressing 1.0–2.5 μg of m1-Sam68 (Fig. 3A, bottom panel). Tagged proteins were well expressed in HaCaT cells and did not alter the expression of endogenous Sam68 (Fig. 3B). Notably, HGF-induced ERK1/2 phosphorylation occurred in vector-transfected HaCaT cells, but basal and stimulated activities were somewhat blunted in cells expressing Myc-tagged WT Sam68 as well as Myc-tagged m1-Sam68 (Fig. 3C), although ERK5 levels and activity were not altered (data not shown). These data are the first to complete a pathway of Met-ERK1/2-Sam68-CD44v5 in HGF-stimulated cell migration and highlight the requirement for phospho-Sam68 in this process.
FIGURE 3.
HGF-induced migration requires Sam68 phosphorylation. A, top panel, schematic representation of MAPK consensus (PXX(S/T)P) sites on Myc-tagged WT-Sam68 and phospho-mutant m1 wherein all eight Thr or Ser residues are replaced by Ala (30). Bottom panel, cells were transiently transfected with 2.5 μg of vector and 1 and 2.5 μg of Myc-tagged wild-type Sam68 or Myc-tagged phospho-mutant m1. After 48 h, cells were assayed for HGF-induced migration in Boyden chambers. * denotes significance (p < 0.05) relative to vehicle and between indicated groups, as determined by unpaired Student's t test. B, Western blots showing Myc-tagged Sam68 and endogenous Sam68 expression; C, HGF-induced activation of ERK1/2. Total ERK1/2 served as a loading control.
HGF Induces Breast Cancer Cell Migration via an ERK5- and Sam68-dependent Mechanism
The above findings in the skin model prompted us to consider HGF signaling in breast cancer cell migration. We first tested CD44v5 protein expression in highly migratory MDA-MB-231 cells. Previous work from our laboratory demonstrated that these cells migrate in response to HGF via a breast tumor kinase (Brk/PTK-6) and ERK5-dependent signaling pathway in which either WT or kinase-inactive Brk mediated HGF-induced ERK5 activation (27). Surprisingly, although CD44s (standard isoform) was readily detected (supplemental Fig. 1, A and B), MDA-MB-231 cells failed to express appreciable CD44v5 protein, either basally or following a time course of HGF treatment (Fig. 4A). Similarly, these cells also lacked CD44v6 expression (data not shown). Therefore, we tested whether HGF-induced MDA-MB-231 cell migration is regulated by ERK1/2 activation and/or Sam68 phosphorylation events. As in HaCaT cells (Fig. 2B), the HGF-induced gel upshift of phosphorylated Sam68 (30 min) was prevented by pretreatment of MDA-MB-231 cells with the MEK1/2 inhibitor U0126 (Fig. 4B). We then tested the role of ERK1/2 signaling in MDA-MB-231 cell migration. HGF-stimulated robust MDA-MB-231 cell migration (4 h) that was significantly blunted in chambers containing U0126 relative to vehicle-treated controls (Fig. 4C). However, in contrast to the complete inhibition of HaCaT cell migration under similar conditions (Fig. 1B), we observed only a partial block in MDA-MB-231 cell migration; the presence of inhibitor alone did not alter basal MDA-MB-231 migration.
FIGURE 4.
MAPK signaling mediates Sam68 phosphorylation and migration in MDA-MB-231 breast cancer cells. A, HaCaT and MDA-MB-231 cells were starved for 18 h and treated with either vehicle or 50 ng/ml HGF for 4 h. CD44v5 expression was determined with specific antibodies. Total p38 served as a loading control. B, MDA-MB-231 cells were starved for 18 h, pretreated with either vehicle or 10 μm MEK1/2 inhibitor (U0126) for 30 min, and subsequently treated with 50 ng/ml HGF for 30 min. Sam68 was immunoprecipitated (IP) from whole cell lysates and then subjected to Western blotting (IB) using Sam68-specific antibodies. Normal rabbit IgG served as a specificity control. Upshifted Sam68 band indicates multisite phosphorylation. C, Boyden chamber migration assays preformed with MDA-MB-231 stimulated with HGF in the presence or absence of MEK1/2 inhibitor, U0126. Inset shows HGF-induced ERK1/2 activation; ERK1/2 protein levels served as loading control. * denotes significance (p < 0.05) relative to vehicle and between indicated groups, as determined by unpaired Student's t test.
Notably, HGF is a potent activator of both ERK1/2 and ERK5 in MDA-MB-231 cells (Fig. 5A). Because of structural similarities in MEK1/2 and MEK5 activation domains, the MEK1/2 inhibitor U0126 also blocks MEK5 at somewhat higher concentrations (35). In this system, U0126 (10 μm) partially inhibited ERK5 activation, although ERK1/2 was completely suppressed (Fig. 5A). To dissociate between ERK1/2 and ERK5 activities, we employed PD0325901 (PD), a second generation small molecule inhibitor with specific activity against MEK1/2 at a concentration of just 10 nm. This inhibitor abolishes MEK5 activation at a distinctly higher (>10 μm) concentration range, allowing us to selectively inhibit ERK1/2 but not ERK5 activity (Fig. 5B). Migration assays were again performed in the presence or absence of HGF. Low or high PD was included in both chambers for sustained inhibition of either ERK1/2 or both ERK1/2 and ERK5 activities. HGF-induced MDA-MB-231 cell migration was unaffected by low PD, although ERK1/2 was completely inhibited by this concentration (10 nm). Interestingly, inhibition of both classes of MAPKs by 10 μm PD, significantly reduced HGF-induced cell migration. Low or high concentrations of PD, in the absence of HGF, did not appreciably alter basal cell migration (Fig. 5C). To confirm that ERK5 is the primary mediator of HGF-induced cell migration in MDA-MB-231 cells, we performed ERK5 knockdown experiments. Control and ERK5-targeted siRNA were transiently transfected into MDA-MB-231 cells, and migration assays were repeated. HGF-induced MDA-MB-231 cell migration was entirely ERK5-dependent (Fig. 5D). ERK1/2 levels or activity did not change under these conditions (data not shown).
FIGURE 5.
ERK5 mediates breast cancer cell migration through ERK5-Sam68 complex dissociation. MDA-MB-231 cells were pretreated with either 10 μm U0126 for 30 min (A) or 10 nm or 10 μm PD (B) and then treated with either vehicle or 50 ng/ml HGF for 15 min. ERK1/2 and ERK5 activation was evaluated using phospho-specific and total specific antibodies, respectively; total ERK1/2 served as a loading control. C, MDA-MB-231 cells were assayed for migration in response to HGF in the presence or absence of either low (10 nm) or high (10 μm) concentrations of PD. D, Boyden migration assays were performed using cells transiently expressing control or ERK5 siRNA (inset shows ERK5 and a nonspecific protein loading control) and stimulated with HGF. * denotes significance (p < 0.05) relative to vehicle and between indicated groups, as determined by unpaired Student's t test. § denotes no statistical difference relative to vehicle. E, cells were treated with either vehicle or 50 ng/ml HGF for 15–30 min. ERK5 was immunoprecipitated (IP) from whole cell lysates using total ERK5-specific antibodies. ERK5 immunoprecipitates were then subjected to Western blotting (IB) with total ERK5 and Sam68-specific antibodies. F, cells were pretreated for 30 min with low (10 nm) or high (10 μm) concentration of PD, followed by 15 min of HGF treatment. ERK5 was immunoprecipitated from whole cell lysates using total ERK5-specific antibodies and Western-blotted with ERK5 and Sam68-specific antibodies. ERK1/2 and ERK5 activation was evaluated using phospho-specific and total specific antibodies, respectively.
We suspected that ERK5 may substitute for ERK1/2 in Sam68-containing protein complexes downstream of Met receptor activation. To test for association of these molecules, ERK5 was immunoprecipitated from lysates of HGF-treated (0–30 min) MDA-MB-231 cells, and endogenous Sam68 was detected using specific antibodies (Fig. 5E). Sam68 and ERK5 associated transiently, peaking at ∼15 min of HGF stimulation. To test the requirement for ERK5 kinase activity in HGF-induced Sam68/ERK5 interactions, we again performed co-immunoprecipitation assays following pretreatment of MDA-MB-231 cells with either low or high PD and HGF (15 min). Basal interaction between Sam68 and ERK5 remained low in untreated cells relative to cells treated with HGF alone (Fig. 5F, compare lanes 1 and 2; and see IgG control lanes). However, regardless of HGF treatment, Sam68-ERK5 complex formation was enhanced in the presence of either low or high PD relative to vehicle-treated cells. Interestingly, Sam68/ERK5 interaction remained sensitive to HGF in the presence of low PD (Fig. 5F, lanes 3 and 4), whereas inhibition of both ERK1/2 and ERK5 MAPK activities using high PD abolished HGF-induced regulation (lanes 5 and 6); yet basal interaction remained high in this condition (Fig. 5F, compare lanes 1 and 5). These data imply that HGF-induced Sam68-ERK5 complexes form in both the absence and presence of HGF but may require MAPK activities for their dissociation (in the absence of HGF).
Finally, to implicate Sam68 action downstream of Met receptor signaling in MDA-MB-231 breast cancer cell migration, we again employed gene silencing using transiently transfected control or Sam68-targeted siRNA. As predicted, HGF induced significant migration in cells expressing control siRNA (Fig. 6A). However, MDA-MB-231 cells expressing Sam68 siRNA failed to migrate toward HGF in Boyden chamber assays. To confirm a functional role for MAPK-directed phosphorylation of Sam68 in this process, Boyden chamber assays were again performed but with cells transiently expressing either WT or m1-Sam68. As above (Fig. 3A), MDA-MB-231 cells were transiently transfected with 1.0–2.5 μg of vector, Myc-tagged WT Sam68, or Myc-tagged phospho-mutant m1-Sam68. HGF significantly induced migration relative to vehicle controls in both vector and Myc-tagged WT Sam68-expressing cells. However, cells expressing m1-Sam68 demonstrated a dose-dependent impairment of HGF-induced migration (Fig. 6B). Equal levels of Myc-tagged Sam68 were expressed in MDA-MB-231 cells; total endogenous Sam68 expression remained unaltered by these manipulations (Fig. 6C). As in HaCat cells (Fig. 3C), HGF-induced ERK1/2 activation was also slightly dampened upon expression of either WT or m1-Sam68 (see “Discussion”).
FIGURE 6.
Sam68 mediates HGF-induced migration through MAPK-directed phosphorylation sites in breast cancer cells. A, MDA-MB-231 cells expressing either control or Sam68 siRNA were assayed for HGF-induced migration. Inset, Western blots showing Sam68 knockdown and p38 loading control. B, MDA-MB-231 breast cancer cells were transiently transfected with 2.5 μg of vector or 1–2.5 μg of Myc-tagged WT- or m1-Sam68 and assayed in Boyden chambers for HGF induced migration. C, expression of Myc-tagged and endogenous Sam68 protein and ERK1/2 activation were detected using specific antibodies; total ERK1/2 served as a loading control. D, MDA-MB-231 cells transiently transfected with 2.5 μg of Myc-tagged WT-, m1-, and m4-Sam68 were subjected to HGF-induced Boyden chamber migration assays. Schematic representations of Myc-tagged WT- and m4-Sam68 are shown. Inset, transfected Sam68 protein levels were detected using Myc tag-specific antibodies. E, HaCaT cells were transiently transfected as in D and assayed for migration. * denotes significance (p < 0.05) relative to vehicle and between indicated groups, as determined by unpaired Student's t test. F, endogenous and Myc-tagged WT- and m4-Sam68 and total ERK5 were visualized by Western blotting using specific antibodies. Slower running (higher migrating) Sam68 bands indicate Myc-tagged proteins.
We were surprised that these breast cancer cells required phospho-Sam68 for HGF-induced cell migration but did not appreciably regulate CD44v5 (Fig. 4A), as clearly occurred in HaCaT cells (Figs. 1 and 2). Sam68 ERK consensus sites, Ser-58, Thr-71, and Thr-84, are required for TPA-induced v5 inclusion (into mature CD44 mRNAs) in T-lymphocytes (30). As a further control for Sam68-dependent but CD44v5-independent migration of MDA-MB-231 (CD44v5 null) cells, we employed the m4 phospho-mutant of Sam68 (containing point mutations at Ser-58, Thr-71, and Thr-84) (Fig. 6D, top panel) (30). Wild-type and m1-Sam68 served as positive and negative controls, respectively. Again (as in Fig. 6B), HGF-induced MDA-MB-231 cell migration was significantly blunted upon m1-Sam68 but not WT Sam68 expression. Expression of m4-Sam68 also failed to alter HGF-induced cell migration, which remained comparable with that observed upon expression of WT-Sam68 (Fig. 6D, bottom panel). Tagged proteins were expressed at roughly equal levels (Fig. 6D, inset) and did not alter the expression of endogenous Sam68 in MDA-MB-231 cells (data not shown). However, contrary to our findings in breast cancer cells, expression of the m4-Sam68 mutant in HaCaT cells completely blocked HGF-induced cell migration, similar to m1-Sam68. Again, expression of WT Sam68 was without effect (Fig. 6E), and transient expression of tagged Sam68 constructs did not alter endogenous Sam68 or ERK5 levels or activity (Fig. 6F). These data confirm the ERK1/2-dependent path to Sam68-mediated regulation of CD44v5 in HaCaT cells and suggest a dominant role for ERK5-dependent phospho-Sam68 (i.e. at sites other than Ser-68, Thr-71, or Thr-84), independently of CD44v5 regulation, in HGF-induced MDA-MB-231 cell migration.
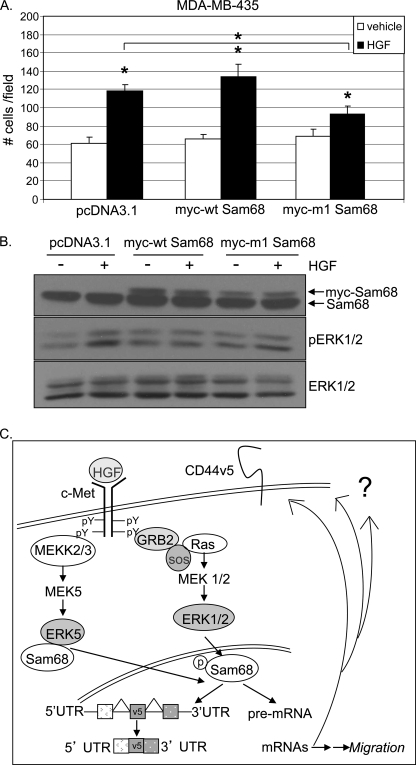
To extend these findings into additional metastatic cancer models, migration assays were performed with MDA-MB-435 cells, a Met+ breast cancer cell line with similarity to melanoma (36). Notably, these cells express negligible levels of CD44v5, yet their HGF-induced migration is ERK1/2-dependent (data not shown). In the presence of HGF, MDA-MB-435 cell migration was significantly increased relative to vehicle controls, in cells expressing either vector or Myc-tagged WT-Sam68. Similar to our results with HaCaT and MDA-MB-231 cells, HGF-induced MDA-MB-435 cell migration was significantly blocked upon expression of phospho-mutant m1-Sam68 (Fig. 7A). Roughly equal levels of Myc-tagged Sam68 protein were expressed in MDA-MB-435 cells; ERK1/2 activation was again very slightly attenuated by both WT and m1 Sam68 (Fig. 7B). These results suggest that Met+ cancer cell migration requires Sam68 phosphorylation, an event that has been linked to the splicing actions of this molecule (30). In addition to CD44v5 (30), Sam68 must target other mRNA species that significantly contribute to HGF-induced breast cancer cell migration, regardless of which ERKs are dominantly activated (discussed below).
FIGURE 7.
Phospho-Sam68 mediates HGF-induced Met+ cancer cell migration. A, MDA-MB-435 cells were transiently transfected as described above with vector, WT-, or m1-Sam68 and assayed for HGF-induced migration. B, Western blot showing transfected and endogenous Sam68 protein expression and MAPK activation. Endogenous and Myc-tagged WT- and m4-Sam68 were visualized by Western blotting using Sam68-specific antibodies. Slower running bands indicate Myc-tagged proteins. ERK1/2 activation was evaluated using phospho-specific antibodies; total ERK1/2 served as a loading control. * denotes significance (p < 0.05) relative to vehicle between indicated groups, as determined by unpaired Student's t test. C, mechanism of action of Sam68 downstream of activated Met receptors. Sam68 is the common effector of HGF-induced migration in keratinocyte and breast cancer cells. Contrary to ERK1/2- and CD44v5-dependent keratinocyte migration, breast cancer cell migration occurs independently of CD44v5 expression via an ERK1/2- or ERK5-dependent mechanism, suggesting a role for phospho-Sam68 in the regulation of splicing of multiple gene targets important for HGF-induced cell migration.
DISCUSSION
Summary
Here, we identified CD44v5 as a target molecule downstream of Met receptor activation that is required for keratinocyte cell migration; activated ERK1/2 and phospho-Sam68 served as key intermediates in this HGF-induced signaling pathway (Fig. 7C). Although Sam68 is a multifunctional molecule, the mechanism of variant exon inclusion into CD44 messages likely involves its intrinsic mRNA splicing activity (24, 30, 37). Selected breast cancer cells also undergo Sam68-dependent migration in response to HGF but via alternate (CD44v5-independent) mechanisms that include either ERK1/2 or ERK5 signaling to phospho-Sam68-mediated splicing of potentially unknown mRNA species to produce alternatively spliced isoforms of key signaling or other regulatory molecules.
Signaling from ERK Family Members to Sam68
Sam68 is a well established ERK1/2 target. In T-lymphoma cells, Thr/Pro phosphorylations detected by specific phospho-Thr/Pro antibodies, and a gel-shifted (phosphorylated) Sam68 fraction, were prevented by inhibition of ERK1/2 activity using the MEK1/2 inhibitor U0126 (30). Accordingly, our data implicate ERK/MAPKs in phosphorylation of Sam68 in both skin (Fig. 2B) and highly migratory (Met+) breast cancer cells (Fig. 4B). In particular, both ERK1/2 and ERK5 are engaged in Sam68 phosphorylation, a major input to cell migration in HaCaT (Figs. 1B and 3A), MDA-MB-231 cells (Figs. 4C and 6B), and MDA-MB-435 cells (Fig. 7A).
ERK5 is a member of the MAPK family, also termed big-MAPK-1 (BMK1). Similar to ERK1/2, ERK5 has been associated with cancer cell proliferation and survival (38, 39), stress-activated signal transduction (40, 41), and activation by growth factor stimuli (38, 39). ERK5 shares high homology with ERK1/2 MAPK, including the amino-terminal half containing the Thr-Glu-Tyr (TEY) activation loop microdomain (42, 43). These related ERKs also possess overlapping substrate specificity (reviewed in Ref. 44). Because of structural similarities, the MEK inhibitor U0126 (10 μm), identified as a MEK1/2-specific inhibitor, also partially inhibits MEK5 (i.e. the direct input to ERK5) activity in a dose-dependent manner (Fig. 5A) (35). In keratinocyte cells, HGF-induced ERK1/2-dependent Sam68-mediated migration is completely prevented by U0126 (Fig. 1B). In this system, U0126 (10 μm) specifically inhibits MEK1/2 but does not affect ERK5 activation (27), indicating that the ERK1/2 MAPK module primarily regulates Sam68 phosphorylation and migration in HaCaT cells (Fig. 2B). In contrast, in breast cancer cells, we observed U0126-induced ERK1/2 inhibition as well as partial inhibition of ERK5, a key input to cell migration in this model (Fig. 4C); this inhibitor (10 μm) also blocked Sam68 phosphorylation (Fig. 4B). Thus, MDA-MB-231 cell migration primarily relies on activation of the ERK5 MAPK module (Fig. 5, C and D). Interestingly, Castro and Lange (27) found that keratinocyte (HaCaT) cell migration shifted to ERK1/2 dependence upon knockdown of ERK5; cell migration actually increased because of compensatory up-regulation of ERK1/2 in the face of ERK5 loss. In contrast, breast cancer cells failed to switch to ERK1/2-dependent migration in similar experiments; their HGF-induced migration required both Brk and ERK5, and these molecules associated in breast cancer cells, as detected by co-immunoprecipitation experiments from HGF-treated cells (27). Here, we have extended the pathway from Met receptor activation to downstream events (ERK activation) leading to Sam68-dependent cell migration. Notably, Sam68 and ERK5 co-purified in response to HGF treatment of MDA-MB-231 cells (Fig. 5E). Our findings corroborate work by Huot et al. (45) who identified large Sam68 complexes composed of ∼40 proteins and regulated by EGF. Enhancement of basal ERK5-Sam68 complex formation and loss of HGF responsiveness when ERK5 is inhibited (Fig. 5F) suggest that these complexes both form and release, in part dependent upon ERK5 activation, prior to cancer cell migration; ERK5-dependent phosphorylation events may favor complex release. Inhibition of ERK1/2 activity may similarly increase basal ERK5/Sam68 aggregation through re-organization of MAPK module components (MEKKs/MEKs/MAPKs), by alteration of one or more kinase and/or scaffolding activities or via altered subcellular localization of module components. Understanding the full complexity of Sam68-containing multiprotein interactions is beyond the scope of this study. However, our data suggest that both ERK1/2 (HaCaT and MDA-MB-435 cells) and ERK5 (MDA-MB-231 cells) act as direct inputs to Sam68 via differential phosphorylation on the multiple MAPK consensus (PXX(S/T)P) sites important for cell migration.
Interestingly, ERK1/2 activation, as measured by phospho-specific antibodies (Fig. 3C), was negatively affected by transient transfection of either WT- or m1-Sam68, suggesting that Sam68 overexpression inhibits Ras-MEK1/2-ERK1/2 activation. Indeed, Sam68 has been reported to form a multiprotein complex with the phosphoprotein associated with glycosphingolipid-enriched microdomain (PAG), Fyn, and RasGAP within lipid rafts on the cell surface, thereby inducing a functional block to Ras activation in stimulated T cells (46). Sam68-induced inhibition of Ras signaling may explain the partial (weak) loss of ERK1/2 activity observed herein (Figs. 3C, 6C, and 7B; see Myc-tagged WT or m1 Sam68 relative to controls). Notably, these events did not correlate with changes in HGF-induced cell migration (Figs. 3A, 6B, and 7A) in that only m1-Sam68 but not WT Sam68 blocked HGF-induced migration. Taken together, our data suggest that Sam68 acts as a convergence point for ERK kinase (ERK1/2 or ERK5) inputs to cell migration. Functional overlap between ERK1/2 and ERK5 is relevant to clinical use of MEK inhibitors for the treatment of metastatic cancers.
Sam68 Is a Multifunctional Protein
The nuclear RNA-binding protein, Sam68, was initially discovered as a substrate of c-Src during mitosis (47–49) and found to interact with a variety of signaling proteins containing Src homology 3 and Src homology 2 domains (Grb2, phospholipase Cγ1, Fyn, p120GAP, Nck, and BRK). These interactions suggest a scaffolding or adaptor function for Sam68 in response to diverse extracellular stimuli (50–54). Recent studies suggest that Sam68 is primarily involved in RNA processing. The Sam68 KH (heterogeneous nuclear ribonucleoprotein K homology) domain, embedded within a larger GSG (GRP33/Sam68/GLD1) domain, mediates homodimerization and RNA binding (55, 56). This domain is essential for prostate cancer cell growth; GFP-Sam68GSG chimeric proteins, encoding the GFP tag sequence fused to the GSG domain of Sam68, interfered with androgen-dependent proliferation and survival (57). In addition to CD44v5 (30), Sam68 is known to regulate mRNA splicing of BCL2L1 (58) and CCND1 (59). More recently, Sam68 has been reported to promote epithelial-to-mesenchymal transition of SW480 (colon adenocarcinoma) cells via ERK1/2-dependent Sam68 phosphorylation and alternative splicing of SRSF1 transcripts (60). Clearly, phosphorylation of Sam68 appears to be an important input to its splicing activity. Simultaneous phosphorylation of Sam68 on three sites (Ser-58, Thr-71, and Thr-84) was required for exon v5 inclusion into mature CD44 mRNAs expressed in T cells (30). Similarly, our data using the m1 and m4 phospho-mutants of Sam68 (Fig. 6, D and E) support this finding in HaCaT cells and imply that differential Sam68 phosphorylation may in part confer splicing selectivity, perhaps in response to regulation by alternate ERKs. Mechanisms of selective mRNA targeting for alternative splicing (i.e. by signaling pathways) remain largely undefined and the topic of a separate study.
Notably, Sam68 appears to possess both tumor-suppressive and oncogenic functions. Overexpression of wild-type or RNA-binding mutant Sam68 in mouse fibroblasts leads to either cell cycle arrest (in G1 phase) or cell transformation (as measured by increased soft agar colony formation), respectively (31). Additionally, Sam68−/− mice present with defects in breast and uterine development, whereas Sam68 haploinsufficiency correlates with delayed mammary tumor onset (62). In breast cancer cells, Sam68 co-localizes with the transcriptional co-activator, cAMP-response element-binding protein-binding protein, and thus may act as an indirect transcriptional repressor, independently of its RNA binding ability (32). On the contrary, in prostate, Sam68 interacts with androgen receptors and acts as a transcriptional co-activator (63). It will be important to understand what function(s) of Sam68 (i.e. regulation of signaling pathways, transcriptional repression or activation, or alternative splicing of mRNAs) mediate changes in normal and cancer cell biology if this molecule is be considered as part of potential targeted cancer therapies.
Mechanism of HGF-induced Migration in Multiple Cell Types
Although studies have been limited, it is clear that Sam68 signaling is complex and employs the use of multiple domains (30, 57, 62, 63). Of note, Sam68-mediated keratinocyte and breast cancer cell migration likely occurs independently of the KH RNA-binding domain. Indeed, phospho-mutant m1-Sam68, which blocked HGF-induced migration (Figs. 3A, 6, B and D, and 7A), harbors mutant Ser/Thr residues localized entirely outside of the KH domain. In support of our findings, Richard and co-workers (33) have previously reported that human cervical cancer cell migration occurs independently of the Sam68 KH domain, but it requires tyrosine phosphorylation within the carboxyl-terminal region that contains a functional nuclear localization signal. Phosphorylation of Sam68 on Tyr-440 has also been reported in MDA-MB-231 cells; upon EGF treatment, breast tumor kinase (Brk) phosphorylates this residue to induce Sam68 nuclear localization and cell cycle progression (64). As discussed above (see Introduction), Brk-dependent signaling events are also relevant to MDA-MB-231 cell migration; gene silencing of Brk inhibited HGF-induced MDA-MB-231 cell migration (27). Notably, MDA-MB-435 cells are Brk-null (27) but still require ERK1/2 and phospho-Sam68 for HGF-induced migration (Fig. 7A). These data suggest that multiple mechanisms of Sam68 regulation are relevant to cancer biology. Phosphorylation on Tyr-440 by Brk (or c-Src) may primarily induce Sam68 nuclear localization required for proliferative responses (64), whereas multisite Ser/Thr phosphorylation downstream of ERK1/2 or ERK5 signaling may dictate the specificity of Sam68-dependent mRNA splicing (Figs. 3A and 6, B, D, and E). We have implicated ERK1/2 or ERK5 as key MAPK family member inputs to Sam68 Ser/Thr phosphorylation, a required step in the pathway leading to HGF-induced (i.e. chemotactic) cellular migration.
In summary, we present a simplified model of our findings (Fig. 7C). Here, we identified Sam68 as a key effector of both keratinocyte and breast cancer cell migration. HGF/Met pathway signaling activates ERK family MAPKs, which serve as direct inputs to Sam68-dependent activation/mRNA splicing. In contrast to skin cells, Met+ breast cancer models did not appear to regulate CD44v5. This result suggests that Sam68 acts as an essential node in cancer cell migration, most likely by targeting other essential (i.e. to migration) spliced gene products. We did not observe regulation of CD44, CCND1, BCL2L1, or SRSF1 mRNAs upon m1-Sam68 expression in MDA-MB-231 cells (data not shown). Future experiments will be aimed at elucidation of Sam68-regulated splice variants of genes involved in HGF-induced migration in cancer cells. Blockade of Sam68, a mediator of alternative splicing and cell migration, may provide a new avenue for therapeutic inhibition of metastatic cancers.
Supplementary Material
Acknowledgments
We thank Nancy Castro for technical assistance; Elizabeth Wattenberg (University of Minnesota School of Public Health) for the HaCaT keratinocyte cell line; Deepali Sachdev (University of Minnesota Dept. of Medicine) for providing MDA-MB-231 cells; James McCarthy (Dept. of Medicine and Pathology, University of Minnesota) for the human CD44 antibody (Cell Signaling); and Harald König (Forschungszentrum Karlsruhe GmbH, Institute for Technology and Genetics, Germany) for kindly providing Myc-tagged WT-, m1, and m4-Sam68 constructs.
This work was supported, in whole or in part, by National Institutes of Health Grant CA107547 from NCI (to C. A. L.). This work was also supported by American Cancer Society RSG TBE-107800 (to C. A. L.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
- HGF
- hepatocyte growth factor
- PD
- PD0325901
- KH
- K homology.
REFERENCES
- 1. Nakamura T., Nawa K., Ichihara A. (1984) Biochem. Biophys. Res. Commun. 122, 1450–1459 [DOI] [PubMed] [Google Scholar]
- 2. Russell W. E., McGowan J. A., Bucher N. L. (1984) J. Cell. Physiol. 119, 183–192 [DOI] [PubMed] [Google Scholar]
- 3. Stoker M., Gherardi E., Perryman M., Gray J. (1987) Nature 327, 239–242 [DOI] [PubMed] [Google Scholar]
- 4. Jiang W., Hiscox S., Matsumoto K., Nakamura T. (1999) Crit. Rev. Oncol. Hematol. 29, 209–248 [DOI] [PubMed] [Google Scholar]
- 5. Birchmeier C., Birchmeier W., Gherardi E., Vande Woude G. F. (2003) Nat. Rev. Mol. Cell Biol. 4, 915–925 [DOI] [PubMed] [Google Scholar]
- 6. Chmielowiec J., Borowiak M., Morkel M., Stradal T., Munz B., Werner S., Wehland J., Birchmeier C., Birchmeier W. (2007) J. Cell Biol. 177, 151–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jiang W. G., Hiscox S. (1997) Histol. Histopathol. 12, 537–555 [PubMed] [Google Scholar]
- 8. Nayeri F., Stromberg T., Larsson M., Brudin L., Soderstrom C., Fosberg P. (2002) J. Dermatol. Treat. 13, 81–86 [DOI] [PubMed] [Google Scholar]
- 9. Nayeri F., Olsson H., Peterson C., Sundqvist T. (2005) J. Dermatol. Sci. 37, 75–85 [DOI] [PubMed] [Google Scholar]
- 10. Nusrat A., Parkos C. A., Bacarra A. E., Godowski P. J., Delp-Archer C., Rosen E. M., Madara J. L. (1994) J. Clin. Invest. 93, 2056–2065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bevan D., Gherardi E., Fan T. P., Edwards D., Warn R. (2004) J. Pathol. 203, 831–838 [DOI] [PubMed] [Google Scholar]
- 12. Nakamura T., Mizuno S., Matsumoto K., Sawa Y., Matsuda H., Nakamura T. (2000) J. Clin. Invest. 106, 1511–1519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Huh C. G., Factor V. M., Sánchez A., Uchida K., Conner E. A., Thorgeirsson S. S. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 4477–4482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ponzo M. G., Lesurf R., Petkiewicz S., O'Malley F. P., Pinnaduwage D., Andrulis I. L., Bull S. B., Chughtai N., Zuo D., Souleimanova M., Germain D., Omeroglu A., Cardiff R. D., Hallett M., Park M. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 12903–12908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Graveel C. R., DeGroot J. D., Su Y., Koeman J., Dykema K., Leung S., Snider J., Davies S. R., Swiatek P. J., Cottingham S., Watson M. A., Ellis M. J., Sigler R. E., Furge K. A., Vande Woude G. F. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 12909–12914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ponta H., Sherman L., Herrlich P. A. (2003) Nat. Rev. Mol. Cell Biol. 4, 33–45 [DOI] [PubMed] [Google Scholar]
- 17. Black D. L. (2003) Annu. Rev. Biochem. 72, 291–336 [DOI] [PubMed] [Google Scholar]
- 18. Günthert U., Hofmann M., Rudy W., Reber S., Zöller M., Haussmann I., Matzku S., Wenzel A., Ponta H., Herrlich P. (1991) Cell 65, 13–24 [DOI] [PubMed] [Google Scholar]
- 19. Stamenkovic I., Aruffo A., Amiot M., Seed B. (1991) EMBO J. 10, 343–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Borland G., Ross J. A., Guy K. (1998) Immunology 93, 139–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Naor D., Sionov R. V., Ish-Shalom D. (1997) Adv. Cancer Res. 71, 241–319 [DOI] [PubMed] [Google Scholar]
- 22. Wu S. T., Sun G. H., Hsieh D. S., Chen A., Chen H. I., Chang S. Y., Yu D. (2003) J. Formos Med. Assoc. 102, 229–233 [PubMed] [Google Scholar]
- 23. Cheng C., Sharp P. A. (2006) Mol. Cell. Biol. 26, 362–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Weg-Remers S., Ponta H., Herrlich P., König H. (2001) EMBO J. 20, 4194–4203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Günthert U. (1993) Curr. Top. Microbiol. Immunol. 184, 47–63 [DOI] [PubMed] [Google Scholar]
- 26. Brown T. A., Bouchard T., St John T., Wayner E., Carter W. G. (1991) J. Cell Biol. 113, 207–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Castro N. E., Lange C. A. (2010) Breast Cancer Res. 12, R60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kermorgant S., Zicha D., Parker P. J. (2004) EMBO J. 23, 3721–3734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. van Weering D. H., Baas P. D., Bos J. L. (1993) PCR Methods Appl. 3, 100–106 [DOI] [PubMed] [Google Scholar]
- 30. Matter N., Herrlich P., König H. (2002) Nature 420, 691–695 [DOI] [PubMed] [Google Scholar]
- 31. Liu K., Li L., Nisson P. E., Gruber C., Jessee J., Cohen S. N. (2000) J. Biol. Chem. 275, 40195–40201 [DOI] [PubMed] [Google Scholar]
- 32. Hong W., Resnick R. J., Rakowski C., Shalloway D., Taylor S. J., Blobel G. A. (2002) Mol. Cancer Res. 1, 48–55 [PubMed] [Google Scholar]
- 33. Huot M. E., Brown C. M., Lamarche-Vane N., Richard S. (2009) Mol. Cell. Biol. 29, 1933–1943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chang L., Karin M. (2001) Nature 410, 37–40 [DOI] [PubMed] [Google Scholar]
- 35. Mody N., Leitch J., Armstrong C., Dixon J., Cohen P. (2001) FEBS Lett. 502, 21–24 [DOI] [PubMed] [Google Scholar]
- 36. Chambers A. F. (2009) Cancer Res. 69, 5292–5293 [DOI] [PubMed] [Google Scholar]
- 37. Konig H., Moll J., Ponta H., Herrlich P. (1996) EMBO J. 15, 4030–4039 [PMC free article] [PubMed] [Google Scholar]
- 38. Kato Y., Tapping R. I., Huang S., Watson M. H., Ulevitch R. J., Lee J. D. (1998) Nature 395, 713–716 [DOI] [PubMed] [Google Scholar]
- 39. Kato Y., Kravchenko V. V., Tapping R. I., Han J., Ulevitch R. J., Lee J. D. (1997) EMBO J. 16, 7054–7066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Abe J., Kusuhara M., Ulevitch R. J., Berk B. C., Lee J. D. (1996) J. Biol. Chem. 271, 16586–16590 [DOI] [PubMed] [Google Scholar]
- 41. Abe J., Takahashi M., Ishida M., Lee J. D., Berk B. C. (1997) J. Biol. Chem. 272, 20389–20394 [DOI] [PubMed] [Google Scholar]
- 42. Zhou G., Bao Z. Q., Dixon J. E. (1995) J. Biol. Chem. 270, 12665–12669 [DOI] [PubMed] [Google Scholar]
- 43. Lee J. D., Ulevitch R. J., Han J. (1995) Biochem. Biophys. Res. Commun. 213, 715–724 [DOI] [PubMed] [Google Scholar]
- 44. Nishimoto S., Nishida E. (2006) EMBO Rep. 7, 782–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Huot M. E., Vogel G., Richard S. (2009) J. Biol. Chem. 284, 31903–31913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Smida M., Posevitz-Fejfar A., Horejsi V., Schraven B., Lindquist J. A. (2007) Blood 110, 596–615 [DOI] [PubMed] [Google Scholar]
- 47. Taylor S. J., Shalloway D. (1994) Nature 368, 867–871 [DOI] [PubMed] [Google Scholar]
- 48. Weng Z., Thomas S. M., Rickles R. J., Taylor J. A., Brauer A. W., Seidel-Dugan C., Michael W. M., Dreyfuss G., Brugge J. S. (1994) Mol. Cell. Biol. 14, 4509–4521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Fumagalli S., Totty N. F., Hsuan J. J., Courtneidge S. A. (1994) Nature 368, 871–874 [DOI] [PubMed] [Google Scholar]
- 50. Richard S., Yu D., Blumer K. J., Hausladen D., Olszowy M. W., Connelly P. A., Shaw A. S. (1995) Mol. Cell. Biol. 15, 186–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Paronetto M. P., Venables J. P., Elliott D. J., Geremia R., Rossi P., Sette C. (2003) Oncogene 22, 8707–8715 [DOI] [PubMed] [Google Scholar]
- 52. Jabado N., Jauliac S., Pallier A., Bernard F., Fischer A., Hivroz C. (1998) J. Immunol. 161, 247–269 [PubMed] [Google Scholar]
- 53. Lawe D. C., Hahn C., Wong A. J. (1997) Oncogene 14, 223–231 [DOI] [PubMed] [Google Scholar]
- 54. Derry J. J., Richard S., Valderrama Carvajal H., Ye X., Vasioukhin V., Cochrane A. W., Chen T., Tyner A. L. (2000) Mol. Cell. Biol. 20, 6114–6126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Chen T., Boisvert F. M., Bazzett-Jones D. P., Richard S. (1999) Mol. Cell. Biol. 10, 3015–3033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lin Q., Taylor S. J., Shalloway D. (1997) J. Biol. Chem. 272, 27274–27280 [DOI] [PubMed] [Google Scholar]
- 57. Busà R., Paronetto M. P., Farini D., Pierantozzi E., Botti F., Angelini D. F., Attisani F., Vespasiani G., Sette C. (2007) Oncogene 26, 4372–4382 [DOI] [PubMed] [Google Scholar]
- 58. Paronetto M. P., Achsel T., Massiello A., Chalfant C. E., Sette C. (2007) J. Cell Biol. 176, 929–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Paronetto M. P., Cappellari M., Busà R., Pedrotti S., Vitali R., Comstock C., Hyslop T., Knudsen K. E., Sette C. (2010) Cancer Res. 70, 229–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Valacca C., Bonomi S., Buratti E., Pedrotti S., Baralle F. E., Sette C., Ghigna C., Biamonti G. (2010) J. Cell Biol. 191, 87–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Deleted in proof.
- 62. Richard S., Vogel G., Huot M. E., Guo T., Muller W. J., Lukong K. E. (2008) Oncogene 27, 548–556 [DOI] [PubMed] [Google Scholar]
- 63. Rajan P., Gaughan L., Dalgliesh C., El-Sherif A., Robson C. N., Leung H. Y., Elliott D. J. (2008) J. Pathol. 215, 67–77 [DOI] [PubMed] [Google Scholar]
- 64. Lukong K. E., Larocque D., Tyner A. L., Richard S. (2005) J. Biol. Chem. 280, 38639–38647 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.