Abstract
Current antiretroviral therapy (ART) provides potent suppression of HIV-1 replication. However, ART does not target latent viral reservoirs, so persistent infection remains a challenge. Small molecules with pharmacological properties that allow them to reach and activate viral reservoirs could potentially be utilized to eliminate the latent arm of the infection when used in combination with ART. Here we describe a cell-based system modeling HIV-1 latency that was utilized in a high-throughput screen to identify small molecule antagonists of HIV-1 latency. A more detailed analysis is provided for one of the hit compounds, antiviral 6 (AV6), which required nuclear factor of activated T cells for early mRNA expression while exhibiting RNA-stabilizing activity. It was found that AV6 reproducibly activated latent provirus from different lymphocyte-based clonal cell lines as well as from latently infected primary resting CD4+ T cells without causing general T cell proliferation or activation. Moreover, AV6 complemented the latency antagonist activity of a previously described histone deacetylase (HDAC) inhibitor. This is a proof of concept showing that a high-throughput screen employing a cell-based model of HIV-1 latency can be utilized to identify new classes of compounds that can be used in concert with other persistent antagonists with the aim of viral clearance.
Keywords: Antiviral Agents, Cooperativity, Drug Action, Gene Regulation, HIV, Lentivirus, HIV Latency, High-throughput Screening
Introduction
The ability of human immunodeficiency virus type 1 (HIV-1) to establish a latent infection results in life-long virus persistence even after long-term antiretroviral therapy (ART).4 The role that latency plays in preventing sustained clearance of the virus infection has become evident in recent years. Patients that have been successfully treated with ART, having undetectable levels of viral RNA (below 50 copies/ml) in the plasma for years, experienced rapid virus rebound upon withdrawal of therapy (1, 2). Moreover, it was found that after T cell activation, virus could be isolated from CD4+ T cells taken from these patients, underscoring the need to eliminate the latently infected cells to eradicate the virus (3–5).
Activation of latent proviruses from infected cells in combination with ART is part of a therapeutic strategy that may lead to the complete elimination of HIV infection. Prior attempts to “flush out” the virus by activation of latently infected resting CD4+ T cells with the administration of IL-2 and/or anti-CD3 monoclonal antibodies were ultimately unsuccessful, probably because of its inability to reach all of the latent viral reservoirs and the toxicity of the regimen (6–10). A more promising approach to complete viral clearance is the use of small molecules with pharmacological properties that allow them to access the viral reservoirs and to specifically reactivate the latent proviruses. The concept of small molecule activation of latent HIV-1 has been tested in a clinical study using the histone deacetylase (HDAC) inhibitor valproic acid (VA) (11). However, it is questionable whether VA alone can be used as a supplement to ART for successful HIV eradication. It was shown that the latent reservoir was still present in HIV-1 patients after long-term ART and simultaneous VA treatment for neurological and psychiatric disorders (12, 13). Other small molecules, such as suberoylanilide hydroxamic acid (SAHA) (14–16), resveratrol (17), and prostratin (18, 19), have been proposed as potential agents for the disruption of HIV latent infection. Prostratin induces the NF-κB activation pathway (20, 21), which puts into question its use in clinical studies because abnormal NF-κB signaling has been related to the pathophysiology of cancer, inflammatory diseases, and neurodegenerative disorders (22–25).
The common shortcoming of the current strategies to disrupt latent infection is their adherence to well known mechanisms of latent HIV reactivation (such as histone remodeling and NF-κB activation) and low-to-medium throughput testing of agents available for research studies (26). It is more efficient to perform a high-throughput screen (HTS) of available large chemical libraries to discover novel agents capable of reactivating latent HIV infection. To discover novel HIV latency activators, we developed a robust cell-based assay adaptable to HTS (27). In this study, we present the optimization and utilization of our latency model in an HTS of a library of small molecules that led to identification of compounds capable of reactivating latent HIV-1. In addition, we present data demonstrating that one of the reactivating compounds requires NFAT activity for early gene expression while affecting RNA stability.
EXPERIMENTAL PROCEDURES
Cells and Reagents
24STNLSG, SupT1, ACH-2, and peripheral blood mononuclear cells (PBMC) were cultured in RPMI 1640 medium (Invitrogen) containing 10% FBS (HyClone), 100 units of penicillin/ml, 100 μg of streptomycin/ml, and 2 mm glutamine. TNFα was obtained from R&D Systems (Minneapolis, MN). VA and trichostatin A were purchased from Sigma. HIV-1 p24 Gag protein ELISA kits were purchased from ZeptoMetrix (Buffalo, NY). SEAP detection kits were purchased from BD Biosciences. SEAP readings were performed using a ViewLux CCD imager (10-s read time), and the plates were imaged to quantify signal output. RNA polymerase II antibody was from Santa Cruz Biotechnology, and NFAT1 antibody, c-Jun antibody, and NFκB p65 antibody were purchased from Abcam. VIVIT and FK506 were purchased from EMD4biosciences and Sigma-Aldrich, respectively.
HTS Data Analysis
The ActivityBase software package (IDBS, Guildford, Surrey, UK) was used to manage HTS data. After completion of a daily screen, data were automatically uploaded into Oracle through ActivityBase protocols that convert the raw data into percentage of activation. Templates were designed for each screen to calculate the activation value for the standard control activator (TNFα) and the Z-factor for each plate. Thus, the reliability of the data was assessed immediately and compared from plate-to-plate and day-to-day. A full statistical analysis was conducted with the final set of data utilizing the software packages from Spotfire (28) and Pipeline Pilot (SciTegic, San Diego, CA) to determine the fitness and the level of statistical significance of the assay. HTS “hits” with statistically significant activity were considered those that produced an activation signal three standard deviations or greater from the mean of the assay run.
Flow Cytometric Analysis
Flow cytometric analysis was performed to determine the percentage of GFP-expressing cells after compound treatment. A total of 1 × 106 cells were washed twice with PBS and resuspended in 1 ml of 0.5% paraformaldehyde in PBS. To analyze the efficiency of quiescent CD4+ T cell enrichment, 1 × 105 cells were incubated for 30 min on ice with 2 μl of fluorescein isothiocyanate (FITC)-labeled anti-CD4+ antibodies (CD4+-FITC) and 4 μl of phycoerythrin (PE)-labeled anti-HLA-DR (HLA/DR-PE) antibodies or 4 μl of CD69-PE. Stained cells were washed in PBS containing 0.1% NaN3 and analyzed by flow cytometry using FACScan (Becton Dickinson) equipped with CellQuest Pro (Macintosh, Sunnyvale, CA).
SEAP Assay
The assay was performed using the Great EscAPe SEAP chemiluminescence detection kit (BD Biosciences) according to the manufacturer's protocol. The relative light units were measured with an MLX microtiter plate luminometer (DYNEX Technologies).
Activity in Primary Resting CD4+ T Cells
Resting CD4+ T cells were purified by first isolating PBMCs on a Ficoll-Hypaque (Sigma) gradient. CD4+ T cells were then isolated by negative selection using Dynabeads® UntouchedTM human CD4+ T cells (Invitrogen) antibody-coated magnetic beads. Activated CD4+ T cells were removed using CD25 and HLA-DR microbeads (Miltenyi, Bergisch Gladbach, Germany). Recovered cells were cultured in RPMI 1640 medium containing 10% FBS. Virus was generated by transfecting 293T cells with transducing plasmid, NLRlucRFP, an HIV-1-based vector containing Renilla luciferase (rluc) and red fluorescent protein (rfp) expression cassettes, pCMV-ΔR8.2 packing vector, and HXB2 expression vector pCMS-IVS-HXB2 by CaPO4 precipitation (16, 29). At 48–72 h after transfection, viral supernatants were filtered and then supplemented with PEG-it (Systems Biosciences, Mountain View, CA) and refrigerated overnight. The viral supernatants were concentrated 100-fold and added to the resting CD4+ T cells, which were then centrifuged at 1,200 × g for 2 h. Three days after infection, cells were treated as indicated for 24 h. Finally, total RNA was extracted using RNAqueous kits (Ambion, Austin, TX). rfp mRNA levels were quantified using a one-step quantitative real-time RT-PCR (qRT-PCR), normalized to β-actin. No-template and no-RT qRT-PCR controls were also performed.
Cellular Proliferation
DNA synthesis, measured as [3H]thymidine incorporation 72 h after compound treatment, was used to assess cell proliferation. PBMCs (2 × 105) were incubated for 8 h with [3H]thymidine (10 μCi/ml) and harvested onto glass fiber filters. Thymidine incorporation into DNA was measured with a liquid scintillation counter.
Cellular Toxicity
Cellular toxicity was determined using Cell Titer GloR reagent (Promega, Madison WI) according to the manufacturer's protocol. The relative light units were measured with MLX microtiter plate luminometer (DYNEX Technologies, Chantilly, VA). Additionally, the percentage of apoptotic cells was determined via flow cytometric analysis of cells using an annexin V-FITC apoptosis detection kit, APO-AF (Sigma), according to the manufacturer's recommendations.
HDAC Activity Assay
SupT1 or THP-1 nuclear extracts (40 μg) were incubated with 1 mm HDAC assay substrate (HDAC assay kit, colorimetric detection, Upstate Biotech Millipore, Temecula, CA) in the absence or presence of 1 μm trichostatin A, 1 mm VA, or 10 μm AV6. Reactions were carried out according to the manufacturer's protocol.
qRT-PCR
Total cellular RNA was isolated from 1 × 106 untreated or treated cells using the RNeasy mini kit according to the manufacturer's instructions (Qiagen). Extracted RNA was subsequently treated with RQ1 DNase kit (Promega) to remove any traces of DNA. Two-step qRT-PCR was performed for relative quantification of viral RNA. cDNA was synthesized with TaqMan reverse transcription reagents and oligo(dT)16 following the protocol supplied by Applied Biosystems (Roche Applied Science). qRT-PCR was performed with SYBR Green PCR master mix and run in a DNA Engine Opticon 2 (MJ Research) detector with Opticon Monitor Analysis software, version 1.4. The following primers were used in the qRT-PCR reactions: EEF1A1, forward, (5′-TAGCTGGATCCGCTGACTTT-3′) and EEF1A1, reverse, (5′-AACAGTACTTGCCCGTGTCC-3′); β-actin, forward, (5′-CTGGAACGGTGAAGGTGACA-3′) and β-actin, reverse, (5′-AAGGGACTTCCTGTAACAATGCA-3′); total HIV1 RNA, forward, (5′-CTGACCTTTGGATGGTGCTA-3′) and total HIV1 RNA, reverse, (5′-TGAAATGCTAGGCGGCTGTC-3′); Tat, forward, (5′-CGAAGAGCTCATCAGAACAGTCA-3′) and Tat, reverse, (5′-CTTCCTCATTGATGGTCTC-3′); Gag, forward, (5′-CGACCAGGACTCGGC-3′) and Gag, reverse, (5′-CTTCCTCATTGATGGTCTC-3′). For each primer set, amplification efficiencies were determined by obtaining a standard curve with 2-fold serial dilutions of cDNA. A dissociation curve was generated for each primer pair to confirm amplification of a single product. The sizes of the amplified products were confirmed by agarose gel electrophoresis, and the specificity of the products was confirmed by sequencing. Reactions were completed in duplicate, and no-template and no-reverse transcriptase controls were included for each primer pair.
qRT-PCR to measure viral mRNA levels in Fig. 4D was performed using tat-specific primers (5′-GCATCTCCTATGGCAGGAAG-3′ and 5′-TTCTATTCCTTCGGGCCTGT-3′). Two hours after treating 5 × 106 J-lat 9.2 cells with AV6 plus or minus inhibitors, total RNA was isolated from each sample followed by qRT-PCR using a combination of the SuperScript® VILOTM cDNA synthesis kit (Invitrogen) and Maxima® SYBR Green (Fermentas). Data were normalized to Ct values obtained from the same samples utilizing β-globin gene primers (5′-CCCTTGGACCCAGAGGTTCT-3′ and 5′-CGAGCACTTTCTTGCCATGA-3′). All the data are averages from three independent samples.
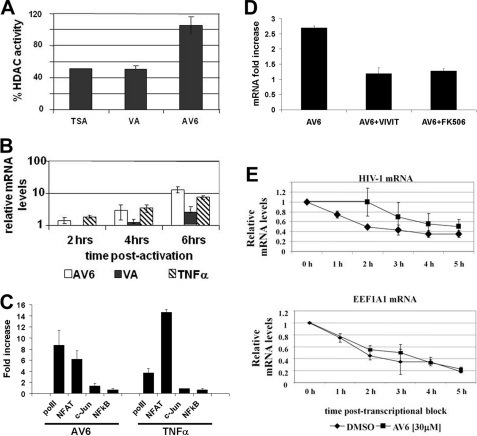
FIGURE 4.
AV6 reactivates latent HIV-1 through a novel mechanism. A, AV6 lacks HDAC inhibition activity. SupT1 nuclear extract (40 μg) was incubated with HDAC assay substrate (see “Experimental Procedures”) in the absence or presence of AV6 (10 μm), trichostatin A (TSA, 1 μm), or VA (1 mm). The percentage of HDAC activity inhibition was determined as ratios of samples containing compound versus no compound sample. These data present results from two independent experiments. B, viral RNA species accumulate shortly after AV6 treatment. qRT-PCR was performed with total RNA isolated from 24STNLSG cells at the indicated time points after stimulation with AV6 (30 μm), VA (1 mm), or TNFα (50 ng/ml). The data show the means and S.D. from two representative experiments. AV6-dependent early mRNA expression requires NFAT. C, chromatin immunoprecipitation assay. J-lat 9.2 cells were treated with AV6 (12 μm) or TNFα (50 ng/ml) for 2 h followed by chromatin immunoprecipitation from each sample with antibody specific for RNA polymerase II (polII), NFAT1, c-Jun, or NFκB p65 and with LTR-specific primers and probe. D, HIV-1 mRNA levels in J-lat 9.2 cells were measured 2 h after treatment with AV6 (12 μm) via quantitative real-time PCR in the presence or absence of the indicated inhibitors (3 μm VIVIT or 20 μm FK506). The -fold change in viral mRNA expression was obtained by normalizing to the mRNA levels at the zero time point. E, AV6 treatment leads to stabilization of viral RNA species. 24STNLSG latent cells (1 × 107) were preincubated at 37 °C with actinomycin D (2 μg/ml) for 1 h followed by incubation with AV6 (30 μm) or 0.5% DMSO. Samples were taken every 20 min, and total RNA was isolated. Relative RNA levels were calculated using the normalized signal of the sample treated with 0.5% DMSO as a reference point. The means and S.D. from two representative experiments are shown. It is noteworthy that a higher concentration of AV6 was used than in other experiments given the shortened time frame of the experiment.
Chromatin Immunoprecipitation
For each sample, 5 × 106 J-lat 9.2 cells were treated employing the Abcam cross-linking chromatin immunoprecipitation (X-ChIP) protocol with slight modification. Briefly, after 6 min of sonication, cell lysates were treated with antibody for NFAT1, c-Jun, RNA polymerase II, or NFκB p65 overnight. Following immunoprecipitation with G-protein agarose beads and washing, chromatin was isolated and reverse-cross-linked with proteinase K overnight. The chromatin was then purified with phenol-chloroform followed by precipitation of DNA fragments with ethanol. Isolate was subjected to quantitative real-time PCR using primers (5′-CCGTCTGTTGTGTGACTCTGGTAA-3′ and 5′-GTCGAGAGATCTCCTCTGGCTTTACT-3) and a probe (5′-/6FAM/TTCGCTTTCAAGTCCCTGTTCGG/Iowa Black FQ/-3′) corresponding to the LTR U5 region. All data were normalized to Ct values obtained from the same samples utilizing β-globin gene primers (5′-CCCTTGGACCCAGAGGTTCT-3′ and 5′-CGAGCACTTTCTTGCCATGA-3′). Each data point represents an average from three separate samples.
RESULTS
Identification of Novel Activators of Latent HIV-1
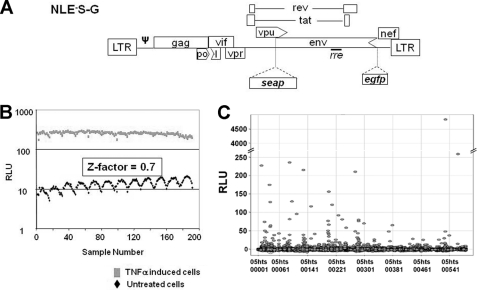
We previously established a human CD4+ T cell latency model in which the lymphoid cell lines harbor a single copy of a latent HIV-1 provirus containing two marker genes that can be used for screening (27). The marker gene secretable alkaline phosphatase (seap) provides a highly sensitive chemiluminescent assay amenable for HTS and, given its position in the viral genome, serves as an indicator of late gene expression (Fig. 1A). These cells, designated 24STNLSG, have a low background of SEAP activity and a high degree of inducibility, which provided excellent assay reliability in a 96-well format as reflected by the Z-factor value (27, 30). It should be noted that the Z-factor is a commonly used measurement of assay performance and reliability, and a score between 0.5 and 1.0 is an indication of an excellent assay. We used the 24STNLSG cell line to establish optimal conditions for latent HIV-1 activation in a 384-well format and obtained a Z-factor of 0.7 when TNFα was used as an activator (Fig. 1B). The excellent assay reliability proved that the system could be utilized in HTS of a small molecule library to identify HIV-1 latency activators. Initially, a validation screening was performed to test the latency system in an HTS format run against a representative set of 3,300 compounds. During the validation screen, 10 plates were tested where each plate carried 64 wells dedicated to negative (0.5% DMSO-treated) and positive (TNFα-induced) controls, with the latter in serial dilution. The screening was carried out at 7.5 μm for each compound, and the SEAP activity was measured 24 h after treatment and resulted in three hits. The low hit rate is not unexpected for HTS to identify activators where usually the highly cytotoxic compounds that contribute to the false positive hits in inhibition assays are eliminated. Another reason for the low hit rate from the HIV-1 latency validation screen could be the short time of compound exposure, which selected only for the strongest and fastest acting inducers of late viral gene expression.
FIGURE 1.
A, schematic representation of the transducing vector, pNE−S-G, used to establish the cell-based model of latent HIV-1, 24STNLSG. Partial deletions of the viral genes are shown by arrowheads. LTR, long terminal repeats; ψ, encapsidation signal; rre, Rev protein recognition element; egfp, enhanced green fluorescent protein. B, Z-factor of the cell-based latency model in 384-well plate format. 24STNLSG (3 × 104/well) cells were seeded in 384-well plates, and both untreated and 2.25 μm TNFα-induced cells were assayed for SEAP activity after 24 h. RLU, relative light units. C, identification of primary hits from the HTS of a small molecule library. The x axis represents the compound identification number. The chart shows the results from eight plates.
Identification of three compounds from the validation screen that could reactivate the latent provirus demonstrated that the system could be scaled up for full HTS. Therefore, HTS was carried out using a chemical library of 200,000 compounds. This chemical library is an accurate reflection of the chemical diversity that is obtainable, and each individual compound in the screening library has at least 25 structurally related “neighbors” that, although not in the library, may also be acquired and examined in hit follow-up. The compounds ultimately chosen for inclusion into the library have also been evaluated to reduce redundancy of scaffold types while permitting rapid analog identification, have been prescreened for pharmacological potential through the elimination of known toxophoric moieties, and have been reviewed to ensure that the collection possessed many of the favorable characteristics of known orally active drugs. The screening process was similar to that of the validation assay, but the compound exposure was extended to 48 h. Fig. 1C represents the varied intensity of SEAP induction by some of the primary hits. HTS hits with statistically significant activity were defined as those compounds that increased reporter gene expression three standard deviations or more from the mean of the assay run. Generally, the cutoff that was used for up-regulation of gene expression was about 2-fold. With a 5-fold cutoff from the mean value of the no-compound controls, a total of 172 primary hits were identified, resulting in a hit rate of ∼0.1%. The primary hits were tested again in a 384-well format assay in a two-point dose response in duplicate. A total of 27 hits were reconfirmed to induce SEAP activity in 24STNLSG cells with a 2-fold cutoff above the background signal. Among those hit compounds, some demonstrated a higher selective index (CC50/EC50), and those reaching 25 hits will be used in consequent steps of chemical modifications aimed at improved pharmacokinetics.
Latent HIV-1 Can Be Reactivated by an HTS Hit Compound Independently of the Site of Provirus Integration
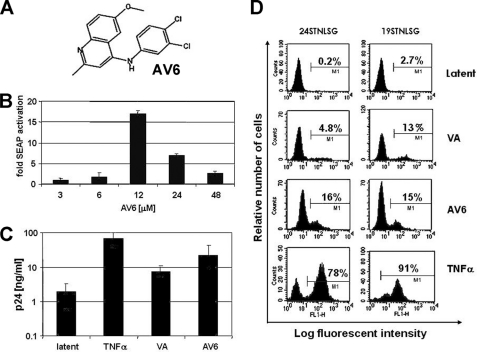
The compound designated AV6 consistently induced HIV-1 production from latently infected cell lines and was chosen from the pool of confirmed HTS hits for more detailed analysis (Fig. 2A). AV6 is a 4-3′,4′-dichloroanilino-6-methoxyquinoline compound. In 24STNLSG cells, AV6 produced dose-dependent activation of latent provirus with maximum induction at 12 μm and activity decreasing at higher concentrations, probably due to increasing cytotoxicity (Fig. 2B and data not shown). Although significantly less potent than TNFα, AV6 achieved a maximal level of induction that was 2–3-fold greater than that produced by optimal concentrations of the HDAC inhibitor VA. This effect was manifested both in 24STNLSG cells and in the ACH-2 model of latency, the latter harboring a replication-competent provirus (31) (Fig. 2C). A quantitative analysis of virus reactivation by AV6 was performed, monitoring GFP production via flow cytometry. As shown in Fig. 2D, the compound was equally efficient at virus activation in two different SupT1-derived clonal cell lines each harboring latent NE−S-G virus integrated at different genomic locations (27), indicating that activation was independent of the site of provirus integration (Figs. 1A and 2D). Taken together, the results obtained from our studies show that AV6 reproducibly acts as an HIV-1 latency antagonist in different model systems.
FIGURE 2.
Secondary assays for latent virus reactivation by AV6. A, the chemical structure of AV6. B, a dose dependence study was performed with AV6. 24STNLSG cells were treated for 48 h in the presence of AV6, at the indicated concentrations in triplicate. The data are expressed as the -fold activation relative to 0.5% DMSO-treated negative control. C, ACH-2 cells were exposed to TNFα (50 ng/ml), VA (1 mm), or AV6 (8 μm) in a 96-well plate for 24 h. Production of HIV-1 Gag p24 protein was measured via ELISA. latent represents an untreated control. Results shown are the means and S.D. of one of two representative experiments. D, the percentage of reactivated 24STNLSG or 19STNLSG cells was evaluated by flow cytometry 48 h after treatment with TNFα (50 ng/ml), VA (1 mm), or AV6 (8 μm).
AV6 Activated Latent HIV in Primary Cells without Inducing Cell Proliferation
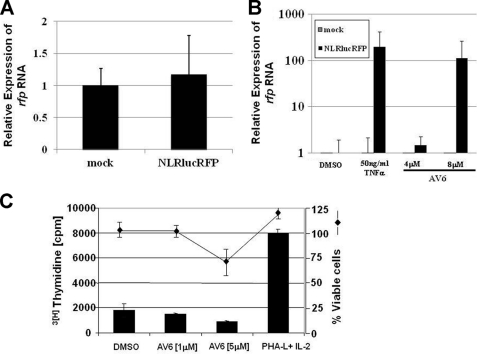
To assay the efficacy of AV6 in primary, infected cells, we utilized resting primary CD4+ T cells infected with the NLRlucRFP, an HIV-1-based vector that contains expression cassettes for Renilla luciferase (rluc) and red fluorescent protein (rfp) (16). This latency system allows one to monitor activation of latent virus using autologous, relevant primary cells. Although researchers do employ patient samples, when doing so, MHC mismatched CD4+ T cells are typically added to the cultures to allow expansion of the virus, so it is unclear whether allogeneic reactivity might contribute to virus activation. Viral particles enveloped with the CXCR4-tropic envelope integrate into the genomes of resting CD4+ T cells and result in a latent infection, as early as 3 days after infection, which can be reactivated by various stimuli including IL-7 and CD3/CD28 stimulation (32–35). PBMCs from whole blood were isolated by Ficoll gradient centrifugation and then subjected to negative selection to isolate resting CD4+ T cells. The cells were mixed with viral supernatant containing PEG-concentrated NLRlucRFP vector virus, displaying the CXCR4-tropic envelope of the HXB2 strain, and spinoculated for 2 h. Three days after infection, the cells were treated with 50 ng/ml TNFα or AV6 at concentrations of 4 or 8 μm for 24 h. Total RNA was extracted from the cells, and the level of rfp RNA was measured by qRT-PCR. The levels of rfp expression in untreated mock-infected cells and untreated NLRlucRFP-infected cells were comparable, showing that the population of resting CD4+ T cells harbors latent virus (Fig. 3A). Comparing the level of rfp mRNA in TNFα- or 8 μm AV6-treated cells with untreated cells revealed approximately a 250- and a 100-fold induction of viral expression, respectively (Fig. 3B). This demonstrates that AV6 activates latent virus in primary cells of a relevant cell type.
FIGURE 3.
AV6 induces latent virus RNA from primary resting CD4+ T cells. Resting CD4+ T cells were isolated from donors and infected with a CXCR4-tropic NLRlucRFP virus. Cells were treated as indicated, and the level of rfp RNA was measured by qRT-PCR. A, level of rfp mRNA in untreated-NLRlucRFP infected cells relative to untreated mock-infected cells. The results were normalized to the expression of β-actin. B, level of rfp mRNA in compound-treated mock- or NLRlucRFP-infected cells relative to DMSO-treated cells. C, effect of AV6 on PBMC proliferation and cell survival. PBMCs isolated from healthy blood donors were cultured in the presence of the indicated concentrations of AV6, 0.2% DMSO, or stimulation provided by PHA-L (4 μg/ml) plus IL-2 (20 units/ml). At day 2 after treatment, cells were pulsed with tritium-labeled thymidine for 8 h and subjected to analysis. Data from triplicate wells from a representative of three experiments are shown. The percentage of viable cells was determined via CellTiterGlo reagent (Promega) where the total ATP pool from cells exposed to different compound concentrations was compared with the ATP amount from cells treated with 0.2% DMSO. The means and S.D. were obtained from three independent experiments.
It has been reported that T cell proliferation simultaneously induces latent HIV (4, 6, 7, 36, 37). To test whether AV6 activated latent virus as a result of host cell proliferation, we performed a set of experiments monitoring de novo DNA synthesis or expression of activation markers in PBMC isolated from healthy blood donors. The experiments were conducted on samples isolated from three different blood donors to examine compound effect upon lymphocytes with varied genetic background and activation profiles. PBMCs were treated with AV6 or phytohaemagglutinin-L (PHA-L) plus IL-2 for 2 days. Data shown in Fig. 3C and Table 1 demonstrate that AV6 did not stimulate cell cycle progression in PBMC and did not alter the expression of activation markers of CD4+ T cells.
TABLE 1.
Expression of activation markers in primary CD4+ T-cells
| Untreated | PHA-L + IL-2 | AV6 | |
|---|---|---|---|
| % | % | % | |
| Ki-67 | 0.1 | 17 | 0.08 |
| CD69 | 1 | 58 | 0.9 |
| HLA-DR | 0.7 | 16 | 0.8 |
AV6 Requires NFAT for Early Viral Gene Expression
Because histone remodeling has a crucial role in regulating HIV promoter activity, there is a growing interest in using HDAC inhibitors in the clinical setting as part of a treatment strategy, along with the use of ART, to eradicate HIV infection. Therefore, we tested whether AV6 affected HDAC activity as compared with the class I and II HDAC inhibitors trichostatin A and VA, which preferentially inhibit class I HDAC (38). As shown in Fig. 4A, AV6 did not inhibit substrate deacetylation by the nuclear proteins. These data indicated that the compound could activate viral gene expression via a different mechanism than that of HDAC inhibitors.
AV6- and VA-treated 24STNLSG cells displayed a difference in the kinetics of viral transcript accumulation. Increased levels of HIV mRNA could be detected shortly after AV6 exposure similar to TNFα treatment in contrast to VA activation, which needed a prolonged time of cell exposure (Fig. 4B). Chromatin immunoprecipitation (ChIP) was performed to examine whether binding of transcription factors, previously reported to be important for HIV expression, to the viral promoter was affected by AV6. As seen, NFAT binding to the promoter was significantly up-regulated after treatment with AV6 (Fig. 4C). If NFAT binding is essential for early viral mRNA expression, an inhibitor of NFAT activity such as VIVIT, which interacts with NFAT, impeding binding to its target DNA transcriptional motif (39), should block early viral gene expression in the presence of AV6. VIVIT did block AV6-dependent mRNA expression (Fig. 4D). It would be anticipated that an inhibitor such as FK506, a calcineurin inhibitor, might also block AV6-dependent viral expression because NFAT function is regulated by calcineurin (40). It was found that FK506 also blocked viral mRNA expression in AV6-treated cells. These results suggest that NFAT is required for AV6 up-regulation of viral gene expression.
Given the early effect of AV6 upon viral gene expression, an experiment was performed to examine whether AV6 affected viral RNA stability, and an experiment was performed in which the activity of RNA polymerase II was inhibited by pretreatment of 24STNLSG cells with actinomycin D followed by incubation with AV6. Samples were collected at the indicated times, and the relative RNA ratio was calculated via qRT-PCR (Fig. 4E). Compound AV6 was able to stabilize the viral RNA levels, extending the half-life by several hours. As a control, the effect of AV6 upon the stability of the housekeeping gene eukaryotic translation elongation factor 1 α1 (EEF1A1) mRNA was determined. AV6 did not stabilize EEF1A1 mRNA (Fig. 4E). However, additional experiments suggested that the stabilizing effects of AV6 might not be specific for HIV-1 RNA because AV6 also stabilized β-actin mRNA (data not shown).
The Novel HIV Latency Activator Cooperates with VA for Enhanced Expression of the Viral Genome
Although VA was shown to activate latent virus in tissue culture, the compound did not have an appreciable effect upon the latent reservoirs of patients (12, 41, 42). Because our preliminary data suggested that AV6 acts at least in part at a posttranscriptional level, which differentiates it from the mechanism of action of VA, we were interested in testing whether the combined treatment with VA, known to activate transcription from the viral promoter, would have an additive effect. AV6 demonstrated cooperation with VA for enhanced activation of latent HIV-1 regardless of the proviral site of integration (Fig. 5, A and B, as compared with Fig. 2D). A marked increase of the genomic unspliced viral RNA species (Gag) was achieved with the combined action of AV6 and VA (Fig. 5C). It is noteworthy that the viral RNA levels were normalized to β-actin mRNA levels, which as noted above are stabilized by AV6, suggesting that the levels of viral RNA induction are being underestimated (Figs. 4B and 5C), yet there is still a measureable increase in viral RNA, probably because of positive feedback from Tat expression. Collectively, these data demonstrated that inducers targeting different aspects of HIV-1 replication could be combined for a greater stimulation of viral gene expression.
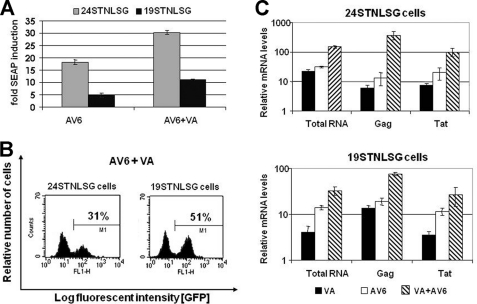
FIGURE 5.
AV6 cooperates with the HDAC inhibitor VA for enhanced induction of the viral gene expression. 24STNLSG and 19STNLSG cells were treated with VA (1 mm), with AV6 (8 μm) alone, or in combination for 24 h. A, the -fold SEAP induction was calculated as a ratio of relative light units obtained from compound-treated cells versus mock-treated (0.5% DMSO) latent cells. The means and S.D. for the relative light units were obtained from three independent experiments. B, the percentage of GFP-expressing cells was determined via flow cytometry. C, qRT-PCR was performed with RNA isolated from cells 24 h after stimulation with AV6 (8 μm) alone or in combination with VA (1 mm). The indicated regions were analyzed. The results were normalized to the expression of β-actin. The data from three representative experiments are shown.
DISCUSSION
It is possible that ART can be combined with a novel class of agents capable of specific reactivation of the latent provirus to eliminate infected cells through the cytopathic effects of actively replicating HIV combined with immune clearance. To initiate the discovery of new antiviral compounds, we developed a cell-based assay that models HIV latency and is amenable to HTS (27). Our latency system was adapted for a 384-well plate format assay yielding excellent reliability, as reflected by the Z-factor values, and was utilized in HTS of a small molecule library of 200,000 compounds. A total of 27 compounds were identified that reproducibly activate latent HIV in cell-based model systems. The low hit rate was expected given that the marker reactivation assay, in contrast to assays for inhibitors, by definition eliminates many false positive hits resulting from cell toxicity. A hit rate of 0.1% reflected a robust and highly selective model of HIV latency that identifies only the most effective activator molecules.
Secondary assays for latent virus reactivation demonstrated that the confirmed HTS hit compounds, as represented by AV6, were equally efficient on alternative cell-based models of HIV-1 latency (Fig. 2). Additionally, AV6 was capable of stimulating virus outgrowth from primary resting CD4+ T cells. Importantly, no alteration of the proliferative status of primary lymphocytes exposed to AV6 was observed because this would likely lead to unacceptable toxic side effects in patients (Fig. 3C and Table 1). Such data provide a compelling demonstration that an HIV-1 latency model based on a lymphoid cell line can be used as a tool for identifying agents capable of activating latent virus from quiescent CD4+ T cells.
There is a good chance that using small molecules to purge latent virus will require the use of two or more inducers of HIV gene expression acting in accord via different mechanisms. First, we were interested in characterizing some of the molecular effects of AV6 to determine what class of molecules it might complement. Some HDAC inhibitors, used in clinical therapies of cancer or neurological disorders, have been shown to stimulate latent HIV (14–16, 43). Thus, we examined whether AV6 also acted as an HDAC inhibitor and found that it did not (Fig. 4A). In addition, a time course was performed to examine HIV-1 RNA expression after AV6 treatment, and it was found that there was a significant increase in viral RNA levels as early as 2 h after treatment (Fig. 4B). One would anticipate that many HIV-1 latency antagonists would exert their effects through transcription initiation. Thus, we monitored whether AV6 affected binding of transcription factors, known to mediate HIV-1 transcription, to the viral promoter. It was found that early after AV6 treatment, NFAT binding was significantly up-regulated (Fig. 4C). Treatment with inhibitors of NFAT action indicated that NFAT is essential for AV6-mediated early viral gene expression (Fig. 4D). The results indicate that AV6 is acting rapidly upon transcription initiation, creating a positive feedback loop with the early viral genes including tat and rev. Interestingly, Bosque and Planelles (44)developed a primary CD4+ T cell model mimicking HIV-1 latency in memory cells and found that NFAT activation was needed for efficient reactivation of latent virus, whereas NFκB was not required. These results suggested that AV6 might act cooperatively with an HDAC inhibitor because its mode of activation is distinct from this class of molecule. Latently infected cells were co-treated with AV6 and VA, which is a class I HDAC inhibitor and a relatively weak virus activator. A strong cooperative effect was observed using the combination of AV6 and VA (Fig. 5). This result leads to the speculation that in vivo therapy, which combines two or more inducers of HIV gene expression acting in accord via different mechanisms, may produce a potent reactivation effect.
Surprisingly, it was also found that AV6 affected the half-life of HIV-1 RNA (Fig. 4E). AV6 treatment resulted in extending the RNA half-life, albeit not exclusively HIV RNA. It is unclear at this juncture whether effects upon RNA stability are essential for the latency antagonist activity of AV6 as is the case for NFAT. Although not within the scope of this study, it will be of interest to determine the specific binding partners of AV6 and to establish whether there is a common pathway affecting both NFAT binding and viral mRNA stability or whether these constitute separate pathways individually affected by AV6.
In summary, we identified compounds that are able to reactivate virus in different latency model systems including primary resting CD4+ T cells and a cell-based assay that models HIV latency in an HTS. These findings strongly support HTS as a promising approach for the identification of novel agents with alternative and possibly more specific therapeutic characteristics for eliminating HIV persistence in the future.
This work was supported, in whole or in part, by National Institutes of Health Grants AI070039 and AI081307 (to J. P. D.). J. D. G., J. B., Z. G., and S. W. P. declare competing financial interests.
- ART
- antiretroviral therapy
- HTS
- high-throughput screen
- AV6
- antiviral 6
- VA
- valproic acid
- PBMC
- peripheral blood mononuclear cells
- SEAP
- secretable alkaline phosphatase
- EEF1A1
- elongation factor 1 α1
- NFAT
- nuclear factor of activated T-cells
- qRT-PCR
- quantitative real-time RT-PCR
- DMSO
- dimethyl sulfoxide
- 6FAM
- 6-carboxyfluorescein.
REFERENCES
- 1. Chun T. W., Davey R. T., Jr., Ostrowski M., Shawn Justement J., Engel D., Mullins J. I., Fauci A. S. (2000) Nat. Med. 6, 757–761 [DOI] [PubMed] [Google Scholar]
- 2. Davey R. T., Jr., Bhat N., Yoder C., Chun T. W., Metcalf J. A., Dewar R., Natarajan V., Lempicki R. A., Adelsberger J. W., Miller K. D., Kovacs J. A., Polis M. A., Walker R. E., Falloon J., Masur H., Gee D., Baseler M., Dimitrov D. S., Fauci A. S., Lane H. C. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 15109–15114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chun T. W., Stuyver L., Mizell S. B., Ehler L. A., Mican J. A., Baseler M., Lloyd A. L., Nowak M. A., Fauci A. S. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 13193–13197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Finzi D., Hermankova M., Pierson T., Carruth L. M., Buck C., Chaisson R. E., Quinn T. C., Chadwick K., Margolick J., Brookmeyer R., Gallant J., Markowitz M., Ho D. D., Richman D. D., Siliciano R. F. (1997) Science 278, 1295–1300 [DOI] [PubMed] [Google Scholar]
- 5. Fondere J. M., Petitjean G., Huguet M. F., Salhi S. L., Baillat V., Macura-Biegun A., Becquart P., Reynes J., Vendrell J. P. (2004) J. Virol. 78, 10536–10542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chun T. W., Engel D., Mizell S. B., Hallahan C. W., Fischette M., Park S., Davey R. T., Jr., Dybul M., Kovacs J. A., Metcalf J. A., Mican J. M., Berrey M. M., Corey L., Lane H. C., Fauci A. S. (1999) Nat. Med. 5, 651–655 [DOI] [PubMed] [Google Scholar]
- 7. Dybul M., Hidalgo B., Chun T. W., Belson M., Migueles S. A., Justement J. S., Herpin B., Perry C., Hallahan C. W., Davey R. T., Metcalf J. A., Connors M., Fauci A. S. (2002) J. Infect. Dis. 185, 61–68 [DOI] [PubMed] [Google Scholar]
- 8. Prins J. M., Jurriaans S., van Praag R. M., Blaak H., van Rij R., Schellekens P. T., ten Berge I. J., Yong S. L., Fox C. H., Roos M. T., de Wolf F., Goudsmit J., Schuitemaker H., Lange J. M. (1999) AIDS 13, 2405–2410 [DOI] [PubMed] [Google Scholar]
- 9. Stellbrink H. J., van Lunzen J., Westby M., O'Sullivan E., Schneider C., Adam A., Weitner L., Kuhlmann B., Hoffmann C., Fenske S., Aries P. S., Degen O., Eggers C., Petersen H., Haag F., Horst H. A., Dalhoff K., Möcklinghoff C., Cammack N., Tenner-Racz K., Racz P. (2002) AIDS 16, 1479–1487 [DOI] [PubMed] [Google Scholar]
- 10. van Praag R. M., Prins J. M., Roos M. T., Schellekens P. T., Ten Berge I. J., Yong S. L., Schuitemaker H., Eerenberg A. J., Jurriaans S., de Wolf F., Fox C. H., Goudsmit J., Miedema F., Lange J. M. (2001) J. Clin. Immunol. 21, 218–226 [DOI] [PubMed] [Google Scholar]
- 11. Lehrman G., Hogue I. B., Palmer S., Jennings C., Spina C. A., Wiegand A., Landay A. L., Coombs R. W., Richman D. D., Mellors J. W., Coffin J. M., Bosch R. J., Margolis D. M. (2005) Lancet 366, 549–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Siliciano J. D., Lai J., Callender M., Pitt E., Zhang H., Margolick J. B., Gallant J. E., Cofrancesco J., Jr., Moore R. D., Gange S. J., Siliciano R. F. (2007) J. Infect. Dis. 195, 833–836 [DOI] [PubMed] [Google Scholar]
- 13. Steel A., Clark S., Teo I., Shaunak S., Nelson M., Gazzard B., Kelleher P. (2006) AIDS 20, 1681–1682 [DOI] [PubMed] [Google Scholar]
- 14. Archin N. M., Espeseth A., Parker D., Cheema M., Hazuda D., Margolis D. M. (2009) AIDS Res. Hum. Retroviruses 25, 207–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Contreras X., Schweneker M., Chen C. S., McCune J. M., Deeks S. G., Martin J., Peterlin B. M. (2009) J. Biol. Chem. 284, 6782–6789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Edelstein L. C., Micheva-Viteva S., Phelan B. D., Dougherty J. P. (2009) AIDS Res. Hum. Retroviruses 25, 883–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Krishnan V., Zeichner S. L. (2004) J. Virol. 78, 9458–9473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Brooks D. G., Arlen P. A., Gao L., Kitchen C. M., Zack J. A. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 12955–12960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Korin Y. D., Brooks D. G., Brown S., Korotzer A., Zack J. A. (2002) J. Virol. 76, 8118–8123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rullas J., Bermejo M., García-Pérez J., Beltán M., González N., Hezareh M., Brown S. J., Alcamí J. (2004) Antivir. Ther. 9, 545–554 [PubMed] [Google Scholar]
- 21. Williams S. A., Chen L. F., Kwon H., Fenard D., Bisgrove D., Verdin E., Greene W. C. (2004) J. Biol. Chem. 279, 42008–42017 [DOI] [PubMed] [Google Scholar]
- 22. Greten F. R., Karin M. (2004) Cancer Lett. 206, 193–199 [DOI] [PubMed] [Google Scholar]
- 23. Karin M. (2006) Nature 441, 431–436 [DOI] [PubMed] [Google Scholar]
- 24. Pikarsky E., Porat R. M., Stein I., Abramovitch R., Amit S., Kasem S., Gutkovich-Pyest E., Urieli-Shoval S., Galun E., Ben-Neriah Y. (2004) Nature 431, 461–466 [DOI] [PubMed] [Google Scholar]
- 25. Kucharczak J., Simmons M. J., Fan Y., Gélinas C. (2003) Oncogene 22, 8961–8982 [DOI] [PubMed] [Google Scholar]
- 26. Yang H. C., Xing S., Shan L., O'Connell K., Dinoso J., Shen A., Zhou Y., Shrum C. K., Han Y., Liu J. O., Zhang H., Margolick J. B., Siliciano R. F. (2009) J. Clin. Invest. 119, 3473–3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Micheva-Viteva S., Pacchia A. L., Ron Y., Peltz S. W., Dougherty J. P. (2005) Antimicrob. Agents Chemother. 49, 5185–5188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ahlberg C. (1999) Drug Discov. Today 4, 370–376 [DOI] [PubMed] [Google Scholar]
- 29. Naldini L., Blömer U., Gallay P., Ory D., Mulligan R., Gage F. H., Verma I. M., Trono D. (1996) Science 272, 263–267 [DOI] [PubMed] [Google Scholar]
- 30. Zhang J. H., Chung T. D., Oldenburg K. R. (1999) J. Biomol. Screen. 4, 67–73 [DOI] [PubMed] [Google Scholar]
- 31. Duh E. J., Maury W. J., Folks T. M., Fauci A. S., Rabson A. B. (1989) Proc. Natl. Acad. Sci. U.S.A. 86, 5974–5978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Agosto L. M., Yu J. J., Dai J., Kaletsky R., Monie D., O'Doherty U. (2007) Virology 368, 60–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Agosto L. M., Yu J. J., Liszewski M. K., Baytop C., Korokhov N., Humeau L. M., O'Doherty U. (2009) J. Virol. 83, 8153–8162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Swiggard W. J., Baytop C., Yu J. J., Dai J., Li C., Schretzenmair R., Theodosopoulos T., O'Doherty U. (2005) J. Virol. 79, 14179–14188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yu D., Wang W., Yoder A., Spear M., Wu Y., Farzan M. (2009) PLoS Pathog. 5, e1000633–e1000633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hermankova M., Siliciano J. D., Zhou Y., Monie D., Chadwick K., Margolick J. B., Quinn T. C., Siliciano R. F. (2003) J. Virol. 77, 7383–7392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Williams S. A., Kwon H., Chen L. F., Greene W. C. (2007) J. Virol. 81, 6043–6056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Göttlicher M., Minucci S., Zhu P., Krämer O. H., Schimpf A., Giavara S., Sleeman J. P., Lo Coco F., Nervi C., Pelicci P. G., Heinzel T. (2001) EMBO J. 20, 6969–6978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Noguchi H., Matsushita M., Okitsu T., Moriwaki A., Tomizawa K., Kang S., Li S. T., Kobayashi N., Matsumoto S., Tanaka K., Tanaka N., Matsui H. (2004) Nat. Med. 10, 305–309 [DOI] [PubMed] [Google Scholar]
- 40. Jain J., McCaffrey P. G., Miner Z., Kerppola T. K., Lambert J. N., Verdine G. L., Curran T., Rao A. (1993) Nature 365, 352–355 [DOI] [PubMed] [Google Scholar]
- 41. Archin N. M., Cheema M., Parker D., Wiegand A., Bosch R. J., Coffin J. M., Eron J., Cohen M., Margolis D. M. (2010) PLoS ONE 5, e9390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Archin N. M., Eron J. J., Palmer S., Hartmann-Duff A., Martinson J. A., Wiegand A., Bandarenko N., Schmitz J. L., Bosch R. J., Landay A. L., Coffin J. M., Margolis D. M. (2008) AIDS 22, 1131–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ylisastigui L., Archin N. M., Lehrman G., Bosch R. J., Margolis D. M. (2004) AIDS 18, 1101–1108 [DOI] [PubMed] [Google Scholar]
- 44. Bosque A., Planelles V. (2009) Blood 113, 58–65 [DOI] [PMC free article] [PubMed] [Google Scholar]