Abstract
The nuclear receptor TR4 is a key regulator for many physiological processes, including growth, development, and metabolism. However, how the transcriptional activity of TR4 is regulated in the absence of ligand(s) remains largely unknown. Here we found that an androgen receptor (AR) coactivator, ARA55, might function as a corepressor to suppress TR4 transactivation. Molecular mechanistic dissection with mutation analysis found that ARA55 could enhance TR4 acetylation at the conserved acetylation sites of lysine 175 and lysine 176 in the DNA-binding domain via recruiting proteins with histone acetyl transferase activity, which might then reduce significantly the TR4 DNA binding activity that resulted in the suppression of TR4 transactivation. These results are in contrast to the classic ARA55 coactivator function to enhance AR transactivation partially via increased AR acetylation in the hinge/ligand-binding domain. Together, these results not only provide a novel functional mechanism showing that acetylation of different nuclear receptors at different domains by coregulator may lead to differential receptor transactivation activity but also provide a new way for small molecules to control TR4 transactivation via altering TR4 acetylation levels, and such small molecules may have potential therapeutic applications in the future.
Keywords: Coregulator Transcription, DNA-Protein Interaction, Nuclear Receptors, Post-translational Modification, Transcription Regulation, TR4, Acetylation
Introduction
The androgen receptor (AR)2-associated protein ARA55 is a nuclear receptor coactivator (1, 2) that belongs to the group III LIM domain-containing protein family (3), which can function as molecular adaptors to stabilize protein complexes (4, 5). First identified as an AR coactivator (1, 2), ARA55 could also enhance the transactivation of other nuclear receptors, such as glucocorticoid receptor (6) and peroxisome proliferator-activated receptor γ (7). ARA55 lacks histone acetyltransferase (HAT) or methyltransferase activity (5). However, ARA55, with four LIM domains within its amino terminus, could still be able to influence acetylation on nuclear receptors via recruitment of histone acetyltransferase-containing coactivators, such as CBP and p300 (5). ARA55 also recruits corepressor complexes to glucocorticoid-responsive promoters in the absence of ligands (7), suggesting that ARA55 may be able to coordinate corepressor release, as well as coactivator recruitment upon glucocorticoid ligand stimulation.
An important mechanism by which ARA55 enhances AR transactivation is that ARA55 serves as a bridge protein to recruit HATs, such as CBP/p300 (8). Acetylation of the histones by HATs promotes chromatin structural changes that result in the promotion of gene transactivation (9). CBP/p300 can also acetylate AR on conserved lysines located in the hinge/ligand-binding domain (LBD). A recent genomic study identified 3600 lysine acetylation sites on 1750 proteins (10), suggesting that protein acetylation could be as common as protein phosphorylation to alter or influence protein function (11). Indeed, almost all of the enzymes involved in human or yeast metabolic processes are acetylated (12, 13). The consequence of non-histone protein acetylation is subtle and variable. It can result in either increasing (14–16) or decreasing (17–20) DNA binding affinity and transcriptional activation (21). Such diverse effects on transcription activities could be dependent on the acetylation sites in different functional domains of nuclear receptors that affect DNA binding affinity, protein stability, and receptor-coregulator interaction (19, 21–24). For example, ARA55 can increase the acetylation of AR in the hinge/LBD that results in the increased AR transactivation (22, 25). In contrast, acetylation of FOXO1 in the DNA-binding domain (DBD) results in reduced DNA binding ability (19).
TR4 is a member of the nuclear receptor superfamily (26–28). TR4 null (TR4−/−) mice have significant growth retardation (29), defects in early embryonic development and stem cell self-renewal and differentiation (30), defects in female reproductive function and maternal behavior (29), impaired cerebella function (31–33), reduced sperm production (34), reduced myelination (35), and defects in foam cell formation (36). TR4 has a strong circadian expression in key metabolic tissues, including adipocytes, liver, and muscle (37, 38) and might function as a master regulator to control glucose and lipid metabolism (39, 40). Identified originally as an orphan nuclear receptor without known ligand(s) in 1994, TR4 emerged as a fatty acid sensor with the finding that its transactivation can be activated by polyunsaturated fatty acids metabolites and thiazolidinediones (36, 41). However, how TR4 transcriptional activity is regulated in the absence of ligands still remains largely unclear.
Here we identified ARA55 as a corepressor of TR4 via increasing acetylation levels of TR4 in the DBD, suggesting that the distinct coregulator effects of ARA55 on AR and TR4 could be partly due to diverse effects of non-histone protein acetylation in the different functional domains.
EXPERIMENTAL PROCEDURES
Chemicals and Reagents
Trichostatin A (TSA) was purchased from Calbiochem. Dihydrotestosterone was from Sigma. Anti-FLAG and anti-His antibodies were purchased from Sigma (Sigma). Anti-acetylated lysine antibody was from Cell Signaling Technology. Anti-CD36 and anti-GAPDH antibodies were from Santa Cruz. Anti-ARA55 antibody was purchased from Santa Cruz. Anti-TR4 antibody (antibody 15) was produced as previously reported (29).
Cell Cultures and Transfections
293T cells, H1299 cells, and mouse hepatoma Hep1-6 cells were purchased from ATCC and maintained in Dulbecco's modified Eagle's medium supplemented with 8% FBS. Transient transfection in cells was performed using Superfect (Qiagen) according to the manufacturer's instructions.
Plasmids
pSG5-AR, pSG5-ARA55, pCMX-TR4, Gal4-TR4LBD, and VP16-ARA55 plasmids were constructed as described previously (48). Full-length TR4 and ARA55 cDNAs were subcloned into pCDNA3-flag expression vector to generate flagTR4 and flagARA55, respectively. Full-length TR4 and ARA55 cDNAs were subcloned into pCDNA4HisMax (Invitrogen) at KpnI and XbaI sites to generate HisTR4 and HisARA55, respectively.
Site-directed Mutagenesis
Site-directed mutagenesis on acetylated lysine residues of pCDNA3-flagTR4 or pCMX-TR4 was performed using the QuikChange XL site-directed mutagenesis kit (Stratagene) following the manufacturer's protocol. Replacements of acetylated lysine residues with alanine, glutamic acid, or arginine were made by using mutagenic primers.
RT-PCR
Total RNA was isolated from Hep1-6 cells using a TRIzol (Invitrogen), and RT reaction was conducted using Superscript III reverse transcriptase enzyme (Invitrogen) following the manufacturer's protocol. Real time PCR was applied using SYBR green PCR mix (Qiagen). The specific primers for CD36 were 5′-CTA TGG GGC TGT CAG TTG TG-3′ (sense) and 5′-CTC CTC CAC TGC TAT CTA TC-3′ (antisense), and those for 18 S rRNA were 5′-TGC CTT CCT TGG ATG TGG TAG-3′ (forward) and 5′-CGT CTG CCC TAT CAA CTT TCG-3′ (reverse).
Coimmunoprecipitation
Coimmunoprecipitation-Western blotting analyses were performed as described previously (54). Briefly, the cells were lysed by radioimmune precipitation assay buffer (1× PBS, 0.5% Nonidet P-40, 0.5% sodium desoxycholate, 0.1% SDS, and 1 mm phenylmethylsulfonyl fluoride). The cell lysates were incubated with antibody overnight at 4 °C with constant rocking; 30 μl of protein A/G beads were then added into each tube and incubated for another 1 h with constant rocking. The samples were eluted from the beads and then subjected to Western blotting.
EMSAs
Nuclear extract preparations and EMSAs were carried out as described previously (55). Briefly, 8 μg of nuclear extract protein was incubated in a reaction solution containing 20 mm Tris-HCl, pH 7.9, 2 mm MgCl2, 1 mm EDTA, 50 mm KCl, 0.5 mm dithiothreitol, 10% glycerol, 0.1% Nonidet P-40, and 2 μg of poly(dI-dC) for 20 min. Then the 32P-end-labeled duplex oligonucleotide (2 × 104 cpm) was added, and the reaction was incubated for another 20 min.
ChIP Assay
The ChIP assays were performed as described previously (55). Anti-FLAG antibody (2.0 μg) was added to the lysate and incubated at 4 °C overnight. The primer pairs for the region of the CD36 promoter were 5′-ACC AGA AAT AGA CCC TTG TGA G-3′ (sense) and 5′-GCT CAC AAG GGT CTA TTT CTG G-3′ (antisense), and the nonspecific primer pairs for the region away from CD36 promoter were 5′-GAC CAT ACC TAC CTC TAC CTA C-3′ and 5′-CAG CAT CTA CTG AAG CAT CC-3′ (antisense).
In Vitro Acetylation Assay
The cell lysates were harvested and immunoprecipitated with anti-FLAG antibody. The immunoprecipitate was separated by SDS-PAGE and blotted with anti-acetylated lysine antibody.
RESULTS
ARA55 Enhances AR Transactivation yet Suppresses TR4 Transactivation
Early studies using TR4−/− revealed a variety of physiological functions of TR4 in growth, development, and metabolism (29, 33–35, 40, 42, 43). In the search for coregulators that can alter TR4 transactivation for the potential therapeutic agents to modulate TR4-mediated physiological functions, we identified ARA55, an AR coactivator, which could also interact with TR4 protein. Interestingly, we found that although ARA55 enhanced AR transactivation in 293T cells (Fig. 1A, left panel) in a ligand-dependent manner (1, 2), ARA55 suppressed TR4 transactivation in the same cell line (Fig. 1A, right panel). We further confirmed these contrasting findings by assaying the effect of ARA55 on the TR4 direct target gene CD36 expression in mouse liver Hep1-6 cell line and found TR4 enhanced CD36 mRNA expression. The addition of ARA55 resulted in the suppression of TR4-mediated CD36 mRNA expression (Fig. 1B). Western blot analysis also confirmed the suppression of TR4-induced CD36 expression by ARA55 at protein levels (Fig. 1C). Together, results from Fig. 1 (A–C) demonstrated that ARA55 coregulator could modulate AR and TR4 nuclear receptors in opposite ways.
FIGURE 1.
ARA55 enhances AR transcriptional activity but inhibits TR4 activity. A, ARA55 enhances AR transcriptional activity but inhibits TR4 transcriptional activity. Left panel, 293T cells were cotransfected with pSG5-AR, MMTV-luc, and pCDNA3-flagARA55 as indicated (the ratio of AR to ARA55 was 1:3) using Superfect according to the manual instructions. After 16 h, the cells were treated with vehicle or dihydrotestosterone for an additional day and then harvested for AR luciferase assay. Right panel, 293T cells were cotransfected with pCMX-TR4, HCR-luc, and pcDNA3-flagARA55 as indicated (the ratio of TR4 to ARA55 was 1:3). After 24 h, the cells were harvested, and TR4 luciferase activity was examined. B, ARA55 reduces the mRNA expression level of CD36, a target gene of TR4. Hep1-6 cells were cotransfected with pCDNA3-flagTR4 and pCDNA4-HisARA55 as indicated. After 24 h, the cells were harvested, and CD36 mRNA levels were examined by real time RT-PCR. 18 S rRNA level was used as the internal control, and the CD36 mRNA expression in control cells (with empty vector) was set as 1. The data represent the means ± S.D. of triplicate samples. C, ARA55 reduces the protein level of CD36. Hep1-6 cells were cotransfected with pCDNA3flagTR4 and pCDNA4HisARA55 as indicated. After 24 h, the cells were harvested, and the protein levels of CD36 as well as TR4 and ARA55 were examined by Western blot analysis. GAPDH served as the loading control. DHT, dihydrotestosterone; IB, immunoblot.
ARA55 Suppresses TR4 Transactivation via Acetylation on the TR4 DBD
Early studies documented well that the acetylation of AR at the hinge/LBD is required for the ARAR55-enhanced AR transactivation (22, 25). ARA55 could recruit HATs to enhance AR transactivation, and ARA55 showed little influence on the transactivation of mutant AR that failed to be acetylated (22, 25).
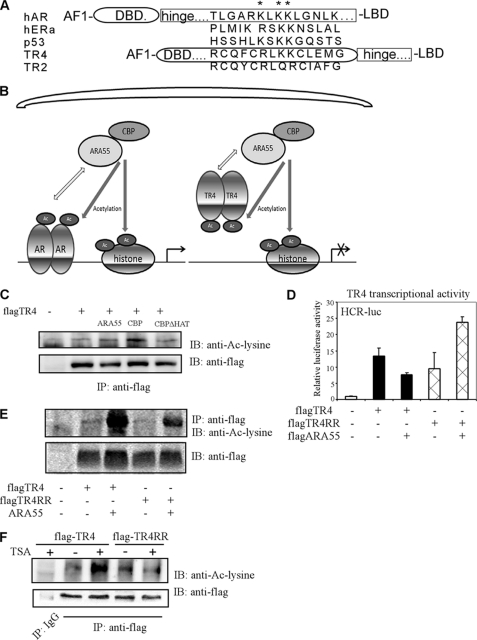
We were interested to see whether the contrasting effects of ARA55 on suppression, instead of promotion of TR4 transactivation could be due to ARA55-mediated acetylation on TR4 at different functional domains. We first examined whether TR4 is an acetylated protein. Sequence analysis found the conserved acetylation motif (RLKK) in many nuclear receptors, including AR, estrogen receptor, and TR4 (Fig. 2A). Interestingly, both AR and estrogen receptor α have their conserved acetylation motif ((R/K)XKK) located in the hinge/LBD, whereas TR4 conserved acetylation motif (RLKK) was found in the DBD. Early studies suggested that acetylation on the DBD of several transcriptional factors, such as FOXO1 (19) and YY1 (20), resulted in the suppression of DNA binding affinity to their target genes. It is therefore possible that acetylation of TR4 on the DBD may also result in the suppression of its transactivation via reduction of DNA binding to its target genes. Here an extremely simplified model to explain this hypothesis is presented in Fig. 2B.
FIGURE 2.
ARA55 suppresses TR4 transactivation via acetylation on the TR4 DBD. A, a sequence comparison of the putative acetylation sites of AR, estrogen receptor α, p53, TR4, and TR2 is shown. TR4 contains potential acetylation motifs in DBD, whereas the AR acetylation motif is in hinge/LBD. TR4 contains the sequence of RLKK, which is consistent with the conserved acetylation motif (R/K)XKK (22). B, the working hypotheses, shown in this extremely simplified model, to explain why ARA55 suppresses activity of TR4 but enhances AR activity. The recruitment of ARA55 brings in HAT activity to acetylate both AR and TR4 but in different domains. TR4 acetylation results in reduced TR4 DNA binding and thus inhibits TR4 activity. C, ARA55 and CBP enhance TR4 acetylation levels. 293T cells were transiently transfected with pCDNA3-flagTR4, and CBP or mutant CBP without HAT activity (CBPΔHAT), and pCDNA4-HisARA55. The cells were harvested, and the cell lysates (500 μg) were immunoprecipitated with anti-FLAG antibody followed by Western blot assay using anti-acetyl-lysine antibody or anti-FLAG antibody. D, TR4 acetylation mutant modulates ARA55 effect on TR4 transcriptional activity. 293T cells were cotransfected with HCR-luc, pcDNA3-flagARA55, and pCDNA3-flagTR4 or pCDNA3-flagTR4RR as indicated (the ratio of TR4 to ARA55 was 1:3). After 24 h, the cells were harvested, and luciferase activities were examined. E, TR4 acetylation mutant impairs ARA55 effect on TR4 acetylation level. 293T cells were cotransfected with pcDNA3-hisARA55, and pCDNA3-flagTR4 or pCDNA3-flagTR4RR as indicated (the ratio of TR4 to ARA55 was 1:9). After 24 h, the cells were harvested, and coimmunoprecipitation assays were performed. F, the HDAC inhibitor TSA enhances TR4 acetylation levels. 293T cells were transiently transfected with pcDNA3-flagTR4 or pCDNA3-flagTR4RR. The cells were then treated with or without 1 nm deacetylase inhibitor TSA for 16 h. The cells were harvested, and the cell lysates (500 μg) were immunoprecipitated with anti-FLAG antibody followed by Western blot assay using anti-acetyl-lysine antibody or anti-FLAG antibody. IP, immunoprecipitation; IB, immunoblot.
To prove this interesting hypothesis, we assayed the influence of ARA55 on TR4 acetylation and transactivation. We found that the addition of ARA55 resulted in the increase of acetylation of wild type TR4 (TR4-WT) (Fig. 2C). We then constructed a TR4 mutant that is resistant to acetylation in DBD yet still maintains positive charges by converting the two lysine residues at 175 and 176 in the conserved acetylated motif sites of TR4 DBD into arginine (TR4-RR). Interestingly, this TR4 DBD mutant TR4-RR significantly impaired the enhanced TR4 acetylation level by ARA55 (Fig. 2E; the ratio of ARA55 to TR4-WT or TR4RR is higher than in Fig. 2C), supporting the hypothesis that ARA55 will modulate TR4 acetylation of the putative acetylation sites in its DBD. Our data also suggested that TR4 might be acetylated in other regions. Consistently, luciferase reporter assay found that adding ARA55 could suppress TR4-WT transactivation but enhance TR4-RR transactivation (Fig. 2D). These results from Fig. 2 (A–E) suggest that ARA55 may suppress TR4 transactivation via reduction of DNA binding by recruiting HATs to acetylate TR4 on the DBD. Failure to acetylate TR4-RR may instead enable ARA55 to induce TR4-RR transactivation via recruitment of HATs to enhance only histone acetylation.
We applied two more approaches to strengthen the above conclusion that TR4 could be acetylated at the TR4 DBD. First, via anti-acetyl lysine antibody assay, we found that the addition of TSA, a HDAC inhibitor, could increase the TR4-WT acetylation. In contrast, TSA showed much less effect on the TR4-RR acetylation (Fig. 2F), suggesting that TR4 could be acetylated at residues 175 and 176 sites of conserved acetylated motif in the TR4 DBD.
We then used another TR4 coregulator CBP to confirm our above finding. Unlike ARA55, which needs to recruit other coregulators with HAT activity, CBP has intrinsic HAT activity (44). Our results showed that TR4 acetylation levels could be enhanced by CBP, but not mutant CBP (CBPΔHAT) that loses HAT activity (Fig. 2C), suggesting that TR4 could be acetylated directly by HATs, such as CBP.
Acetylation of TR4 Suppresses TR4 Transactivation via Reduction of Its DNA Binding Affinity to Its Target Gene
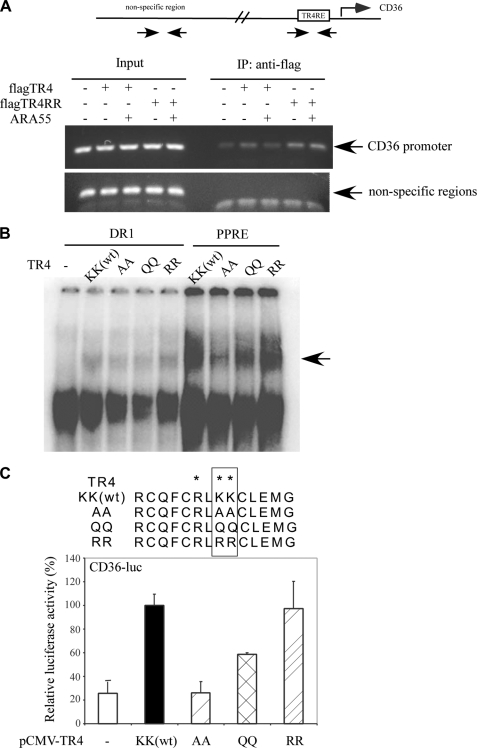
To dissect the molecular mechanism by which acetylation of TR4 in DBD results in the suppression of TR4 transactivation, we examined whether ARA55 could suppress TR4 transcriptional activity via suppression of TR4 DNA binding affinity. Using mammalian two-hybrid (Fig. 3A) and coimmunoprecipitation assays (Fig. 3B), we first proved that ARA55 could directly interact with TR4. We then preformed ChIP assay to determine whether ARA55 could influence TR4 binding to the target gene CD36 promoter in vivo. As shown in Fig. 4A, TR4 bound to CD36 promoter in vivo, and the addition of ARA55 resulted in the suppression of this in vivo binding. In contrast, the addition of ARA55 did not affect TR4-RR binding to CD36 promoter. We then applied EMSA to test whether acetylation influenced TR4 binding to its target genes. As shown in Fig. 4B, both TR4-WT and TR4-RR could bind to TR4 response elements (either DR1 or PPRE). In contrast, the other two TR4 mutants TR4-AA and TR4-QQ, in which we replaced lysine with either alanine or glutamine to mimic the constitutive acetylation form by neutralizing the positive charges within the DBD, bound only weakly to the TR4 response elements. These contrasting results suggested that the neutralization of the lysines basic residues of acetylation motif in the DBD via acetylation may result in the suppression of TR4 DNA binding affinity to its target gene promoters. As expected, the consequence of reduced DNA binding affinity in TR4-AA or TR4-QQ might then result in the suppression of TR4 transactivation (Fig. 4C).
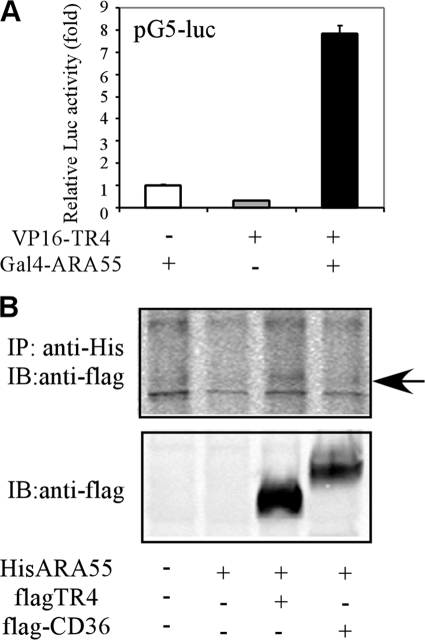
FIGURE 3.
ARA55 interacts with TR4 directly. A, ARA55 interacts with TR4 as examined by mammalian two-hybrid assay. 293T cells were cotransfected with pCMX-VP16-TR4 and/or pCMX-Gal4-ARA55 as indicated. The reporter was pG5-luciferase, and the internal control was pRL-TK. The activity of Gal4-ARA55 was set as 1. B, ARA55 interacts with TR4 as examined by coimmunoprecipitation assay. 293T cells were cotransfected with pCDNA4-HisARA55, pCDNA3flagTR4, and pCDNA3flagCD36, as indicated for 1 day. The cells were then harvested, and the cell lysates were immunoprecipitated with anti-His antibody followed by Western blot assay using anti-FLAG antibody. IP, immunoprecipitation; IB, immunoblot.
FIGURE 4.
Acetylation of TR4 suppresses TR4 transactivation via reduction of its DNA binding ability to its target gene. A, ARA55 reduces TR4 but not TR4RR binding to CD36 promoter in vivo. The ChIP assays were performed in Hep1-6 cells transfected with TR4 or/and ARA55. Anti-FLAG antibody was added to the lysate and incubated at 4 °C overnight. The primer pairs used in the PCR span the region of the CD36 promoter or the nonspecific region. B, TR4 acetylation mutants reduce the binding ability of TR4 to DNA. The 32P-labeled probes DR1 (first through fifth lanes) and PPRE (sixth through ninth lanes) were incubated with or without in vitro translated TR4 protein or TR4 acetylation mutant protein. The complexes were resolved in 4.5% polyacrylamide gels. The specific complex is indicated by an arrow. C, TR4 acetylation mutants reduce TR4 transcriptional activity. 293T cells were cotransfected with CD36-luc, pCMX-TR4, or different pCMX-TR4 acetylation mutant plasmids. After 24 h, the cells were harvested, and luciferase activities were examined. IP, immunoprecipitation.
To further confirm that acetylation of TR4-DBD might contribute to ARA55-mediated suppression of TR4 physiological functions, we assayed the changes of mRNA and protein expression of TR4 target gene CD36 in the TR4 and TR4 mutants. The addition of ARA55 resulted in the suppression of CD36 mRNA and protein expression. In contrast, the addition of ARA55 resulted in TR4-RR-mediated CD36 induction at both mRNA (Fig. 5A) and protein (Fig. 5B) expressions in Hep1-6 cells. Together, results from Figs. 4 and 5 clearly demonstrate that the addition of ARA55 to interact with TR4 might lead to increased TR4 acetylation in the DBD, which might then result in the suppression of TR4-mediated physiological function.
FIGURE 5.
Physiological consequences of TR4 acetylation. A, TR4 acetylation mutant blocks ARA55 effect on the mRNA expression levels of CD36, a target gene of TR4. Hep1-6 cells were cotransfected with pCDNA3-flagTR4 and pCDNA4-flagARA55 as indicated. After 24 h, the cells were harvested, and CD36 mRNA levels were examined by real time RT-PCR. The internal control was 18 S rRNA, and the mRNA expression in control cells (with empty vector) was set as 1. The data represent the means ± S.D. of triplicate samples. B, TR4 acetylation mutant blocks ARA55 effect on the protein expression levels of CD36, a target gene of TR4. Hep1-6 cells were cotransfected with pCDNA3flagTR4 and pCDNA4-flagARA55 as indicated. After 48 h, the cells were harvested, and protein levels of CD36 as well as TR4 and ARA55 were examined by Western blot analysis. GAPDH served as a loading control.
DISCUSSION
Here we revealed that TR4 transactivation could be modulated by acetylation and deacetylation without adding its ligands. Our results are in contrast to the classic coactivator role of ARA55 that promoted AR transactivation via increased acetylation levels. The diverse yet opposite effects of acetylation on the transactivation of different nuclear receptors is believed to be largely dependent on the acetylation sites on the different domains (21). AR is acetylated on lysines located in the hinge/LBD. It was reported that AR acetylation might lead to modulated AR ubiquitination and recruitment of coactivators and corepressors (22, 25). In contrast, TR4 can be acetylated at two conserved lysines that are located within the DBD. Early studies found that acetylation of several transcription factors on the DBD all resulted in a decrease of their DNA binding activity, such as FOXO1 and YY1 (19, 20). Here we showed that these two basic lysine residues in the TR4 DBD might also contribute to the interaction of TR4 with negatively charged phosphate residues in the minor groove of the DNA. Acetylation of TR4 DBD thus reduces TR4 DNA binding affinity by neutralization of the positive charges of these two lysine residues. To our knowledge, TR4 is the first nuclear receptor found to be acetylated in the DBD.
The finding that ARA55 suppressed wild type TR4 but enhanced TR4 acetylation mutant activity suggested that TR4 acetylation plays a dominant role in ARA55-mediated TR4 transactivation. However, we do not rule out other mechanisms by which ARA55 suppresses TR4 transcriptional activities. Coregulators may be able to modulate nuclear receptor transactivation in many ways, for example, via regulating nuclear receptor dimerization, protein stability, and competing with other limited cellular resources (45). Modulation of TR4 transactivation via acetylation could be vital for TR4 in vivo functions. Although lipid ligand/activators can activate TR4 transactivation (36, 41), it remains important to understood how TR4 transactivation is regulated in the absence of ligand(s). FOXO3a (46) and C/EBP (39) could modulate TR4 activity via directly binding to the TR4 promoter. Post-translational modifications, including phosphorylation (47) and acetylation, could modulate TR4 activity in the receptor level.
Early studies using TR4−/− mice revealed that TR4 is essential for a variety of pathological-physiological processes (29, 33–35, 42, 48). Our findings here showed that TR4 can be modulated via acetylation, which provides a platform to utilize the small molecules that can modulate TR4 acetylation to control the diverse biological functions. Currently, several clinical trials using small molecule HDAC inhibitors are in the process of being tested for their effects on cancer, brain disorders, heart diseases, and aging (49–53). HDAC inhibitors have been successful in many clinical trials but also faced significant challenges, such as cardiac side effects (51). TR4 is essential in many physiological processes and shows both beneficial and detrimental effects in animal models. HDAC inhibitors increase TR4 acetylation levels to suppress TR4 activity in vitro. However, it remains to be determined how different HDAC inhibitors affect TR4 activity in vivo in different tissues. HDAC inhibitors have a broad range of targets, with both histones and non-histone proteins, and thus may affect TR4 transactivation in a cell- or tissue-specific manner. TR4 acetylation may also respond differentially to different stress or stimuli or different HDAC inhibitors in different tissues. It is also possible that TR4 acetylation may affect TR4 phosphorylation, TR4-coregulator interactions, or protein stability. Taken together, our results may warrant further study of small molecules or HDAC inhibitors that can influence TR4 acetylation in a tissue-specific manner to battle various TR4-mediated diseases.
This work was supported, in whole or in part, by National Institutes of Health Grant DK06312. This work was also supported by a George Whipple Professorship Endowment and Taiwan Department of Health Clinical Trial and Research Center of Excellence Grant DOH99-TD-B-111-004 (to China Medical University, Taiwan).
- AR
- androgen receptor
- HAT
- histone acetyltransferase
- CBP
- cAMP-responsive element-binding protein-binding protein
- LBD
- ligand-binding domain
- DBD
- DNA-binding domain
- TSA
- trichostatin A
- HDAC
- histone deacetylase.
REFERENCES
- 1. Fujimoto N., Yeh S., Kang H. Y., Inui S., Chang H. C., Mizokami A., Chang C. (1999) J. Biol. Chem. 274, 8316–8321 [DOI] [PubMed] [Google Scholar]
- 2. Rahman M. M., Miyamoto H., Lardy H., Chang C. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 5124–5129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Heitzer M. D., DeFranco D. B. (2007) Steroids 72, 218–220 [DOI] [PubMed] [Google Scholar]
- 4. Jones C. A., Nishiya N., London N. R., Zhu W., Sorensen L. K., Chan A. C., Lim C. J., Chen H., Zhang Q., Schultz P. G., Hayallah A. M., Thomas K. R., Famulok M., Zhang K., Ginsberg M. H., Li D. Y. (2009) Nat. Cell Biol. 11, 1325–1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Heitzer M. D., DeFranco D. B. (2006) Nucl. Recept. Signal. 4, e019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Guerrero-Santoro J., Yang L., Stallcup M. R., DeFranco D. B. (2004) J. Cell. Biochem. 92, 810–819 [DOI] [PubMed] [Google Scholar]
- 7. Drori S., Girnun G. D., Tou L., Szwaya J. D., Mueller E., Xia K., Shivdasani R. A., Spiegelman B. M. (2005) Genes Dev. 19, 362–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Heitzer M. D., DeFranco D. B. (2006) Mol. Endocrinol. 20, 56–64 [DOI] [PubMed] [Google Scholar]
- 9. Wang Z., Zang C., Cui K., Schones D. E., Barski A., Peng W., Zhao K. (2009) Cell 138, 1019–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Choudhary C., Kumar C., Gnad F., Nielsen M. L., Rehman M., Walther T. C., Olsen J. V., Mann M. (2009) Science 325, 834–840 [DOI] [PubMed] [Google Scholar]
- 11. Smith K. T., Workman J. L. (2009) Nat. Biotechnol. 27, 917–919 [DOI] [PubMed] [Google Scholar]
- 12. Zhao S., Xu W., Jiang W., Yu W., Lin Y., Zhang T., Yao J., Zhou L., Zeng Y., Li H., Li Y., Shi J., An W., Hancock S. M., He F., Qin L., Chin J., Yang P., Chen X., Lei Q., Xiong Y., Guan K. L. (2010) Science 327, 1000–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang Q., Zhang Y., Yang C., Xiong H., Lin Y., Yao J., Li H., Xie L., Zhao W., Yao Y., Ning Z. B., Zeng R., Xiong Y., Guan K. L., Zhao S., Zhao G. P. (2010) Science 327, 1004–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gu W., Roeder R. G. (1997) Cell 90, 595–606 [DOI] [PubMed] [Google Scholar]
- 15. Yuan Z. L., Guan Y. J., Chatterjee D., Chin Y. E. (2005) Science 307, 269–273 [DOI] [PubMed] [Google Scholar]
- 16. Marzio G., Wagener C., Gutierrez M. I., Cartwright P., Helin K., Giacca M. (2000) J. Biol. Chem. 275, 10887–10892 [DOI] [PubMed] [Google Scholar]
- 17. Lührs H., Hock R., Schauber J., Weihrauch M., Harrer M., Melcher R., Scheppach W., Bustin M., Menzel T. (2002) Int. J. Cancer 97, 567–573 [DOI] [PubMed] [Google Scholar]
- 18. Munshi N., Merika M., Yie J., Senger K., Chen G., Thanos D. (1998) Mol. Cell 2, 457–467 [DOI] [PubMed] [Google Scholar]
- 19. Matsuzaki H., Daitoku H., Hatta M., Aoyama H., Yoshimochi K., Fukamizu A. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 11278–11283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yao Y. L., Yang W. M., Seto E. (2001) Mol. Cell. Biol. 21, 5979–5991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Glozak M. A., Sengupta N., Zhang X., Seto E. (2005) Gene 363, 15–23 [DOI] [PubMed] [Google Scholar]
- 22. Fu M., Rao M., Wang C., Sakamaki T., Wang J., Di Vizio D., Zhang X., Albanese C., Balk S., Chang C., Fan S., Rosen E., Palvimo J. J., Janne O. A., Muratoglu S., Avantaggiati M. L., Pestell R. G. (2003) Mol. Cell. Biol. 23, 8563–8575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cui Y., Zhang M., Pestell R., Curran E. M., Welshons W. V., Fuqua S. A. (2004) Cancer Res. 64, 9199–9208 [DOI] [PubMed] [Google Scholar]
- 24. Subramanian C., Opipari A. W., Jr., Bian X., Castle V. P., Kwok R. P. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 4842–4847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fu M., Wang C., Wang J., Zhang X., Sakamaki T., Yeung Y. G., Chang C., Hopp T., Fuqua S. A., Jaffray E., Hay R. T., Palvimo J. J., Jänne O. A., Pestell R. G. (2002) Mol. Cell. Biol. 22, 3373–3388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chang C., Da Silva S. L., Ideta R., Lee Y., Yeh S., Burbach J. P. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 6040–6044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lee Y. F., Lee H. J., Chang C. (2002) J. Steroid Biochem. Mol. Biol. 81, 291–308 [DOI] [PubMed] [Google Scholar]
- 28. Giguère V. (1999) Endocr. Rev. 20, 689–725 [DOI] [PubMed] [Google Scholar]
- 29. Collins L. L., Lee Y. F., Heinlein C. A., Liu N. C., Chen Y. T., Shyr C. R., Meshul C. K., Uno H., Platt K. A., Chang C. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 15058–15063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shyr C. R., Kang H. Y., Tsai M. Y., Liu N. C., Ku P. Y., Huang K. E., Chang C. (2009) Endocrinology 150, 2454–2462 [DOI] [PubMed] [Google Scholar]
- 31. Kim Y. S., Harry G. J., Kang H. S., Goulding D., Wine R. N., Kissling G. E., Liao G., Jetten A. M. (2010) Cerebellum 9, 310–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chen Y. T., Collins L. L., Chang S. S., Chang C. (2008) Cerebellum 7, 9–17 [DOI] [PubMed] [Google Scholar]
- 33. Chen Y. T., Collins L. L., Uno H., Chang C. (2005) Mol. Cell. Biol. 25, 2722–2732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mu X., Lee Y. F., Liu N. C., Chen Y. T., Kim E., Shyr C. R., Chang C. (2004) Mol. Cell. Biol. 24, 5887–5899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang Y., Chen Y. T., Xie S., Wang L., Lee Y. F., Chang S. S., Chang C. (2007) Mol. Endocrinol. 21, 908–920 [DOI] [PubMed] [Google Scholar]
- 36. Xie S., Lee Y. F., Kim E., Chen L. M., Ni J., Fang L. Y., Liu S., Lin S. J., Abe J., Berk B., Ho F. M., Chang C. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 13353–13358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yang X., Downes M., Yu R. T., Bookout A. L., He W., Straume M., Mangelsdorf D. J., Evans R. M. (2006) Cell 126, 801–810 [DOI] [PubMed] [Google Scholar]
- 38. Bookout A. L., Jeong Y., Downes M., Yu R. T., Evans R. M., Mangelsdorf D. J. (2006) Cell 126, 789–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Liu N. C., Lin W. J., Yu I. C., Lin H. Y., Liu S., Lee Y. F., Chang C. (2009) Endocrine 36, 211–217 [DOI] [PubMed] [Google Scholar]
- 40. Liu N. C., Lin W. J., Kim E., Collins L. L., Lin H. Y., Yu I. C., Sparks J. D., Chen L. M., Lee Y. F., Chang C. (2007) Diabetes 56, 2901–2909 [DOI] [PubMed] [Google Scholar]
- 41. Tsai N. P., Huq M., Gupta P., Yamamoto K., Kagechika H., Wei L. N. (2009) Biochim. Biophys. Acta 1789, 734–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tanabe O., McPhee D., Kobayashi S., Shen Y., Brandt W., Jiang X., Campbell A. D., Chen Y. T., Chang C., Yamamoto M., Tanimoto K., Engel J. D. (2007) EMBO J. 26, 2295–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kim E., Yang Z., Liu N. C., Chang C. (2005) Biochem. Biophys. Res. Commun. 328, 85–90 [DOI] [PubMed] [Google Scholar]
- 44. Chan H. M., La Thangue N. B. (2001) J. Cell Sci. 114, 2363–2373 [DOI] [PubMed] [Google Scholar]
- 45. McKenna N. J., O'Malley B. W. (2002) Cell 108, 465–474 [DOI] [PubMed] [Google Scholar]
- 46. Li G., Lee Y. F., Liu S., Cai Y., Xie S., Liu N. C., Bao B. Y., Chen Z., Chang C. (2008) Endocrinology 149, 3490–3499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Huq M. D., Gupta P., Yi N. P., Wei L. N. (2006) Mol. Cell. Proteomics 5, 2072–2082 [DOI] [PubMed] [Google Scholar]
- 48. Kim E., Xie S., Yeh S. D., Lee Y. F., Collins L. L., Hu Y. C., Shyr C. R., Mu X. M., Liu N. C., Chen Y. T., Wang P. H., Chang C. (2003) J. Biol. Chem. 278, 46919–46926 [DOI] [PubMed] [Google Scholar]
- 49. Close P., Creppe C., Gillard M., Ladang A., Chapelle J. P., Nguyen L., Chariot A. (2010) Cell. Mol. Life Sci. 67, 1255–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lavu S., Boss O., Elliott P. J., Lambert P. D. (2008) Nat. Rev. Drug Discov. 7, 841–853 [DOI] [PubMed] [Google Scholar]
- 51. Kazantsev A. G., Thompson L. M. (2008) Nat. Rev. Drug Discov. 7, 854–868 [DOI] [PubMed] [Google Scholar]
- 52. McKinsey T. A., Kass D. A. (2007) Nat. Rev. Drug Discov. 6, 617–635 [DOI] [PubMed] [Google Scholar]
- 53. Bolden J. E., Peart M. J., Johnstone R. W. (2006) Nat. Rev. Drug Discov. 5, 769–784 [DOI] [PubMed] [Google Scholar]
- 54. Xie S., Lin H. K., Ni J., Yang L., Wang L., di Sant'Agnese P. A., Chang C. (2004) Prostate 60, 61–67 [DOI] [PubMed] [Google Scholar]
- 55. Yang L., Xie S., Jamaluddin M. S., Altuwaijri S., Ni J., Kim E., Chen Y. T., Hu Y. C., Wang L., Chuang K. H., Wu C. T., Chang C. (2005) J. Biol. Chem. 280, 33558–33565 [DOI] [PubMed] [Google Scholar]