Abstract
Investigation of helper T cell markers in HTLV-1-transformed cell lines demonstrated that HuT-102 has an IL-9-producing Th17 phenotype. We confirmed the vital role of retinoic acid-related orphan receptor C, a Th17 transcription factor, in the expression of IL-17. Interferon regulatory factor 4 (IRF4), a transcription factor overexpressed in all HTLV-1-infected cells, regulated IL-17 and IL-9 concomitantly. We further demonstrated a novel pathway for the regulation of Tax-induced cytokines, IL-9 and IL-6, through TAK1-mediated nuclear accumulation of c-Rel. A microarray analysis for IRF4 knocked down HuT-102 cells showed a significant up-regulation in the set of genes related to Th1, mainly IFN-γ and several transcription factors. T-bet and IRF1, but not STAT1 and IRF9, participated in counteracting the inhibitory effect of IRF4 on the production of IFN-γ. Finally, suppression of both IRF4 and c-Rel resulted in the reduced proliferation. Collectively, these findings indicate that TAK1-c-Rel and IRF4 pathways play distinct roles in the maintenance of IL-9-producing Th17 phenotype of HTLV-1-transformed cells.
Keywords: Interferon, Interleukin, NF-kB Transcription Factor, Signal Transduction, Viral Immunology, HTLV-1, IRF4, T Helper Cell
Introduction
Human T cell lymphotropic virus 1 (HTLV-1)2 infects 20 million people worldwide with 3% developing adult T cell leukemia (ATL), and a further 0.25–3% developing an inflammatory disease of the CNS known as HTLV-1-associated myelopathy/tropical spastic paraparesis (1, 2). ATL is an aggressive proliferation of mature activated CD4+ T cells, usually showing very poor prognosis for treatment (3, 4). Although the antiviral combination therapy with IFN-α and zidovudine (AZT) is considered a treatment for ATL, patients frequently suffer relapse. This relapse emphasizes the need for new therapeutic approaches and strategies.
Clonal expansions of HTLV-1 result from the expression of the viral transactivator protein Tax, which is thought to be a key molecule of ATL onset. Tax has many pathological functions such as virus replication, immortalization of host cells, and the activation of several transcriptional factors and signal transduction molecules (5–7). We also have shown previously Tax-dependent constitutive activation of TAK1-MAPK and TAK1-IRF3 pathways (8, 9).
IRF4, which is preferentially expressed in lymphoid cells, was first identified as a transcription factor that negatively regulates the activity of IFN-regulated genes and TLR signaling (10, 11). In 2007, Ramos et al. (12) showed that either IRF4 or c-Rel was overexpressed in antiviral-resistant ATL cells. On the other hand, IRF4 is reported to be emerging as a critical regulator of T-helper cell (Th) differentiation, playing an important role in both Th2 and Th17 development by controlling cytokine expression and apoptosis (13, 14).
Th1-, Th2-, and T regulatory cell-associated cytokines were shown previously to be detected in the serum from HTLV-1-infected patients (15). On the other hand, a study of T cells showed a close relationship between HTLV-1-associated myelopathy/tropical spastic paraparesis and both multiple sclerosis and experimental autoimmune encephalomyelitis lesions, which are also known as being pathological indicators for the presence of Th17 (16, 17). In a 2004 study, ATL cells were suggested to be derived from T regulatory cells after the detection of FOXP3 gene transcription in 47% of ATL cases (18). In the same year, one year before the proposal of Th17 as a new T helper lineage, Dodon et al. (19) showed that Tax induces IL-17 gene expression. From the previous data, it is clear that the phenotype for ATL is a matter of debate.
In this study, we managed to identify the T cell lineages involved in HTLV-1. Subsequently, we explored the role of both IRF4 and c-Rel in the expression of pivotal cytokines in this phenotype and proliferation. We found that IRF4 preferentially maintains the axis of IL-17–IL-9 production against IFN-γ production.
EXPERIMENTAL PROCEDURES
Antibodies and Reagents
Antibodies against IRF1, IRF3, IRF4, IRF9 (p48), p50, p52, p65, RelB, c-Rel, RORγt (RORC), STAT1, STAT2, proliferating cell nuclear antigen, lamin B, α-tubulin, and β-actin were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). STAT3, phospho-STAT1 (Tyr-701), phospho-STAT2 (Tyr-690), and phospho-p65 (Ser-536) antibodies were obtained from Cell Signaling Technology (Danvers, MA).
Cell Culture and Transfection
Jurkat and HTLV-1-transformed cells were cultured in RPMI 1640 supplemented with 10% FCS, 100 units/ml penicillin, and 100 μg/ml streptomycin at 37 °C in 5% CO2. HuT-102 cells were stably transfected with pSUPER.gfp_neo vectors (OligoEngine, Seattle, WA) to express shRNAs against human TAK1 or firefly luciferase, as described previously (9).
RNA Interference
Cells were transfected with siRNA using the Amaxa electroporation system. IRF1, IRF3, IRF4, IRF9, STAT1, c-Rel, RORC, Tax, and T-bet siRNAs were designed at and purchased from Invitrogen. Luc siRNA with a two-nucleotide overhanging at the 3′-end of the sequence was synthesized by Hokkaido System Science (Sapporo, Japan). The target sequences are summarized in supplemental Table S1.
Cell Proliferation Assay
HuT-102 cells transfected with siRNAs against Luc, IRF4, c-Rel, or both IRF4 and c-Rel were harvested. Viable cells were counted microscopically using trypan exclusion assay. The statistical significance of cell proliferation was calculated by performing Turkey-Kramer test, and p values < 0.01 were regarded as significant.
Immunoblotting
Whole cell lysates, cytoplasmic extracts, and nuclear extracts prepared as described previously (20), resolved by SDS-PAGE, and transferred to an Immobilon-P nylon membrane (Millipore, Bedford, MA). The membrane was treated with BlockAce (Dainippon Pharmaceutical Co. Ltd., Suita, Japan) overnight at 4 °C and probed with primary antibodies, as described above. Antibodies were detected using horseradish peroxidase-conjugated anti-rabbit, anti-mouse, anti-goat, and anti-sheep IgG (DakoCytomation, Glostrup, Denmark) and visualized with the ECL system (GE Healthcare).
Immunoprecipitation
Cell lysates prepared as described previously (21) were immunoprecipitated with anti-STAT1 antibody. The immunoprecipitates were immunoblotted as described above.
Plasmid DNA
pcDNA-IRF1 expression vector was kindly provided by Dr. Mark Perrella (Brigham and Women's Hospital, Boston, MA). Transfection was performed using the Amaxa electroporation system.
DNA Microarray
Total RNA was extracted from cells using RNAeasy Mini Kit (Qiagen, Valencia, CA). Gene expression was analyzed using a GeneChip® system with Human Genome Array U133 plus 2.0 (Affymetrix, Santa Clara, CA) as described previously (22). In this study, six arrays were used: two for HuT-siLuc cells, two for HuT-siIRF4 cells, and two for HuT-siIRF3 cells (positive counter control). A fold change value of >2 (up-regulated) or <0.5 (down-regulated) was considered to be biologically important. The statistical significance of the fold change was calculated for two groups by performing a Student's t test, and p values < 0.05 were regarded as significant. The microarray results were deposited in the GEO Database (accession no. 22036).
Real-time RT-PCR
Total RNAs was prepared using the RNeasy Mini kit (Qiagen). First-strand cDNA was synthesized by SuperScript II reverse transcriptase (Invitrogen). The cDNA was amplified quantitatively using SYBR Premix Ex Taq (Takara Bio, Otsu, Japan). The primer sequences are summarized in supplemental Table S2. Real-time quantitative RT-PCR was performed using a Prism 7300 sequence detection system (Applied Biosystems, Foster City, CA). All data were normalized to β-actin mRNA. The data shown are representative of at least three independent experiments.
ELISA
The DuoSet® ELISA development system for human IL-17 was purchased from R&D Systems. Briefly, each cell line (1 × 106 cells/ml) was cultured in RPMI1640 supplemented with 10% FCS, 100 units/ml penicillin, and 100 μg/ml streptomycin. Cells were left to reach confluency by incubating at 37 °C in 5% CO2. After centrifugation, supernatants were collected and used in the analysis as described by the manufacturer.
RESULTS
HuT-102 Is an IL-9-producing Th17 Phenotype
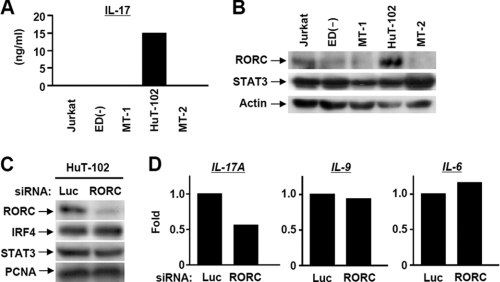
Based on previous data designating possible CD4+ phenotypes for HTLV-1-infected cells, we used two Tax-negative HTLV-1-infected cell lines (ED40515(−) and MT-1), and two Tax-positive cell lines (HuT-102 and MT-2) for a Th phenotype screening. Jurkat leukemic T cells were used as HTLV-1-free control lymphocytes. We performed real-time RT-PCR of the main cytokines and transcription factors involved in Th1, Th2, Th17, and T regulatory CD4+ cells. HuT-102 cells, in contrast to other cell lines, showed a characteristic phenotype of IL-9-producing Th17 cells. The results showed, similar to classic Th17 cells, high levels of IL-17 and RORC and moderate levels of STAT3 and IL-23. Of note, IL-9 and IL-6 were significantly expressed in HuT-102 as well as MT-2 Tax-positive cell lines. On the other hand, IRF4 was expressed highly in the HuT-102 as well as other cell lines (Table 1). As IL-17 and RORC are considered to be the hallmark cytokine and transcription factor, respectively, for the presence of Th17, we tended to confirm the RT-PCR data for both of them. We utilized an ELISA assay to estimate the absolute cytokine content of IL-17, showing high cytokine production by HuT-102 cells (15 ng/ml), whereas in contrast, other cell lines secreted slightly detectable amounts of IL-17 (48, 34, 23, and 35 pg/ml for Jurkat, ED, MT-1, and MT-2, respectively) (Fig. 1A). In agreement with this, HuT-102 cells displayed substantial expression of RORC, whereas STAT3 showed an almost identical pattern of expression among the four cell lines (Fig. 1B). On the other hand, we and others have previously confirmed the preferential high expression levels of IRF4 in all HTLV-1-infected cells (9). To further corroborate the crucial role of RORC, we transfected cells with a specific siRNA for RORC (Fig. 1C). The knockdown of RORC caused selective down-regulation of IL-17 expression (Fig. 1D). Meanwhile, IRF4, STAT3, IL-6, and IL-9 were unchanged (Fig. 1, C and D). These results confirmed the critical role of RORC in the maintenance of the HuT-102 Th17 phenotype.
TABLE 1.
Expression of genes involved in Th cell differentiation in HTLV-1-transformed T cell lines
Shown is the fold relative to Jurkat cells (*, relative to ED40515(-) cells because Jurkat cells did not express). −−−−, < 0.01; −−−, > 0.01–0.1; −−, > 0.1–0.25; -, > 0.25–0.67; ±, >0.67–1.5; +, > 1.5–4; ++, > 4–10; +++, > 10–100; ++++, > 100.
| Gene | Role in CD4 T cells | Tax negative |
Tax positive |
||
|---|---|---|---|---|---|
| ED40515(-) | MT-1 | HuT-102 | MT-2 | ||
| Th cell cytokines | |||||
| IFN-γ | Main Th1 cytokine | ± | −−−− | − | ++++ |
| IL-4 | Main Th2 cytokine | −−−− | −−−− | + | +++ |
| IL-6 | Induction of Th17 and Treg | −−− | + | ++++ | ++++ |
| IL-9 | Cytokine produced by Th17 and Th9 | − | −−−− | ++++ | ++++ |
| IL-17* | Main Th17 cytokine | ± | −−−− | ++++ | −−−− |
| IL-23 | Induction of Th17 | + | − | + | ± |
| TGF-β1 | Induction of Th17 and Treg | ± | + | −− | −− |
| Transcription factors | |||||
| FoxP3 | Treg master regulator | − | + | − | ++++ |
| GATA3 | Th2 master regulator | −− | − | −− | −− |
| IRF1 | Role in Th1 | − | −− | −− | ± |
| IRF4 | Differentiation of Th2 and Th17 | ++++ | ++++ | ++++ | ++++ |
| RORC | Th17 master regulator | + | — | ++ | −− |
| STAT1 | Differentiation of Th1 | + | + | + | ++ |
| STAT3 | Differentiation of Th17 | + | − | ± | + |
| T-bet | Th1 master regulator | +++ | − | + | +++ |
FIGURE 1.
HuT-102 cells show a Th17-like phenotype and respond to RORC knockdown. A, absolute concentration (ng/ml) of IL-17 in the supernatant collected from HTLV-1-transformed cell culture was determined by ELISA. B, protein expression of RORC, STAT3, and actin in HuT-102 cells was determined by Western blot. C, HuT-102 cells were transfected with siRNAs against RORC and Luc. At 60 h post-transfection, protein expression of RORC, IRF4, STAT3, and PCNA was determined by Western blotting. D, effects of RORC knockdown on the expression of IL-17A, IL-9, and IL-6 were examined by real-time RT-PCR.
IRF4 and c-Rel Differentially Regulated IL-17, IL-9, and IL-6 in HuT-102
Ramos et al. (12) reported previously that both IRF4 and c-Rel are expressed in ATL cells derived from antiviral-resistant patients. This fact, in addition to the reported role of IRF4 in Th17, raised our interest to investigate the role of IRF4 and c-Rel in HuT-102 cells. To this end, we transfected HuT-102 cells with siRNAs against IRF4 or c-Rel and confirmed the selective knockdown of the target proteins (Fig. 2, A and C). The knockdown of IRF4 caused both IL-17 and IL-9 down-regulation (Fig. 2B), whereas c-Rel knockdown caused both IL-9 and IL-6 down-regulation (Fig. 2D). Given the reported regulation of IL-6 and IL-9 by Tax in HTLV-1-infected cells (23, 24), one could argue that c-Rel regulation of IL-9 and IL-6 is done by regulating Tax. To rule out this possibility, we checked for the effect of c-Rel knockdown on Tax expression and demonstrated that c-Rel does not control Tax expression level (supplemental Fig. S1). In contrast, Tax knockdown caused the down-regulation of c-Rel (supplemental Fig. S1). Of note, we confirmed the down-regulation of IL-9 and IL-6 by Tax knockdown (supplemental Fig. S1).
FIGURE 2.
Distinct roles of IRF4 and c-Rel in the expression of IL-17, IL-9, and IL-6. A and C, effects of siRNAs against IRF4 (A) or c-Rel (C) on the protein expression of IRF4 and c-Rel by Western blot. B and D, effects of IRF4 (B) and c-Rel (D) siRNAs on the expression of IL-17A, IL-9, and IL-6 were examined by real-time PCR.
Role of TAK1 in c-Rel-dependent Regulation of IL-9 and IL-6
Several reports have previously pointed out the possible regulation of c-Rel by a Tax oncoprotein (25). But whether the regulation of IL-9 and IL-6 is a simple Tax/c-Rel regulatory pathway or whether it would involve other factors is still an unanswered question. Recently, our group has reported the Tax-dependent constitutive TAK1 activation in HTLV-1-infected cells (9). To this end, we managed to gain further insight into the possible involvement of TAK1 in the Tax/c-Rel pathway. In that context, we checked for the expression of IL-9 and IL-6 in HuT-102 cells stably transfected with a TAK1 shRNA vector (HuT-shTAK1 cells). As expected, TAK1 knockdown caused both IL-9 and IL-6 down-regulation (Fig. 3A). To uncover a possible link between TAK1 and subsequently c-Rel in regulating IL-9 and IL-6, we investigated the role of TAK1 in the control of NF-κB pathways. Our results showed slight down-regulation of total protein expression level for both p52 and c-Rel in response to TAK1 knockdown (Fig. 3B). We further elucidated this effect by fractionating the cells obtained on harvesting into cytoplasmic and nuclear extracts. We found that c-Rel and p100, the precursor of p52, were not changed in the cytoplasmic fractions, whereas nuclear fractions showed the significant down-regulation of both c-Rel and the active p52 (Fig. 3C). Collectively, it was clear that Tax regulates IL-9 and IL-6 through the TAK1/c-Rel pathway. To uncover whether IL-9, commonly regulated by IRF4 and c-Rel, is controlled by separate or common pathways, we used HuT-shTAK1 and control HuT-shLuc cells and further transfected them with siRNA for either luciferase or IRF4. We noticed that the knockdown of both TAK1 and IRF4 induced an additional reduction of IL-9 expression, demonstrating that IRF4 and TAK1/c-Rel might regulate IL-9 independently (Fig. 3D). Due to the important role of both IRF4 and c-Rel in regulating cytokine production, we checked for the effect of knockdown of both c-Rel and IRF4 on cell proliferation. Our results showed a significant reduction in proliferation of double knocked down cells (Fig. 3E).
FIGURE 3.
TAK1 is the upstream regulator of c-Rel and controls IL-9 independently from IRF4. A, the expression of TAK1, IL-17A, IL-9, and IL-6 in HuT-shTAK1 and HuT-shLuc cells were examined by real-time RT-PCR. B and C, whole cell lysates (B) or cytoplasmic/nuclear extracts (C) prepared from HuT-shLuc, and HuT-shTAK1 cells were immunoblotted with antibodies for NF-κB/Rel subunits. D, HuT-shLuc and HuT-shTAK1 cells were transfected with siRNAs for IRF4 or Luc. At 60 h post-transfection, IL-9 expression was examined by RT-PCR. E, effects of IRF4 and c-Rel siRNAs on cell proliferation was examined in a colorimetric WST-1 assay. Data are the mean ± S.D. of triplicate determinations. The statistical significance of differences between groups was calculated by applying Tukey-Kramer method of analysis. *, p < 0.01 was statistically significant.
IRF4 Knockdown Up-regulates Th1-related Genes
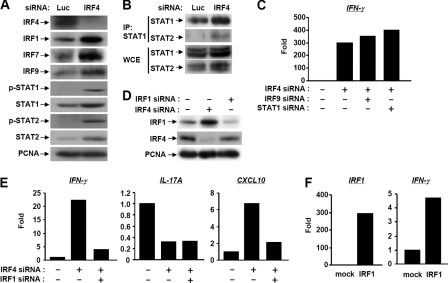
In addition to the significant role in regulating the two vital lineage-specific cytokines, IL-17 and IL-9, we have previously reported that IRF4 counteracted TAK1-IRF3-mediated expression of interferon-inducible genes (9). Therefore, to understand the comprehensive roles of IRF4 in HuT-102 cells, we performed microarray analysis for genome-wide screening of IRF4-regulated genes. We used IRF3 as a positive counter control to confirm the expected effect of IRF4 on interferon-inducible genes. The microarray showed the up-regulation of a set of important genes related to T helper cell development, especially Th1 (Table 2). Some of those genes were related directly to IFN-γ production, as IRF1, IL18RAP, Spp1, and other served as main Th1 regulators, namely, T-bet and STAT1 (26–29). We confirmed the microarray results for several important candidates in our study by real-time PCR (Table 2). On the other hand, the set of interferon-inducible genes were shown to be up-regulated by IRF4 knockdown and down-regulated by IRF3 knockdown (supplemental Fig. S2). To generalize our finding on other cell lines, we knocked down IRF4 in both the Tax-negative ED40515(−) and the Tax-positive MT-2 cell lines and found the same pattern of IFN-γ up-regulation as in HuT-102 cells (Fig. 4A). Moreover, we compared the expression of IFN-γ in HuT-102 cells transfected with siRNAs against IRF4, RORC, c-Rel, or luciferase control. The results demonstrated specific up-regulation of IFN-γ only after IRF4 knockdown (Fig. 4A). We also confirmed the effect of IRF4 knockdown in ED40515(−) cells by showing the up-regulation of both T-bet and IRF1, and the down-regulation of IL-9 (supplemental Fig. S3). Collectively, these results strongly confirm the selective role of IRF4 against IFN-γ even in the absence of high IL-17 production, as in the case of ED40515(−) or MT-2 cells. To assess the pivotal role of the Th1 cell-specific transcription factor T-bet on IFN-γ production, we displayed the effect of T-bet siRNA on the specific down-regulation of IFN-γ (Fig. 4B). T-bet knockdown alone, as expected, caused the down-regulation of basal expression of IFN-γ (supplemental Fig. S4).
TABLE 2.
Selected Th cell-related genes up-regulated by IRF4 siRNA
An asterisk indicates main IFN-γ related genes.
| Unigene | Gene name | Ratios (siIRF4/siLuc) |
|
|---|---|---|---|
| Microarray | RT-PCR | ||
| Chemokines/cytokines and their receptors | |||
| Hs.72918 | CCL1 | 4.58 | |
| Hs.75498 | CCL20 | 2.14 | |
| Hs.1349 | CSF2 | 6.44 | |
| Hs.78913 | CX3CR1 | 7.53 | |
| Hs.89690 | CXCL3 | 2.00 | |
| Hs.77367 | CXCL9 | 10.41 | |
| Hs.632586 | CXCL10 | 6.55 | 17.91 |
| Hs.632592 | CXCL11 | 8.98 | |
| Hs.856 | IFNG* | 48.92 | 195.91 |
| Hs.158315 | IL18RAP* | 2.21 | |
| Hs.635723 | IL7R | 36.04 | |
| Hs.654459 | TNFRSF9 | 2.16 | |
| Hs.478275 | TNFSF10 | 3.11 | |
| Hs.525157 | TNFSF13B | 4.22 | |
| Hs.546295 | XCL1 | 28.24 | |
| Signal transduction/transcription factors | |||
| Hs.436061 | IRF1* | 2.80 | 3.86 |
| Hs.166120 | IRF7 | 3.25 | |
| Hs.1706 | IRF9* | 2.39 | |
| Hs.656213 | JAK2 | 1.90 | |
| Hs.498570 | PRKCQ | 4.10 | |
| Hs.708051 | STAT1* | 2.44 | 2.57 |
| Hs.530595 | STAT2* | 2.52 | 2.50 |
| Hs.272409 | T-bet* | 2.48 | 1.77 |
| Others | |||
| Hs.576612 | ENTPD1 | 2.39 | |
| Hs.369039 | PHF11 | 4.73 | 4.44 |
| Hs.130759 | PLSCR1 | 2.30 | |
| Hs.313 | SPP1* | 2.44 | |
FIGURE 4.
IFN-γ is specifically controlled by IRF4. A, HuT-102, ED40515(−), and MT-2 cells were transfected with siRNAs against IRF4, c-Rel, RORC, and Luc. At 60 h post-transfection, IFN-γ mRNA expression was examined by RT-PCR. B, HuT-102 cells were transfected with siRNAs against IRF4 and T-bet. IRF4, T-bet, IFN-γ, and IL-17A mRNAs were quantified by real-time RT-PCR.
IRF1 Counteracts Effect of IRF4 on IFN-γ
Consistent with the microarray results; Western blotting showed an up-regulation of transcription factors related to IFN-γ production, including IRF1, IRF9, STAT1, and STAT2 (Fig. 5A). Due to the up-regulation of the set of genes that constitute IFN-stimulated gene factor 3 complex (IRF9, STAT1, and STAT2), we confirmed the formation of IFN-stimulated gene factor 3 complex using immunoprecipitation technique for STAT1. Our results clearly demonstrated the co-precipitation of STAT1 and STAT2 heterodimer and their up-regulation by IRF4 knockdown (Fig. 5B). Eventually, we showed that the use of either STAT1 or IRF9 siRNA concomitantly with IRF4 siRNA did not repress IFN-γ up-regulated by IRF4 knockdown (Fig. 5C). The results indicated that IFN-γ production is independent of STAT1 or STAT2.
FIGURE 5.
IRF4 control IFNγ by an independent pathway through IRF1. A, HuT-102 cells were transfected with IRF4 siRNA. The up-regulation of Th1-related proteins was confirmed by Western blot. B, formation of the STAT1-STAT2 heterodimer was confirmed by immunoprecipitation. Whole cell lysates (WCE) were immunoprecipitated with anti-STAT1 antibody, and then the immunoprecipitates (IP) were immunoblotted with anti-STAT1 and STAT2 antibodies. C, neither STAT1 nor IRF9 knockdown reversed the up-regulation of IFN-γ in response to IRF4 knockdown in HuT-102 cells. D and E, HuT-102 cells were transfected with IRF4 and IRF1 siRNAs. Protein expression of IRF1 and IRF4 was determined by Western blot (D). IFN-γ, CXCL10, and IL-17A mRNAs were quantified by real-time RT-PCR (E). F, HuT-102 cells were transfected with an empty vector (mock) or pcDNA-IRF1. At 60 h post-transfection, the expressions of IRF1 and IFN-γ mRNAs were examined by real-time RT-PCR.
Kano et al. (26) reported that IRF1 contributes to the IFN-γ-IL-12 signaling axis and Th1 versus Th17 differentiation of CD4+ T cells. The effect of IRF1 knockdown on down-regulation of basal IFN-γ expression was confirmed as shown in supplemental Fig. S4. We primarily confirmed that IRF4 knockdown induced IRF1 expression at both mRNA and protein levels (Table 2 and Fig. 5D). IRF1 did counteract the effect of IRF4 on both IFN-γ and CXCL10, but not IL-17, indicating that IRF4 controls IL-17 and IFN-γ independently (Fig. 5E). In addition, IRF1 overexpression induced the expression IFN-γ (Fig. 5F). We reported previously that interferon-inducible genes, including CXCL10, whose expression is maintained by the Tax-dependent constitutive activation of the TAK1-IRF3 pathway, is down-regulated by IRF4 in HuT-102 cells (9). To examine the role of IRF3 in IFN-γ production, we performed either single or double siRNA transfection for IRF4 and IRF3. The up-regulation of IFN-γ by IRF4 knockdown was not reversed by IRF3 knockdown, elucidating that the effect on IFN-γ is independent of constitutive activation of IRF3 (supplemental Fig. S2).
DISCUSSION
In this study, we described HuT-102 as an IL-9-producing Th17 phenotype using microarray, RT-PCR, and Western blot. Our evidence was based on the expression of IL-17, RORC, STAT3, and IRF4, in addition to the microarray, which highlighted the expression of CCR6, Ahr, BATF, IL-23R, and RORα (data not shown). On the other hand, other cell lines utilized in our comparative study did not show a clear phenotypic character. Nevertheless, MT-2 cells showed a sophisticated model, expressing a variety of genes related to totally divergent CD4+ phenotypes. This might raise an interest in the possibility of finding different CD4+ phenotypes in the future. The presentation of HuT-102 cells, in contrast to other used cell lines, with a clear T helper cell phenotype was a cornerstone for selecting HuT-102 cells mainly for our study. Moreover, being a Tax-positive cell line universalized the results obtained to be similar to fresh cells from ATL, which has the same morphological and biochemical phenotype as cells that express Tax (4). On the other hand, the main findings of the study were supported by experiments done on ED40515(−) and/or MT-2 cell lines to make our evidence stronger (Fig. 4A and supplemental Fig. S3). Low expression of Th1-specific transcription factors or cytokines was also shown in HuT-102 cells, raising the question for the extent of plasticity of T helper cells.
We confirmed the potential role of RORC in maintaining IL-17. Furthermore, we determined the role of IRF4 in controlling lineage specific cytokines IL-17 and IL-9, whereas c-Rel contributed to the regulation of IL-6 and IL-9. In Table 1, it appears that RORC, T-bet, Foxp3, in addition to IRF1 patterns of expression, are related either positively or negatively with the expression of both IL-17 and/or IFN-γ, and yet, according to our findings, we assume that IRF4 regulates at least both T-bet and IRF1 and subsequently IFN-γ. On the other hand, the regulation of IL-17 was not only confirmed by our knockdown experiments of IRF4 but rather was mentioned previously (13). Collectively, it is difficult to argue whether IRF4 directly controls cytokine expression or not. We would consider RORC as a master regulator of IL-17, whereas IRF4 might act as a gear that fine-tunes the cytokine profile either directly or through several intermediates such as IRF1 and T-bet. In other words, IRF4 can act as player within a multiplayer network controlling cytokine expression.
The double knockdown of IRF4 and c-Rel and their subsequent significance on reducing cell proliferation were further elucidated. One of the possibilities for this reduction in cell proliferation might be due to inducing apoptosis. Although c-Rel was shown to be required for transcriptional activation of IL-2 (31), it is unknown as to how c-Rel participates in the proliferation of HTLV-1-infected cells. We claim that c-Rel might have essential roles in controlling cell growth through regulating IL-6 and IL-9. On the other hand, another group has recently reported the essential role of IRF4 in the development of Th9 cells (32). This report further enhanced the impact of our findings regarding the possible role of IRF4 in the regulation of HTLV-1 pathogenicity. Altogether, IRF4 is now believed to be involved in the development of all currently known Th cell subsets through regulation of the hallmark cytokines IFN-γ, IL-17, IL-4, and recently, IL-9.
Tax protein was reported previously to induce the activation of canonical and non-canonical NF-κB/Rel pathways. In that study, Tax-induced constitutive activation of p65 was shown to subsequently activate the transcription of the c-Rel gene (33). Currently, we identify TAK1 as the upstream regulator of c-Rel; nevertheless, a role for TAK1 in p65 activation was not identified (8). Thus, we believe that the regulation of c-Rel and p52 by TAK1 involves a more complex mechanism that may identify a possible role for TAK1 in a non-canonical NF-κB pathway. On the other hand, we previously reported that TAK1 was involved in the regulation of IRF3 in addition to p38 and JNK MAPKs (8, 9). In this regard, we emphasized using siRNA transfection that IRF3 and MAPKs were not involved in inducing neither IL-9 nor IL-6 (data not shown).
In the current study, we reported for the first time in a Th17 phenotype that IRF4 knockdown causes up-regulation of Th1 transcription factors/cytokines. Most importantly, the high up-regulation of IFN-γ production was demonstrated in HuT-102 cells and similarly in other HTLV-1 cell lines after IRF4 knockdown. One of the major limitations of knockdown experiments using siRNA is the possible off-target effect. To rule out this limitation, we conducted experiments concurrently using two different IRF4 siRNA sequences. Both sequences specifically down-regulated IL-17 and ultimately up-regulated IFN-γ (data not shown). On the other hand, we also confirmed our main knockdown finding by performing IRF1 overexpression experiments that up-regulated IFN-γ. An interesting point for discussion was the extremely high up-regulation of IFN-γ in the ED40515(−) cell line, whereas the MT-2 cell line showed relatively low up-regulation compared with both HuT-102 and ED40515(−) cell lines. One possible explanation for this is the high basal expression level of IFN-γ in MT-2 cells. It is worth mentioning here that a model of HTLV-1 Tax-transgenic mice deficient in IFN-γ has enhanced tumorigenesis, highlighting the functionally important role of IFN-γ as a possible solution for HTLV-1 pathogenesis (34). We performed our study to show that the inhibition of IFN-γ by IRF4 is dependent on IRF1 but not IFN-stimulated gene factor 3 complex or IRF3. Presumably, we analyzed the IFN-inducible gene, CXCL10, to confirm the role of IRF1 in the IRF4-IFN-γ axis. This mechanistic finding can serve as an additional tuning factor for IFN-γ.
As a conclusion, we have shown that the modulatory effect of IRF4 knockdown in HuT-102 appears to be a “death by a thousand cuts” because numerous IRF4 target genes play crucial roles in the modulation of HuT-102 cells. Doors opened by the microarray in this study are worth further future investigation. The effect of IRF4 knockdown on the up-regulation of Th1 transcription factors and cytokines included a wide range of regulators, such as T-bet, IRF1, STAT1, PHF11, and others. From another point of view, an interesting set of chemokines, namely CXCL9, CXCL10, and CXCL11, which are also related to Th1 cell accumulation, were up-regulated in response to IRF4 knockdown (35, 36). On the other hand, the regulatory effect for IRF4 on IL-9 expression was confirmed by the concomitant expression of muc5ac, which is reported previously to be directly stimulated by IL-9 (37). Overall, the role of IRF4 in inducing a Th1 response could negatively regulate both Th17 and Th9.
The response of HTLV-1 to a combined therapy of IFN-α/AZT was a popular topic in several studies (12, 38–40). According to our findings, it seems that an approach for molecular targeting of IRF4 and c-Rel as well as with antiviral therapy can serve as a powerful treatment tool for AZT-resistant patients. As a matter of concern, it is worth mentioning that the phenotypes of IRF4-deficient mice are strictly limited to the immune system, including defects in the differentiation of plasma cells and certain dendritic cell subsets, as well as in lymphocyte activation. Notably, mice lacking one allele of IRF4 are phenotypically normal (41), yet an average of 50% knockdown of IRF4 mRNA and protein was sufficient to kill myeloma cell lines (30). Thus, a therapeutic window could exist in which IRF4-directed therapy would kill IRF4-addicted malignant cells while sparing normal cells.
Supplementary Material
Acknowledgment
We are grateful to Dr. M. Maeda for the generous gift of MT-1 cells.
This work was supported in part by Grants-in-aid for Scientific Research (C) 19590063 and Young Scientist Start-up 20890078 from the Ministry of Education, Culture, Sports, Science, and Technology (Japan) and a grant from the Takeda Science Foundation.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2 and Figs. S1–S4.
- HTLV-1
- human T cell lymphotropic virus type 1
- Th
- T helper cell
- TAK1
- transforming growth factor-β-activated kinase 1
- IRF
- IFN regulatory factor
- RORC
- retinoic acid-related orphan receptor C
- ATL
- adult T cell leukemia
- Luc
- luciferase.
REFERENCES
- 1. Osame M., Izumo S., Igata A., Matsumoto M., Matsumoto T., Sonoda S., Tara M., Shibata Y. (1986) Lancet. 2, 104–105 [DOI] [PubMed] [Google Scholar]
- 2. Gessain A., Barin F., Vernant J. C., Gout O., Maurs L., Calender A., de Thé G. (1985) Lancet. 2, 407–410 [DOI] [PubMed] [Google Scholar]
- 3. Hinuma Y., Komoda H., Chosa T. (1982) Int. J. Cancer. 29, 631–635 [DOI] [PubMed] [Google Scholar]
- 4. Bazarbachi A., Ghez D., Lepelletier Y., Nasr R., de Thé H., El-Sabban M. E., Hermine O. (2004) Lancet. Oncol. 5, 664–672 [DOI] [PubMed] [Google Scholar]
- 5. Grassmann R., Aboud M., Jeang K. T. (2005) Oncogene 24, 5976–5985 [DOI] [PubMed] [Google Scholar]
- 6. Sun S. C., Yamaoka S. (2005) Oncogene 24, 5952–5964 [DOI] [PubMed] [Google Scholar]
- 7. Matsuoka M., Jeang K. T. (2007) Nat. Rev. Cancer 7, 270–280 [DOI] [PubMed] [Google Scholar]
- 8. Suzuki S., Singhirunnusorn P., Mori A., Yamaoka S., Kitajima I., Saiki I., Sakurai H. (2007) J. Biol. Chem. 282, 25177–25181 [DOI] [PubMed] [Google Scholar]
- 9. Suzuki S., Zhou Y., Refaat A., Takasaki I., Koizumi K., Yamaoka S., Tabuchi Y., Saiki I., Sakurai H. (2010) J. Biol. Chem. 285, 4441–4446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yamagata T., Nishida J., Tanaka S., Sakai R., Mitani K., Yoshida M., Taniguchi T., Yazaki Y., Hirai H. (1996) Mol. Cell. Biol. 16, 1283–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Negishi H., Ohba Y., Yanai H., Takaoka A., Honma K., Yui K., Matsuyama T., Taniguchi T., Honda K. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 15989–15994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ramos J. C., Ruiz P., Jr., Ratner L., Reis I. M., Brites C., Pedroso C., Byrne G. E., Jr., Toomey N. L., Andela V., Harhaj E. W., Lossos I. S., Harrington W. J., Jr. (2007) Blood 109, 3060–3068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Brüstle A., Heink S., Huber M., Rosenplänter C., Stadelmann C., Yu P., Arpaia E., Mak T. W., Kamradt T., Lohoff M. (2007) Nat. Immunol. 8, 958–966 [DOI] [PubMed] [Google Scholar]
- 14. Honma K., Kimura D., Tominaga N., Miyakoda M., Matsuyama T., Yui K. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 15890–15895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Inagaki A., Ishida T., Ishii T., Komatsu H., Iida S., Ding J., Yonekura K., Takeuchi S., Takatsuka Y., Utsunomiya A., Ueda R. (2006) Int. J. Cancer. 118, 3054–3061 [DOI] [PubMed] [Google Scholar]
- 16. Hara H., Morita M., Iwaki T., Hatae T., Itoyama Y., Kitamoto T., Akizuki S., Goto I., Watanabe T. (1994) J. Exp. Med. 180, 831–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Furuzawa-Carballeda J., Vargas-Rojas M. I., Cabral A. R. (2007) Autoimmun. Rev. 6, 169–175 [DOI] [PubMed] [Google Scholar]
- 18. Karube K., Ohshima K., Tsuchiya T., Yamaguchi T., Kawano R., Suzumiya J., Utsunomiya A., Harada M., Kikuchi M. (2004) Br. J. Haematol. 126, 81–84 [DOI] [PubMed] [Google Scholar]
- 19. Dodon M. D., Li Z., Hamaia S., Gazzolo L. (2004) J. Gen. Virol. 85, 1921–1932 [DOI] [PubMed] [Google Scholar]
- 20. Sakurai H., Suzuki S., Kawasaki N., Nakano H., Okazaki T., Chino A., Doi T., Saiki I. (2003) J. Biol. Chem. 278, 36916–36923 [DOI] [PubMed] [Google Scholar]
- 21. Singhirunnusorn P., Ueno Y., Matsuo M., Suzuki S., Saiki I., Sakurai H. (2007) J. Biol. Chem. 282, 12698–12706 [DOI] [PubMed] [Google Scholar]
- 22. Takasaki I., Takarada S., Fukuchi M., Yasuda M., Tsuda M., Tabuchi Y. (2007) J. Cell. Biochem. 102, 1472–1485 [DOI] [PubMed] [Google Scholar]
- 23. Yamashita I., Katamine S., Moriuchi R., Nakamura Y., Miyamoto T., Eguchi K., Nagataki S. (1994) Blood 84, 1573–1578 [PubMed] [Google Scholar]
- 24. Chen J., Petrus M., Bryant B. R., Phuc Nguyen V., Stamer M., Goldman C. K., Bamford R., Morris J. C., Janik J. E., Waldmann T. A. (2008) Blood 111, 5163–5172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Suzuki T., Hirai H., Yoshida M. (1994) Oncogene 9, 3099–3105 [PubMed] [Google Scholar]
- 26. Kano S., Sato K., Morishita Y., Vollstedt S., Kim S., Bishop K., Honda K., Kubo M., Taniguchi T. (2008) Nat. Immunol. 9, 34–41 [DOI] [PubMed] [Google Scholar]
- 27. Morinobu A., Kanno Y., O'Shea J. J. (2004) J. Biol. Chem. 279, 40640–40646 [DOI] [PubMed] [Google Scholar]
- 28. Chabas D., Baranzini S. E., Mitchell D., Bernard C. C., Rittling S. R., Denhardt D. T., Sobel R. A., Lock C., Karpuj M., Pedotti R., Heller R., Oksenberg J. R., Steinman L. (2001) Science 294, 1731–1735 [DOI] [PubMed] [Google Scholar]
- 29. Jansson M., Panoutsakopoulou V., Baker J., Klein L., Cantor H. (2002) J. Immunol. 168, 2096–2099 [DOI] [PubMed] [Google Scholar]
- 30. Shaffer A. L., Emre N. C., Lamy L., Ngo V. N., Wright G., Xiao W., Powell J., Dave S., Yu X., Zhao H., Zeng Y., Chen B., Epstein J., Staudt L. M. (2008) Nature 454, 226–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Köntgen F., Grumont R. J., Strasser A., Metcalf D., Li R., Tarlinton D., Gerondakis S. (1995) Genes Dev. 9, 1965–1977 [DOI] [PubMed] [Google Scholar]
- 32. Staudt V., Bothur E., Klein M., Lingnau K., Reuter S., Grebe N., Gerlitzki B., Hoffmann M., Ulges A., Taube C., Dehzad N., Becker M., Stassen M., Steinborn A., Lohoff M., Schild H., Schmitt E., Bopp T. (2010) Immunity 33, 192–202 [DOI] [PubMed] [Google Scholar]
- 33. Sun S. C., Elwood J., Béraud C., Greene W. C. (1994) Mol. Cell. Biol. 14, 7377–7384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mitra-Kaushik S., Harding J., Hess J., Schreiber R., Ratner L. (2004) Blood 104, 3305–3311 [DOI] [PubMed] [Google Scholar]
- 35. Sallusto F., Lanzavecchia A., Mackay C. R. (1998) Immunol. Today 19, 568–574 [DOI] [PubMed] [Google Scholar]
- 36. Yoshie O., Imai T., Nomiyama H. (2001) Adv. Immunol. 78, 57–110 [DOI] [PubMed] [Google Scholar]
- 37. Longphre M., Li D., Gallup M., Drori E., Ordoñez C. L., Redman T., Wenzel S., Bice D. E., Fahy J. V., Basbaum C. (1999) J. Clin. Invest. 104, 1375–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bazarbachi A., El-Sabban M. E., Nasr R., Quignon F., Awaraji C., Kersual J., Dianoux L., Zermati Y., Haidar J. H., Hermine O., de Thé H. (1999) Blood 93, 278–283 [PubMed] [Google Scholar]
- 39. Bazarbachi A., Nasr R., El-Sabban M. E., Mahé A., Mahieux R., Gessain A., Darwiche N., Dbaibo G., Kersual J., Zermati Y., Dianoux L., Chelbi-Alix M. K., de Thé H., Hermine O. (2000) Leukemia 14, 716–721 [DOI] [PubMed] [Google Scholar]
- 40. Hermine O., Dombret H., Poupon J., Arnulf B., Lefrère F., Rousselot P., Damaj G., Delarue R., Fermand J. P., Brouet J. C., Degos L., Varet B., de Thé H., Bazarbachi A. (2004) Hematol. J. 5, 130–134 [DOI] [PubMed] [Google Scholar]
- 41. Mittrücker H. W., Matsuyama T., Grossman A., Kündig T. M., Potter J., Shahinian A., Wakeham A., Patterson B., Ohashi P. S., Mak T. W. (1997) Science 275, 540–543 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.