Abstract
O-Acetyl-ADP-ribose (OAADPr), produced by the Sir2-catalyzed NAD+-dependent histone/protein deacetylase reaction, regulates diverse biological processes. Interconversion between two OAADPr isomers with acetyl attached to the C-2″ and C-3″ hydroxyl of ADP-ribose (ADPr) is rapid. We reported earlier that ADP-ribosylhydrolase 3 (ARH3), one of three ARH proteins sharing structural similarities, hydrolyzed OAADPr to ADPr and acetate, and poly(ADPr) to ADPr monomers. ARH1 also hydrolyzed OAADPr and poly(ADPr) as well as ADP-ribose-arginine, with arginine in α-anomeric linkage to C-1″ of ADP-ribose. Because both ARH3- and ARH1-catalyzed reactions involve nucleophilic attacks at the C-1″ position, it was perplexing that the ARH3 catalytic site would cleave OAADPr at either the 2″- or 3″-position, and we postulated the existence of a third isomer, 1″-OAADPr, in equilibrium with 2″- and 3″-isomers. A third isomer, consistent with 1″-OAADPr, was identified at pH 9.0. Further, ARH3 OAADPr hydrolase activity was greater at pH 9.0 than at neutral pH where 3″-OAADPr predominated. Consistent with our hypothesis, IC50 values for ARH3 inhibition by 2″- and 3″-N-acetyl-ADPr analogs of OAADPr were significantly higher than that for ADPr. ARH1 also hydrolyzed OAADPr more rapidly at alkaline pH, but cleavage of ADP-ribose-arginine was faster at neutral pH than pH 9.0. ARH3-catalyzed hydrolysis of OAADPr in H218O resulted in incorporation of one 18O into ADP-ribose by mass spectrometric analysis, consistent with cleavage at the C-1″ position. Together, these data suggest that ARH family members, ARH1 and ARH3, catalyze hydrolysis of the 1″-O linkage in their structurally diverse substrates.
Keywords: ADP-ribosylation, Chromatin, Enzyme Mechanisms, Enzyme Turnover, Sirtuins, ADP-ribosylhydrolase, O-Acetyl-ADP-ribose, Poly(ADP-ribose)
Introduction
Mono-ADP-ribosylation is a post-translational modification, in which the ADP-ribose (ADPr)5 moiety of NAD is transferred to an acceptor protein (1). This modification serves as the mechanism by which several bacterial toxins (e.g. Pseudomonas exoenzyme S, cholera toxin, diphtheria toxin) exert their effects on mammalian cells (2, 3). Mammalian cells also produce endogenous ADP-ribosyltransferases that catalyze reactions similar to the bacterial toxins, specifically, the ADP-ribosylation of arginine residues in proteins (4). In addition, mammalian cells possess hydrolases that cleave the ADPr-protein linkage, releasing ADPr and regenerating the unmodified protein (5, 6). An ADP-ribosyl(arginine) hydrolase, termed ARH1, catalyzes in a stereospecific manner, hydrolysis of the α-linkage of arginine-ribose found in ADP-ribosyl(arginine)-protein to ADPr and (arginine)-protein (7, 8), consistent with the regulation of ADP-ribosyl(arginine)-protein levels by opposing activities of transferases and hydrolases, participating in an ADP-ribosylation cycle (4, 9).
Three known members (ARH1–3) of the ARH family of proteins are similar in molecular size (∼39 kDa) and amino acid sequence (10). As noted above, ARH1 catalyzes the hydrolysis of ADP-ribose-arginine and also hydrolyzes ADP-ribose linkages to guanidine. The reaction is stereospecific, and only the α-anomer at the C-1″ position of ADP-ribose-arginine is hydrolyzed (8). ARH1 also hydrolyzed the stereospecific C-1″-C-2′ linkage in poly(ADP-ribose). The ARH1 reaction is inhibited by ADP-ribose, but not by phosphoribose, suggesting that the catalytic site recognizes the adenosine moiety (8). The fact that only the guanidine group of arginine is necessary for hydrolysis further supports a critical role for the ADP-ribose as opposed to the arginine in catalysis (8). ARH2, which has considerable structural similarity to ARH1, has no reported activity. In this regard, the vicinal aspartate residues that were critical for ARH1 activity are replaced by an aspartate-asparagine, which may explain the lack of activity.
We previously reported that ARH3 possesses poly(ADPr) glycohydrolase activity (10). The linkage hydrolyzed by ARH3 and ARH1 in poly(ADP-ribose) is also stereospecific at the C-1″ position with the α-anomer present in the polymer (Fig. 1). As with the ARH1-catalyzed reaction, ADP-ribose is a potent inhibitor. ARH3 also hydrolyzed O-acetyl-ADP-ribose (OAADPr) in a Mg2+-dependent manner to produce ADPr, which inhibited these two ARH3-catalyzed reactions (10, 11). Both poly(ADPr) and OAADPr participate in important biological pathways. Poly(ADPr) is involved in carcinogenesis, DNA repair, and cell differentiation (12–14). OAADPr is the product of NAD+-dependent protein deacetylation reactions catalyzed by Silent information regulator 2 (Sir2) proteins that contribute to diverse operations such as gene silencing, DNA repair, chromosomal stability, and lifespan extension (12, 13). OAADPr may act independently of protein deacetylation in the stabilization of chromatin, formation of silencing complexes, ion-channel gating, and energy metabolism (15–19). Thus, ARH3 can participate in distinct signal transduction pathways that involve poly(ADPr) and OAADPr.
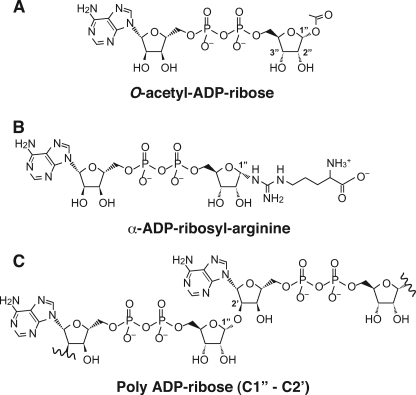
FIGURE 1.
Schematic diagram of substrates hydrolyzed by ARH1 and ARH3. A, structure of OAADPr with position of 1″-O-acetyl-linkage noted. B, structure of α-ADPr-arginine. C, Structure of poly(ADPr) (C1″-C2′).
During the Sir2-catalyzed reaction, 2″-OAADPr has been reported to be the reaction product released by the enzyme (20, 21) (see Fig. 1 for structures). After release, 2″-OAADPr rapidly interconverts at pH 7.5 with 3″-OAADPr, with which it exists in equilibrium, via acetyl migration, in a ratio of about 1:1 (20, 21). In contrast, 2″- and 3″-N-acetyl-ADPr analogs of OAADPr do not undergo acetyl migration, although they do mimic OAADPr binding to macro H2A1.1 (22).
Both ARH1 and ARH3 appear to catalyze stereospecific hydrolysis of ADP-ribose-arginine and poly(ADP-ribose), respectively. In both cases, the linkage subject to hydrolysis is at the C-1″ position (Fig. 1). In addition, both the ARH1- and ARH3-catalyzed reactions are inhibited by ADP-ribose. For ARH1, ADP-ribose, but not phosphoribose, is an inhibitor, and ADP-ribose-arginine, but not phosphoribose-arginine, is a substrate, demonstrating that the adenosine moiety is important for substrate recognition. Therefore, the ribose linkage to arginine is insufficient for substrate recognition. We observed that ARH1 as well as ARH3 hydrolyzed O-acetyl-ADP-ribose.
We thought it unlikely that ARH3 and ARH1 would hydrolyze the 1″-linkage in poly(ADPr) and ADP-ribose-arginine, respectively, and also act on 2″- or 3″-linkage in OAADPr. Further, in our current studies, ADP-ribose was a potent inhibitor, but not 2″-, 3″-N-acetyl-ADP-ribose. In addition, ARH1 hydrolyzed the C-1″ derivative of ADP-ribose(arginine) as well as OAADPr, suggesting that the C-1″ position is the preferred site of cleavage. Here, we propose that, analogous to its poly(ADPr)glycohydrolase activity, ARH3 hydrolyzes 1″-OAADPr, rather than the 2″- or 3″-isomer, and that 1″-, 2″-, and 3″-OAADPr exist in equilibrium at neutral and basic pH values, thus generating 1″-OAADPr for hydrolysis by ARH3. These studies demonstrate that ARH1 and ARH3 show a similar preference for substrates and that hydrolysis proceeds with attack at the C-1″ position. Prior studies that showed the presence of C-2″ and C-3″ O-acetyl-ADP-ribose are in agreement with our current findings that at neutral pH, the equilibrium favors these two isomers and not C-1″ O-acetyl-ADP-ribose.
EXPERIMENTAL PROCEDURES
Materials
β-NAD, ADPr, DTT, TFA, and acetonitrile were purchased from Sigma. Adenine [U-14C]NAD (252 mCi (1 Ci = 37 GBq)) was from GE Healthcare. Acetylhistone peptide H3 (acetylated at Lys-14) was from Millipore. SIRT1 was from Enzo Biochem (New York, NY). [18O]Water was from Cambridge Isotope Laboratories, Inc. (Andover, MA). Recombinant human ARH3 and ARH1 were prepared as described previously (10, 23).
Generation and Analysis of O-Acetyl-[14C]ADP-ribose and Other Small Molecules
O-Acetyl-[14C]ADP-ribose was prepared and purified as described by Ono et al. (11). Briefly, 100 μm [14C]β-NAD (2,500 cpm/pmol) and acetylhistone peptide H3 (100 μg) were incubated for 4 h at 30 °C with SIRT1 (25 units, 6.1 μg) in 50 mm Tris-HCl (pH 7.0) buffer containing 2.7 mm KCl, 1 mm MgCl2, and 0.2 mg of BSA (total volume, 200 μl). Product and substrate were separated on a Vydac C18 column (4.6 mm × 250 mm; W. R. Grace & Co., Columbia, MD) using reverse-phase high-performance liquid chromatography (RP-HPLC) (Hewlett-Packard, series 1100, with a diode array spectrophotometric detector set at 259 nm). Isocratic elution (1 ml/min) with 100% buffer A (0.05% (v/v) TFA in water) from 0 to 5 min was followed by a linear gradient to 60% buffer A and 40% buffer B (0.05% (v/v) TFA in acetonitrile) from 5 to 45 min. Eluate fractions (30 s, 500 μl) were collected for quantification of radioactivity using liquid scintillation counting (TriCarb 1600TR, PerkinElmer Life Sciences). Eluted solvent was fractioned each 30 s (500 μl) using fraction collector and was subjected to a liquid scintillation counter (Liquid Scintillation Analyzer TriCarb 1600TR, PerkinElmer Life Sciences) for measurement of radioactivity.
Hydrolysis of O-Acetyl-ADP-ribose Catalyzed by ARH3 or ARH1
Samples of 1 μm OAADPr and purified recombinant ARH3 (1.5 pmol) or ARH1 (375 pmol) were incubated in 50 mm potassium phosphate buffer (pH 5.0, 7.0, and 9.0), 10 mm MgCl2, and 5 mm DTT (total volume, 200 μl) for 60 min at 30 °C. Substrate and products were separated by RP-HPLC, as described above.
Identification of Isomers of O-Acetyl-ADP-ribose
For identification of OAADPr isomers, 1 μm OAADPr in 50 mm potassium phosphate (pH 9.0), 10 mm MgCl2, and 5 mm DTT (total volume, 200 μl) were incubated (5 min at room temperature), before separation of isomers using RP-HPLC and analysis by MALDI-TOF mass spectrometry as described previously (11). Briefly, before mass spectrometry, 10 mg of α-cyano-4-hydroxycinnamic acid (Bruker Daltonics, Billerica, MA) was mixed in 1 ml of water/acetonitrile (50:50, v/v) containing TFA. Then, 40 pmol of OAADPr in 1 μl was added to 1 μl of matrix solution (described above) on a stainless steel target plate and air-dried. Negative ion MALDI-TOF mass spectra were acquired using the 4700 Proteomics analyzer (Applied Biosystems, Foster City, CA) operated in reflection mode.
Kinetics of ARH3 Hydrolysis
Rates of OAADPr hydrolysis by purified recombinant ARH3 (1.5 pmol) were determined using the indicated concentrations of OAADPr in 50 mm potassium phosphate (pH 7.0), 10 mm MgCl2, and 5 mm DTT (total volume, 200 μl) incubated for 20 min at 30 °C. ADPr was quantified by RP-HPLC; Km and Vmax were calculated from Lineweaver-Burk double reciprocal plots.
Interconversion of O-Acetyl-ADP-ribose Isomers
To assess interconversion of OAADPr isomers at pH 9.0, 3″-OAADPr (peak A), was purified by RP-HPLC. 3″-OAADPr in water containing 0.05% (v/v) TFA (total volume, 200 μl) or in 50 mm potassium phosphate (pH 9.0), 10 mm MgCl2, and 5 mm DTT (total volume, 200 μl) was incubated (5 min, 30 °C) before separation using RP-HPLC.
Nonenzymatic Hydrolysis of O-Acetyl-ADP-ribose
To evaluate nonenzymatic hydrolysis of OAADPr, 1 μm OAADPr in 50 mm potassium phosphate (pH 9.0 or 7.0), 10 mm MgCl2, and 5 mm DTT (total volume, 200 μl) was incubated for the indicated time at 30 °C, before separation of substrate and products by RP-HPLC, as described above.
Preparation of [3H]O-Acetyl-ADP-ribose and N-Acetyl-ADP-ribose Analogs
[3H]OAADPr was generated enzymatically using published methods with [3H]acetylated histone H3 peptides (21, 24). Synthesis of N-acetyl-ADPr analogs was performed as described (22).
Charcoal Binding Assay
[3H]Acetate produced by ARH3-catalyzed hydrolysis of [3H]OAADPr was quantified using a charcoal binding assay (24). Reaction mixtures (200 μl) containing 50 mm potassium phosphate (pH 7.0), 10 mm MgCl2, 5 mm DTT, 16.7 pmol of purified human ARH3, 12.5 μm [3H]OAADPr, and either 2″- or 3″-N-acetyl-ADPr, as indicated, were incubated for 30 min at 30 °C. Reactions were terminated by the addition to a 50-μl charcoal slurry (one-third volume of charcoal solid in two-thirds volume of PBS, pH 7.0). Nonenzymatic [3H]acetate production without ARH3 was used as a control. Charcoal was sedimented by centrifugation, and 200 μl of supernatant containing [3H]acetate was transferred to a clean tube. After centrifugation to remove residual charcoal, 195 μl of the supernatant was transferred to scintillation vials containing counting solution. Supernatant radioactivity represents [3H]acetate produced by ARH3-catalyzed hydrolysis of [3H]OAADPr (1 mol/mol). Difference between amounts of [3H]acetate (pmol) accumulated with and without (control) ARH3 was plotted against inhibitor concentration, and data were fit to an inhibition equation using KaleidaGraph (Synergy Software, Reading, PA) to calculate IC50.
Hydrolysis of α-ADP-ribosyl-arginine Catalyzed by ARH1
ADP-ribosyl-[14C]arginine was generated and purified as described by Kato et al. (23). α-ADP-ribosyl-arginine was isolated by RP-HPLC. Assays containing 25 μm α-ADP-ribosyl-arginine and purified recombinant ARH1 (3 pmol) in 50 mm potassium phosphate (pH 5.0, 7.0, or 9.0), 10 mm MgCl2, and 5 mm DTT (total volume, 200 μl) were incubated for the indicated time at 30 °C. Substrate and products were separated by RP-HPLC, as described above, using Affi-Gel boronate columns (Bio-Rad) (23).
18O Incorporation into ADP-ribose
Assays containing OAADPr (5 nmol), purified recombinant ARH3 (1 nmol in 100 μl), or heat-inactivated ARH3 (1 nmol, incubated at 95 °C for 20 min) in 50 mm pyridine-formic acid (pH 7.0) and 1 mm MgCl2 with [18O]water (total volume, 100 μl) were incubated (30 min, 30 °C) before the addition of 1 ml of 100% methanol and analysis. Negative ion electrospray ionization mass spectra were acquired using the Shimadzu LCMS-2020 (Shimadzu, Kyoto, Japan). Ion chromatograms were analyzed with Shimadzu LCMSsolution software.
RESULTS
pH-dependent Hydrolysis of OAADPr by ARH3
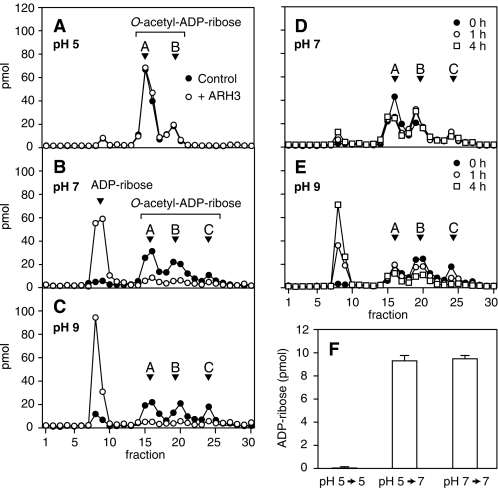
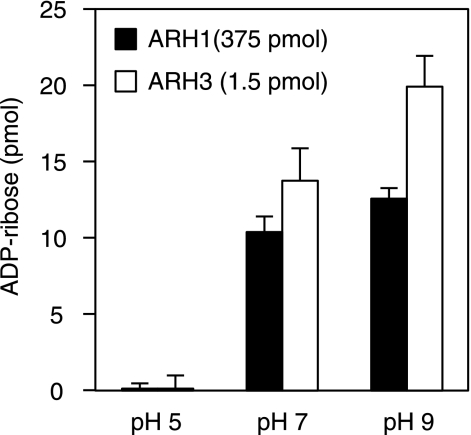
We compared ARH3 hydrolysis of OAADPr (produced from [14C]NAD and acetylated histone H3 peptide by human SIRT1 (see “Experimental Procedures”) at pH 5.0, 7.0, and 9.0. At 5.0, RP-HPLC analysis revealed two peaks of OAADPr, consistent with 2″- (Fig. 2A, peak B) and 3″-OAADPr (Fig. 2A, peak A) (20, 21). At pH 5.0, OAADPr hydrolysis in the presence of ARH3 was not above background (Fig. 2A). After incubation with ARH3 at pH 7.0, the remaining substrate contained not only 2″- and 3″-OAADPr, but also an additional isomer (Fig. 2B, peak C). In OAADPr hydrolase assays containing ARH3 at pH 7.0, 119 pmol of OAADPr, containing all three isomers, were hydrolyzed, with accumulation of 127 pmol of ADPr (Fig. 2B). At pH 9.0, ARH3 activity was greater than that at pH 7.0, and amounts of all three isomers were reduced, with the expected increase in ADPr (Fig. 2C). Peak C, not observed at pH 5.0, was present at low levels at pH 7.0 and higher abundance at pH 9.0 (Fig. 2, A–C). To assess nonenzymatic OAADPr hydrolysis and determine whether the greater activity at pH 9.0 was enzymatic or nonenzymatic, OAADPr was incubated at pH 9.0 and 7.0. At pH 9.0 (Fig. 2E), nonenzymatic OAADPr hydrolysis was clearly faster than at pH 7.0 (Fig. 2D), and all three peaks were reduced. After nonenzymatic OAADPr hydrolysis, ARH3 activity at pH 9.0 was still higher than at pH 5.0 or pH 7.0 (subtracted nonenzymatic hydrolysis of OAADPr was significant only at pH 9.0). The mass of 558 m/z nonenzymatic product (data not shown) was consistent with ADPr. As shown in Fig. 2A, there was no ARH3 activity at pH 5.0. Two possible causes of deactivation of ARH3 at pH 5.0 were (i) denaturation of ARH3 due to low pH, and (ii) a pH below the optimal range for OAADPr hydrolysis by ARH3. To assess denaturation, ARH3 was incubated at pH 5.0 for 30 min and then at pH 7.0 for 5 min, revealing dramatically greater hydrolase activity than that at pH 5.0 (Fig. 2F). Activity of ARH3 that had been incubated at pH 5.0 for 30 min followed by 5 min of incubation at pH 7.0 was the same as that measured at pH 7.0 (Fig. 2F), indicating that the lower activity at pH 5.0 was not due to irreversible ARH3 denaturation. Thus, ARH3 activity correlated with the relative abundance of peak C at different pH values, and all three isomers were decreased by ARH3 activity, consistent with their interconversion.
FIGURE 2.
pH-dependence of O-acetyl-ADP-ribose hydrolysis by ARH3. A–E, [14C]OAADPr (1 μm) (total volume, 200 μl) was incubated at pH 5.0 (A), 7.0 (B), or 9.0 (C) with (open circles) or without (filled circles) recombinant human ARH3 (1.5 pmol) for 1 h at 30 °C before separation of substrate and products as described under “Experimental Procedures.” Nonenzymatic hydrolysis of [14C]OAADPr was assayed in the same conditions at pH 7.0 (D) and 9.0 (E). Substrates and products were separated by RP-HPLC and radioactivity quantified by liquid scintillation counting. F, 1 μm [14C]OAADPr was incubated with ARH3 (1.5 pmol), 50 mm potassium phosphate (pH 5.0), 10 mm MgCl2, 5 mm DTT (total volume, 200 μl) for 35 min at pH 7.0 or 30 min at pH 5.0 (30 °C) followed by 5 min at pH 7.0 (pH was adjusted by the addition of 100 mm dipotassium hydrogen orthophosphate). Substrates and products were quantified as described above. The experiment was repeated three times with similar results.
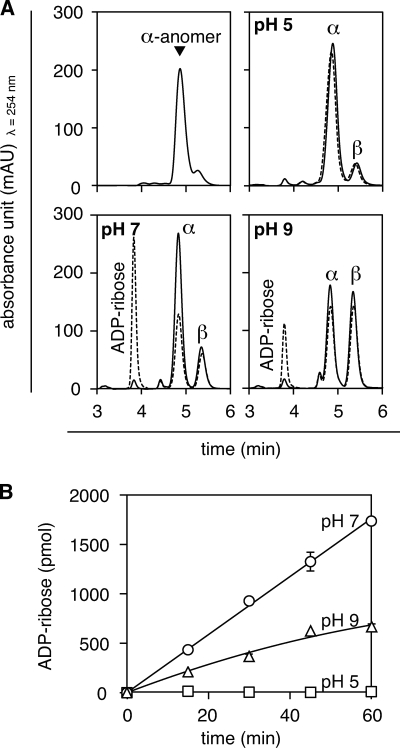
Interconversion of O-Acetyl-ADP-ribose Isomers
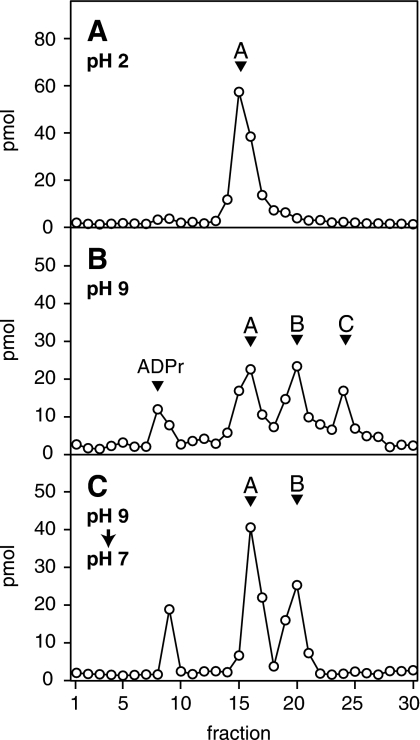
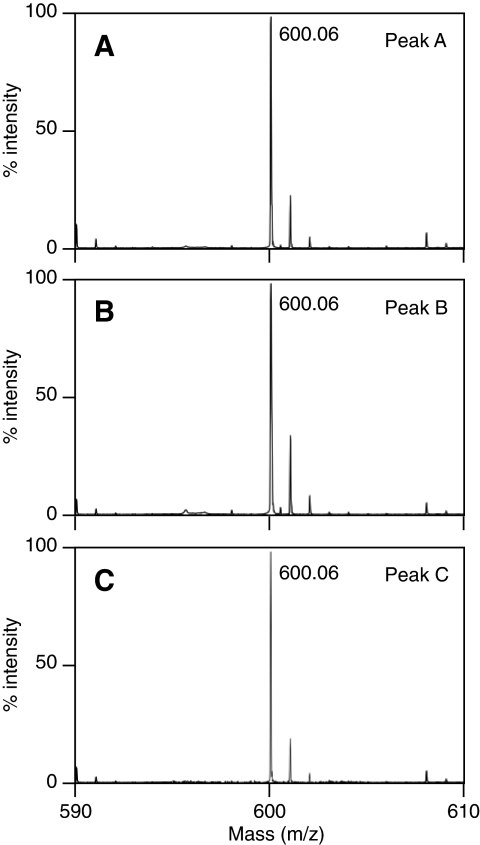
Because the activity and pH-dependence of ARH3 correlated with the relative abundance of peak C, we further characterized that material. Peak A (3″-OAADPr) predominated under pH 2.0 (Fig. 3A), whereas at pH 9.0 three peaks (A, B, and C) were present (Fig. 3B) even if isolated peak C was used as the starting material. Upon addition of 0.05% (v/v) TFA to a solution of OAADPr after being incubated at pH 9.0, peak C was completely converted to peaks A and B (Fig. 3C). This indicated that peak C was in equilibrium with peaks A and B, and, importantly, formation of peak C was reversible. The data are consistent with pH-dependent interconversion of the OAADPr isomers. The mass of each of the three peaks was 600 m/z (Fig. 4, A–C), consistent with peak C being an isomer of OAADPr, with acetate perhaps at the 1″-position.
FIGURE 3.
Interconversion of O-acetyl-ADP-ribose isomers. A, OAADPr (peak A) was separated using RP-HPLC (elution time consistent with 3″-OAADPr) in 200 μl of water containing 0.05% (v/v) TFA as described under “Experimental Procedures.” B, purified 3″-OAADPr in reaction mix described in Fig. 1 at pH 9.0 was maintained at pH 9.0 and separated using RP-HPLC as described. C, purified 3″-OAADPr (pH 9.0) was incubated for 5 min at 30 °C followed by the addition of 0.05% (v/v) TFA and incubation for an additional 10 min at 30 °C before RP-HPLC analysis. The experiment was repeated three times with similar results.
FIGURE 4.
Mass spectrometry of O-acetyl-ADP-ribose isomers. Separated isomers (A, B, and C) of OAADPr (1 μm) like that shown in Fig. 1C were subjected to MALDI-TOF mass spectrometry to reveal a peak at 600 m/z, consistent with the predicted mass of OAADPr, in each sample. The experiment was repeated three times with similar results.
Inhibition of ARH3 Activity by ADP-ribose, 2″-N-Acetyl-ADP-ribose, and 3″-N-Acetyl-ADP-ribose
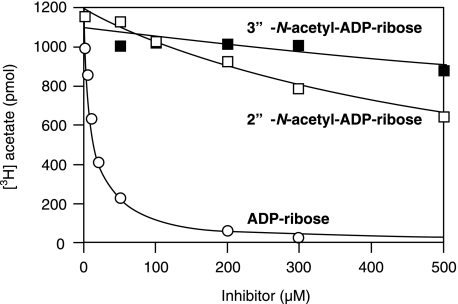
Inhibition of ARH3 hydrolysis of [3H]OAADPr (Km = 4.3 ± 0.3 μm) by 2″- and 3″-N-acetyl-ADPr was assessed by quantification of the [3H]acetate product. The IC50 values of 2″-N-acetyl-ADPr and 3″-N-acetyl-ADPr were ∼60 and ∼200-fold that of ADPr (Table 1 and Fig. 5). Neither 2″- nor 3″-N-acetyl-ADPr was a substrate for ARH3 (data not shown). If the preferred substrate of ARH3 is 1″-OAADPr, it is predicted that ADPr would be better accommodated within the active site than either 2″- or 3″-OAADPr. Therefore, inhibition data are consistent with ARH3 preferentially binding to and hydrolyzing 1″-OAADPr.
TABLE 1.
Calculated IC50 values for ARH3 of ADP-ribose, 2″-N-acetyl-ADP-ribose, and 3″-N-acetyl-ADP-ribose
Assays (30 min, 30 °C) as described under “Experimental Procedures” contained 16.7 pmol of human ARH3. [3H]Acetate was quantified using a modified charcoal binding assay as described under “Experimental Procedures.” [3H]Acetate data (Fig. 4) were plotted and fit to an inhibition equation using KaleidaGraph to calculate IC50.
| Substrate | IC50 |
|---|---|
| mm | |
| ADP-ribose | 0.012 ± 0.003 |
| 2″N-Acetyl-ADP-ribose | ≥0.71 |
| 3″-N-Acetyl-ADP-ribose | ≥2.3 |
FIGURE 5.
ADP-ribose, 2″-N-acetyl-ADP-ribose, and 3″-N-acetyl-ADP-ribose inhibition of ARH3 activity. Human ARH3 (83.5 nm; 16.7 pmol) and 12.5 μm [3H]OAADPr with ADPr (open circles), 2″-N-acetyl-ADPr (open squares), or 3″-N-acetyl-ADPr (filled squares) were incubated for 30 min at 30 °C as described in Fig. 1 before quantification of [3H]acetate as described under “Experimental Procedures.”
pH Dependence of Hydrolysis of O-Acetyl-ADP-ribose and ADP-ribose-arginine by ARH1
Given that the reactions catalyzed by ARH3 and ARH1 are stereospecific at the C-1″ position and prefer the α-anomer of poly(ADP-ribose) (ARH3, ARH1) and ADP-ribose-arginine (ARH1), we hypothesized that ARH1 would also hydrolyze O-acetyl-ADP-ribose at the C-1″ position, and we expected that for ARH1, as for ARH3, conditions favoring the abundance of the C-1″ would enhance hydrolysis. In contrast, hydrolysis of ADP-ribose-arginine by ARH1 should not depend on the alkaline conditions that favor the C-1″ form of O-acetyl-ADP-ribose. Indeed, effects of pH on OAADPr hydrolase activity of ARH1 and ARH3 were similar (Fig. 6). As the hydrolysis we saw with ARH1 was less than a single turnover, the observed pH dependence of OAADPr hydrolysis was not a steady-state phenomenon, whereas differences with ARH3 were at steady state, thus no direct comparisons of rate are possible.
FIGURE 6.
pH-dependence of O-acetyl-ADP-ribose hydrolysis by ARH3 and ARH1. Human ARH 3 (1.5 pmol) or ARH1 (375 pmol) was incubated for 5 min with 1 μm OAADPr at the indicated pH as in Fig. 4 before RP-HPLC separation and quantification of ADPr. Blank data without enzyme were subtracted from each of these points. The experiment was repeated three times with similar results.
ARH1 also hydrolyzes ADPr-arginine, which exists as both α- and β-anomers of the C-1″-guanidine linkage. The α-anomer predominated at pH 5.0 and amounts of α- and β-anomers were similar at pH 9.0, consistent with pH-dependent anomerization (Fig. 7A). ARH1 activity was minimal at pH 5.0, but in contrast to the findings with OAADPr, its hydrolysis of ADPr-arginine at pH 7.0 appeared greater than that at pH 9.0 (Fig. 7). At pH 7.0, anomerization was relatively slow, and preferential hydrolysis of the α-anomer by ARH1 was seen with preservation of β-anomer. At pH 9.0, anomerization was relatively rapid, with depletion of both α- and β-anomers, because hydrolysis of α-anomer was accompanied by continuing anomerization. ARH1 activities at pH 7.0 and 9.0 were constant with time (Fig. 7B), and rates of ARH1-catalyzed hydrolysis correlated with the abundance of the α-anomer at the 1″-position for ADPr-arginine. Thus, as expected, hydrolysis of O-acetyl-ADP-ribose and ADP-ribose-arginine appears to be dependent in part on pH conditions that favor the substrate form of the molecule.
FIGURE 7.
pH-dependence of α-ADP-ribosyl-arginine hydrolysis by ARH1. A, 25 μm α-ADP-ribosyl-[14C]arginine was incubated in 50 mm potassium phosphate buffer (pH 7.0), 10 mm MgCl2, and 5 mm DTT for 1 h at 30 °C with (dashed lines) or without (solid lines) recombinant human ARH1 (3 pmol) before RP-HPLC separation of substrate and products. Substrate and products were detected by absorbance at 254 nm. B, α-ADP-ribosyl-[14C]arginine was incubated as in Fig. 6A, with ARH1 (3 pmol) for the indicated time at 30 °C before quantification of ADPr. Results were similar in three experiments.
18O Incorporation into ADP-ribose
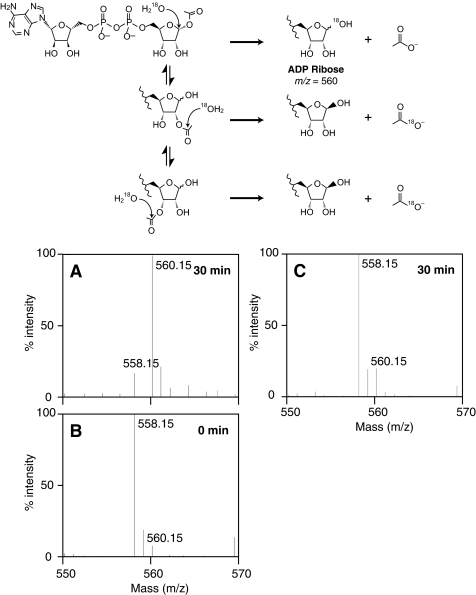
To obtain evidence that ARH3 cleaved OAADPr at the C-1″ position, OAADPr was hydrolyzed by ARH3 in H218O. The reaction product, ADPr, was analyzed by mass spectrometry to quantify 18O incorporation into ADPr. The major ADPr product (85% of total 558.15 + 560.15 m/z) of ARH3-catalyzed hydrolysis incorporated one 18O (560.15 m/z) (Fig. 8A). Reactions with heat-inactivated ARH3 yielded nonenzymatically hydrolyzed ADPr (m/z = 558.15) with no significant 18O incorporation above natural abundance (Fig. 8B). As expected, a 30-min reaction of heat-inactivated ARH3 displayed some nonenzymatic 18O incorporation (14% of total 558.15 + 560.15 m/z) (Fig. 8C). When the [18O]ADPr sample formed in Fig. 8A was acidified by 0.1% formic acid and incubated for 5 min, a [18O] ADPr (560.15 m/z) was diminished, with a corresponding increase of ADPr (558.15 m/z) (data not shown). These data suggested that ARH3 catalyzes attack of [18O]water at the 1″-carbon of OAADPr, with incorporation of 18O into ADPr at the 1″-position. Since only the 1″-position is exchangeable, the 18O must be at the 1″-position.
FIGURE 8.
ARH3-catalyzed 18O labeling of ADPr. A, OAADPr (5 nmol) was incubated with ARH3 (1 nmol) in 40 mm pyridine buffer adjusted to pH 7.0 with formic acid, 1 mm MgCl2, and 90% H218O for 30 min at 30 °C. Then the mixture was diluted with methanol and subjected to mass spectrometry as described under “Experimental Procedures.” B and C, m/z = 558.15 corresponds to ADPr, and m/z = 560.15 to ADPr containing 18O atom. 18O incorporation was measured by the relative peak heights at each m/z. As a control, ADPr was incubated with heat-denatured ARH3 in the same mixture for 0 min (B) to confirm the natural distribution of heavy isotope or 30 min (C) to assess nonenzymatic exchange of the 1″-hydroxyl at 30 °C. Detection and measurement of 18O incorporation are described under “Experimental Procedures.” The experiment was repeated three times with similar results. The proposed reaction mechanisms for 18O incorporation are indicated in the upper panel.
DISCUSSION
Herein, we found not only 2″- and 3″-OAADPr at pH 9.0, but also substantial quantities of a third peak, of mass (600 m/z) that was consistent with 1″-OAADPr. At acidic pH 5.0, 3″-OAADPr predominated, whereas at higher pH, all three forms of OAADPr were observed. Rapid interconversion of 2″- and 3″-OAADPr had been reported (21), and the amount of peak C at pH 9.0 was greater than that at pH 7.0, whereas peak C was absent at pH 5.0. All three peaks appeared to be in rapid equilibrium at pH 9.0. ARH3 hydrolysis of OAADPr at pH 9.0 was significantly faster than that at pH 7.0. Thus, ARH3 activity correlated with peak C concentration, although other factors related to protein stability and active site chemistry may also affect hydrolase activity at acidic or alkaline pH.
ARH3 activity was not inhibited by d-ribose 5-phosphate, AMP, ADP, or β-NAD (11). ADPr was an effective inhibitor, likely due to competition for substrate binding within the ARH3 catalytic active site (11). In contrast to ADPr (11), 2″- and 3″-N-acetyl-ADPr were poor ARH3 inhibitors. These data suggested that the 2″- and 3″-acetyl-ribose rings were, relative to ADPr, accommodated poorly in the ARH3 active site, as might be expected if the preferred substrate were 1″-OAADPr.
ARH3 hydrolyzes not only OAADPr, but also poly(ADPr) (10, 11). However, ARH3-catalyzed generation of ADPr from OAADPr was significantly faster than that from poly(ADPr) (11). Poly(ADPr) is a homopolymer of ADPr molecules, linked by ribose-ribose (C-1″ to C-2′) glycosidic bonds in a linear or branched structure (25, 26). Again, ARH3 catalyzes hydrolysis at the C-1″ position within poly(ADPr).
ARH3 and ARH1 molecules share substantial amino acid sequence similarity (10) and stereospecificity (6–8). ARH1 catalyzes hydrolysis of the α-anomer of ADP-ribosyl-arginine at the ribose C-1″ position (6–8). ARH1 also cleaved OAADPr to produce ADPr but at <1% the rate of ARH3 (11). In the experiments reported here, ARH1 activity was pH-dependent; pH 7.0 was preferred for ADPr-arginine hydrolysis, whereas pH 9.0 was optimal for OAADPr hydrolysis. These data are consistent with the hypothesis that the apparent pH optimum for ARH1 is, in part, determined by substrate availability.
If ARH3 hydrolyzed OAADPr at C-1″, 18O should be incorporated into the OAADPr product at the 1″-hydroxyl. Otherwise, 18O attack occurred on a carbon atom of an acetyl group, resulting in no 18O incorporation into ADPr (scheme in Fig. 8). Indeed, one 18O was incorporated into ADPr (560.15 m/z) in contrast to the result of control experiments with inactive ARH3, which showed basal uncatalyzed 18O incorporation. In addition, when ADPr (560.15 m/z) was acidified in natural abundance water, 18O exchanged out rapidly, in agreement with previous research (27). These data are consistent with OAADPr hydrolysis catalyzed by ARH3 occurring at the C-1″ position.
In summary, we report that three isomers of OAADPr are in pH-dependent equilibrium: under acidic conditions 3″-OAADPr is most abundant, 3″- and 2″-OAADPr predominate at neutral pH, and a third isomer that is consistent with 1″-OAADPr predominates in a basic milieu. ARH3 activity with OAADPr as substrate was optimal at pH 9.0, where the putative 1″-isomer is the greatest. ARH3 activity was not observed at pH 5.0, where the 3″-isomer predominates. Consistent with the conclusion that hydrolysis occurs at the 1″-position of OAADPr, 2″- and 3″-N-acetyl-ADPr were much less effective inhibitors of ARH3 activity than ADPr. Moreover, the alkaline pH optimum for OAADPr hydrolysis was similar for ARH1 and ARH3, but ARH1 cleavage of ADPr-arginine, which does not have 2″-and 3″-isomers, was optimal at neutral pH. Based on these data, we propose that ARH3 specifically hydrolyzes the 1″-OAADPr isomer, i.e. a bond similar to those hydrolyzed by ARH3 in poly(ADPr). As the OAADPr produced in the Sir2-catalyzed NAD-dependent histone/protein deacetylase reaction is reported to participate in several biological processes, including formation of silencing complexes, ion channel gating, and energy metabolism (15–19), ARH3 may influence several signaling pathways through degradation of OAADPr.
Acknowledgments
We thank Dr. Martha Vaughan for helpful discussions and critical review of the manuscript, Drs. Rong-Fong Shen and Angel M. Aponte (Proteomics Core Facility, NHLBI, National Institutes of Health) for assistance with mass spectrometry experiments, and Linda A. Stevens for assistance with the HPLC experiments.
This work was supported, in whole or in part, by the Intramural Research Program, National Institutes of Health, NHLBI.
- ADPr
- ADP-ribose
- ARH
- ADP-ribosylhydrolase
- OAADPr
- O-acetyl-ADP-ribose
- RP-HPLC
- reverse phase-HPLC.
REFERENCES
- 1. Williamson K. C., Moss J. (1990) in ADP-ribosylating Toxins and G Proteins: Insights into Signal Transduction (Moss J., Vaughan M. eds) pp. 493–510, American Society for Microbiology, Washington, D.C: [DOI] [PubMed] [Google Scholar]
- 2. Fishman P. H. (1990) in ADP-ribosylating Toxins and G Proteins: Insights into Signal Transduction (Moss J., Vaughan M. eds) pp. 127–140, American Society for Microbiology, Washington, D.C: [DOI] [PubMed] [Google Scholar]
- 3. Krueger K. M., Barbieri J. T. (1995) Clin. Microbiol. Rev. 8, 34–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Moss J., Zolkiewska A., Okazaki I. (1997) Adv. Exp. Med. Biol. 419, 25–33 [DOI] [PubMed] [Google Scholar]
- 5. Moss J., Tsai S. C., Adamik R., Chen H. C., Stanley S. J. (1988) Biochemistry 27, 5819–5823 [DOI] [PubMed] [Google Scholar]
- 6. Moss J., Stanley S. J., Nightingale M. S., Murtagh J. J., Jr., Monaco L., Mishima K., Chen H. C., Williamson K. C., Tsai S. C. (1992) J. Biol. Chem. 267, 10481–10488 [PubMed] [Google Scholar]
- 7. Moss J., Jacobson M. K., Stanley S. J. (1985) Proc. Natl. Acad. Sci. U.S.A. 82, 5603–5607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Moss J., Oppenheimer N. J., West R. E., Jr., Stanley S. J. (1986) Biochemistry 25, 5408–5414 [DOI] [PubMed] [Google Scholar]
- 9. Okazaki I. J., Moss J. (1998) J. Biol. Chem. 273, 23617–23620 [DOI] [PubMed] [Google Scholar]
- 10. Oka S., Kato J., Moss J. (2006) J. Biol. Chem. 281, 705–713 [DOI] [PubMed] [Google Scholar]
- 11. Ono T., Kasamatsu A., Oka S., Moss J. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 16687–16691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Diefenbach J., Bürkle A. (2005) Cell. Mol. Life Sci. 62, 721–730 [DOI] [PubMed] [Google Scholar]
- 13. Masutani M., Nakagama H., Sugimura T. (2005) Cell. Mol. Life Sci. 62, 769–783 [DOI] [PubMed] [Google Scholar]
- 14. Nguewa P. A., Fuertes M. A., Valladares B., Alonso C., Pérez J. M. (2005) Prog. Biophys. Mol. Biol. 88, 143–172 [DOI] [PubMed] [Google Scholar]
- 15. Kustatscher G., Hothorn M., Pugieux C., Scheffzek K., Ladurner A. G. (2005) Nat. Struct. Mol. Biol. 12, 624–625 [DOI] [PubMed] [Google Scholar]
- 16. Liou G. G., Tanny J. C., Kruger R. G., Walz T., Moazed D. (2005) Cell 121, 515–527 [DOI] [PubMed] [Google Scholar]
- 17. Borra M. T., O'Neill F. J., Jackson M. D., Marshall B., Verdin E., Foltz K. R., Denu J. M. (2002) J. Biol. Chem. 277, 12632–12641 [DOI] [PubMed] [Google Scholar]
- 18. Grubisha O., Rafty L. A., Takanishi C. L., Xu X., Tong L., Perraud A. L., Scharenberg A. M., Denu J. M. (2006) J. Biol. Chem. 281, 14057–14065 [DOI] [PubMed] [Google Scholar]
- 19. Tong L., Lee S., Denu J. M. (2009) J. Biol. Chem. 284, 11256–11266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sauve A. A., Celic I., Avalos J., Deng H., Boeke J. D., Schramm V. L. (2001) Biochemistry 40, 15456–15463 [DOI] [PubMed] [Google Scholar]
- 21. Jackson M. D., Denu J. M. (2002) J. Biol. Chem. 277, 18535–18544 [DOI] [PubMed] [Google Scholar]
- 22. Comstock L. R., Denu J. M. (2007) Org. Biomol. Chem. 5, 3087–3091 [DOI] [PubMed] [Google Scholar]
- 23. Kato J., Zhu J., Liu C., Moss J. (2007) Mol. Cell. Biol. 27, 5534–5543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Borra M. T., Denu J. M. (2004) Methods Enzymol. 376, 171–187 [DOI] [PubMed] [Google Scholar]
- 25. Miwa M., Ishihara M., Takishima S., Takasuka N., Maeda M., Yamaizumi Z., Sugimura T., Yokoyama S., Miyazawa T. (1981) J. Biol. Chem. 256, 2916–2921 [PubMed] [Google Scholar]
- 26. Ferro A. M., Oppenheimer N. J. (1978) Proc. Natl. Acad. Sci. U.S.A. 75, 809–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Smith B. C., Denu J. M. (2006) Biochemistry 45, 272–282 [DOI] [PMC free article] [PubMed] [Google Scholar]