Abstract
Excision of the bacterial group II intron RmInt1 has been demonstrated in vivo, resulting in the formation of both intron lariat and putative intron RNA circles. We show here that the bulged adenosine in domain VI of RmInt1 is required for splicing via the branching pathway, but branch site mutants produce small numbers of RNA molecules in which the first G residue of the intron is linked to the last C residue. Mutations in the coordination loop in domain I reduced splicing efficiency, but branched templates clearly predominated among splicing products. We also found that a single substitution at the EBS3 position (G329C), preventing EBS3-IBS3 pairing, resulted in the production of 50 to 100 times more RNA molecules in which the 5′ and 3′ extremities were joined. We provide evidence that these intron molecules may correspond to both, intron circles linked by a 2′-5′ phosphodiester bond, and tandem, head-to-tail intron copies.
Keywords: Polymerase Chain Reaction (PCR), Reverse Transcription, Ribonuclear Protein (RNP), Ribozyme, RNA Splicing, Mobile Genetic Elements, Group II Introns, Retroelements
Introduction
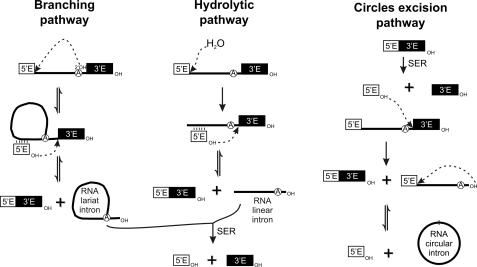
Group II introns are large catalytic RNAs with a conserved secondary structure consisting of six domains arranged around a central wheel (1). Group II introns may self-splice via several different pathways (Fig. 1). The main splicing pathway in group II introns involves two sequential trans-esterification reactions. In the first, the 2′-hydroxyl of a bulged adenosine near the end of the intron, in domain VI, triggers a nucleophilic attack on the 5′ splice site. In the second reaction, the 2′–5′ branched product is released as a lariat when the free 3′-OH at the end of the 5′ exon attacks the 3′ splice junction joining the two exons (2–5). In ai5γ, a yeast mitochondrial group IIB intron, the functional group II intron branch site consists of the bulged adenosine, the wobble pairs (G-U) flanking this residue and, possibly, the purine-enriched sequence in domain VI on the strand opposite the bulged nucleotide (6–8). Recent studies have reported the presence, within domain I of group IIB introns, of a branch site receptor known as a coordination loop (9), which is required for trans-esterification (8), but these findings have been called into question (10). This coordination loop has been identified as a key element for exon ligation, as it positions the exon and intron binding sequences, the EBS3-IBS3 base pair and the EBS1-IBS1 helix, in an appropriate conformation for reaction (9). There is also an alternative splicing pathway, the first step of which involves hydrolysis. The ligated exons and linear intron are then generated in a standard second step (11–13). An additional excision reaction, in which the intron is excised as a putative circle, has also been proposed for some group II introns (14–16). In this case, prior release of the 3′ exon from the precursor molecules, presumably via a trans-splicing mechanism, seems to be required for the generation of a putative 2′-5′ ligated circular intron. The origin and function of such intron circles in vivo remains unknown.
FIGURE 1.
Pathways of group II intron splicing. Linear molecules are shown as solid unbranched lines, lariat molecules are shown as branched lines, and intron circles are shown as circles. Exon sequences are shown as white (5′ exon) or black (3′ exon) boxes. The mechanisms of the different splicing pathways are described in the text.
RmInt1 is a mobile subclass IIB3 intron (17). It has also been assigned to phylogenetic bacterial class D (17, 18) on the basis of the internally encoded (ORF within DIV) reverse transcriptase-maturase (intron-encoded protein (IEP)). This group II intron is excised, both in vivo and in vitro, as an intron lariat (16, 19). No hydrolytic pathway has yet been described in vivo for RmInt1, but the excision of this intron in a linear form has been demonstrated in vitro (19). We have also reported that the RmInt1 group II intron is also excised as putative intron circles in vivo (16).
In this study, we assessed the relevance of the branch point adenosine and the coordination loop, including 3′ exon binding site (EBS3),5 for in vivo excision of the bacterial group II intron RmInt1 and analyzed the production of intron forms in which the 5′ and 3′ extremities of the intron are joined together by a covalent bond. Our findings provide new insights into the possible origin of excised circular intron molecules.
EXPERIMENTAL PROCEDURES
Bacterial Strains, Media, and Growth Conditions
Sinorhizobium meliloti RMO17 was used to characterize the in vivo activities of RmInt1 because this rhizobial strain has no copy of RmInt1 in its genome (20). Escherichia coli DH5α was used for the construction of mutants and cloning. S. meliloti was cultured in TY medium at 28 °C, whereas E. coli strains were grown in LB medium at 37 °C. For plasmid maintenance, we added the following antibiotics as required: 200 μg/ml kanamycin for rhizobia and 50 μg/ml kanamycin for E. coli; 200 μg/ml ampicilin and 10 μg/ml tetracycline, for both types of bacteria.
RmInt1 and Mutant Derivatives
pKGEMA4T7 is a construct similar to pKGEMA4 (20), containing an RmInt1-ΔORF intron with short flanking exons −20/+5 and the IEP expressed from a position upstream from the 5′ exon, but with a phage T7 promoter inserted into the XhoI site engineered in domain IV of the ribozyme. pKG4dVT7 is a mutant derivative of pKGEMA4T7 in which the catalytic triad GTT is replaced by CGA in domain V of the RmInt1 ribozyme. The dVI-ΔA764, dVI-A764C, dVI-pU, EBS3C, EBS3U, EBS3A, and CL2 point mutants were generated by site-directed mutagenesis with the Altered Sites II in Vitro Mutagenesis pAlter-1 System (Promega), with the changes introduced into pALΔORFT7. We generated pALΔORFT7 by inserting the SacI fragment containing ΔORFT7 from pKGEMA4T7 into pAlter-1. The oligonucleotides used for mutagenesis are listed in supplemental Table 1. The final constructs were generated by replacing the ΔORF in pKGEMA4 with a ΔORFT7-containing fragment generated by SacI digestion of the mutated pALΔORFT7 plasmids.
pKG4CL1T7 was constructed by two-step PCR (16), with the Triple MasterTM PCR system (Eppendorf). Two pairs of primers were designed for amplification of the 5′ and 3′ sections of the ribozyme: the SacBbr and CL/DN primers were used to generate the 270-bp upstream fragment; the 435-bp downstream fragment was generated by amplification with the CL/UP and pT7R primers. The mutagenic primers contained an overlapping region of 27 bp and introduced the AGCCC sequence in place of GAGGA at ribozyme positions 222–226. The second PCR step was performed with SacBbr and pT7R primers and generated a 678-bp fragment. The PmlI/XhoI-digested PCR product was inserted in place of the wild-type fragment in pKGEMA4T7. The double mutant was obtained with the same protocol as for pKG4CL2T7, but with pALΔORF-CL1T7 used as the template for mutagenesis.
All six γ-γ′ mutants were obtained by PCR with the AccuprimeTM PCR System (Invitrogen). γ′G was generated by one round of PCR with the 5Q_CAT and γ′G primers. The γC mutant was generated by a two-step PCR in which the 5Q_CAT and γC-rv primers were used to amplify the 5′ region of the ribozyme and the 3′ region was obtained by amplification with the γC-fw and 6QR_LACZ primers. In the second round of PCR, a mixture of these two PCR products was subjected to amplification with the 5Q_CAT and 6QR_LACZ primers. The exchanged γC-γ′G mutant was generated by two-step PCR with the 5Q_CAT and γC-rv primers (5′ region) and the γC-fw and γ′G primers (3′ region). The next round of PCR was performed with the 5Q_CAT and γ′G primers. The EBS3C+γC-γ′G combined mutant was obtained by using an EBS3C mutant ribozyme as the PCR template. All of the mutated ribozymes were then inserted, as PmlI-BlnI fragments, into pICGter-E1E2, which consists of short flanking exons (−20/+5, exon 1 and exon 2, respectively) fused in-frame with the N terminus of the lacZ gene under the control of a constitutive promoter and capped with a custom-designed Rho-independent transcription terminator (21). Mutants derived from pICG required IEP trans-complementation, so we produced this protein constitutively from pKGIEP (22).
RNA Isolation
RNA was extracted from cultures of free-living S. meliloti strain RMO17 containing plasmids carrying the wild-type or mutant RmInt1, as described previously (16). The bacterial cells present in 10 ml of a culture with an A600 of 0.6 units were lysed in the presence of proteinase K (in 1.4% SDS, 4 mm EDTA, 125 μg proteinase K) for 10 min at 65 °C. DNA was eliminated by incubation with 50 units of RNase-free DNase I (Qiagen) and subsequent phenol (pH 4.5)/chloroform/isoamylalcohol extraction. Finally, the RNA was precipitated and resuspended in 25 μl of nuclease-free water.
We minimized the amount of DNA present in the RNA samples by using the RNeasy Mini Kit (Qiagen). RNeasy technology makes it possible to purify high quality RNA rapidly, due to the combination of the stringency of guanidine-isothiocyanate lysis and the efficiency of silica-membrane purification. RNA was isolated according to the manufacturer's instructions. Biological samples were lysed and homogenized. Ethanol was added, and the lysate was loaded onto the RNeasy silica membrane, which efficiently binds RNA, making it possible to wash off all the other contaminants. High quality RNA was eluted in 30 μl of water. This technology efficiently removes most of the DNA without the need for DNase treatment, but certain applications are sensitive to very small amounts of DNA. We therefore removed any residual trace amounts of DNA by RNase-free DNase I digestion (Qiagen) followed by subsequent silica-membrane purification and elution in 30 μl of nuclease-free water. The absence of DNA was verified by PCR.
Primer Extension Assay
Primer extension reactions were performed as described previously (16). We annealed 15 μg of total RNA with 0.2 pmol (300,000 cpm) of (5′-32P)-labeled P primer in 10 mm PIPES (pH 7.5), 400 mm NaCl. This mixture was heated at 85 °C for 5 min, rapidly cooled to 60 °C, and then allowed to cool slowly to 45 °C, over a period of 10 min. Extension reactions were carried out by reverse transcription with AMV reverse transcriptase (Roche Applied Science) at 42 °C for 60 min. Samples were resolved by electrophoresis in a denaturing 6% polyacrylamide gel and quantified with the Quantity One software package (Bio-Rad). The excision efficiency was normalized to the WT levels and calculated according to the intensity of the 97-nt product to compare the results between the different plasmid construct used along the experiment.
RT-PCR
RT-PCR was performed as described previously (16). First-strand cDNA synthesis was initiated by annealing 6.5 μg of total cellular RNA and 25 pmol of a specific primer in the presence of 12.5 mmol dNTPs. We used Ect1 or ICI oligonucleotides for cDNA synthesis. We then added 5× first strand buffer, 0.2 mol DTT, 30 units of RNAguardTM RNase inhibitor (GE Healthcare), and 400 units of SuperScript II RNase H− reverse transcriptase (Invitrogen), and the reaction mixture was incubated at 42 °C for 120 min. We used one-fifteenth of the products of this reaction as a template for PCR with 15 pmol of LL primer, which was designed to be complementary to a sequence 59 nt from the 3′ end of RmInt1, 15 pmol of 5′-radiolabeled P primer (∼150,000 cpm), 6.25 mmol dNTP mix, 50 mm HEPES (pH 7.9), 1.5 mm MgCl2, 50 mm KCl, and 1 unit of Taq polymerase in a final volume of 25 μl. The RT-PCR products (1 μl) were resolved by electrophoresis in a denaturing 6% polycrylamide gel.
The bands subsequently were analyzed by sequencing at least eight different clones. Before sequencing, cloned PCR products derived from templates in which the intron extremities were linked together were selected by PCR with the Iq3′ and P primers.
Quantitative Real-time RT-PCR
We generated the first-strand cDNA from 10 μg of S. meliloti RMO17 total RNA carrying various plasmids, with 12.5 pmol of ICI oligonucleotide used as the primer and 300 units of Superscript II reverse transcriptase RNase H− (Invitrogen), in a final volume of 10 μl (16). We carried out qPCR in the iCycler iQ system (Bio-Rad). We used a 1:10 dilution of the cDNA product, with the addition of 0.2 mm of each specific oligonucleotide primer, SYBR Green I (dilution of 1:10,000) and 0.5 units of Platinum Taq polymerase (Invitrogen), according to the manufacturer's instructions. The following real-time PCR cycling conditions were used for all quantifications: 3 min at 95 °C, followed by 40 cycles of 15 s at 95 °C, 30 s at 68 °C, and 15 s at 72 °C and a final extension phase for 3 min at 72 °C. The templates in which the 5′ and 3′ extremities of the intron were joined together were amplified with the Ect1 and Iq3′ primers. Iq3′ contains the last six nucleotides of the RmInt1 ribozyme, resulting in the amplification principally of the above molecules, with branched (lariat) molecules amplified much less efficiently (additional information in supplemental Fig. 1). Standard dilutions of known quantities of plasmid DNA were included in each run for the generation of quantification curves and the evaluation of amplification efficiency. RNA samples that had not been treated with reverse transcriptase were included to check for DNA contamination. Negative controls in which the template was omitted were also included. In some experiments, amplification was also carried out with the 5Q_CAT and P primers, demonstrating the presence of similar precursor levels in each RNA sample (data not shown). Template amplification was assayed in duplicate in each PCR run, for at least three RNA preparations. A melting curve analysis was performed in all PCR runs to check that there were no primer dimers (data not shown).
RESULTS
Branch Site, Coordination Loop, and EBS3 Mutants
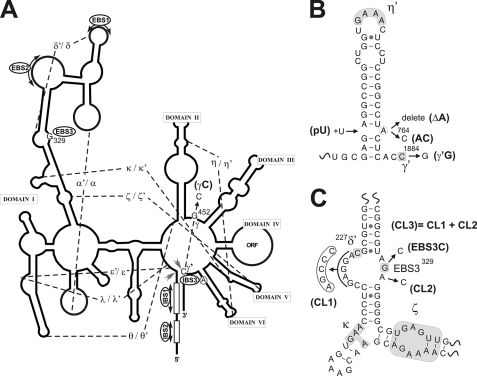
We evaluated the role of the predicted coordination loop, including the EBS3, and the branch site adenosine (Fig. 2A), in RmInt1 excision reaction in vivo, by constructing a series of mutants in pKGEMA4T7. This plasmid contains an RmInt1-ΔORF intron and short flanking exons −20/+5, with a phage T7 promoter inserted in domain IV and the IEP expressed from a position upstream from the 5′ exon. Three mutations were introduced into the branch site: deletion of the adenosine (ΔA764), its replacement by a C residue, and the pairing of this residue with a uridine residue inserted on the other strand of DVI (Fig. 2B), hereafter referred to as ΔA, A764C, and pU, respectively. We also introduced mutations into the coordination loop: one mutation affecting either side of the loop sequence and a third mutation affecting both sides (Fig. 2C), equivalent to the coordination loop mutants generated for in vitro studies in ai5γ (8). The 5′ side loop mutation (CL1) includes nucleotides 222–226 and results in the replacement of a GAGGA sequence with AGCCC. This mutation prevents base pairing between the two sides of the loop. The mutation on the 3′ side of the loop sequence (CL2) involved the replacement of the A in position 330 with a C residue. A combined mutant (CL3) in which both sides of the loop were affected was also generated. Finally, G329 (EBS3) was replaced separately with C, U, and A residues, preventing EBS3-IBS3 base pairing. None of these mutations altered the identity of nucleotide δ′ (C227) that presumably form a canonical pair with nucleotide δ (G266), immediately 5′ of EBS1, interaction that facilitate base pairing between the 5′ exon and the intron (23).
FIGURE 2.
Mutagenic approach to investigation of the RmInt1 excision mechanism. A, schematic secondary structure of the RmInt1 ribozyme is represented as a solid line, with tertiary contacts indicated by dashed lines and Greek letters; solid gray arrows show the 5′ and 3′ splice sites, and the circled A represents the branch point adenosine in domain VI. The γG position, located in the J2/3 region, and EBS3G are highlighted. B, diagram of domain VI, showing the set of mutants analyzed: branch point deletion (ΔA), bulged A-to-C transversion (A764C), adenosine pairing (pU), and intron last nucleotide C-to-G transversion (γ′G). C, schematic diagram of the coordination loop; tertiary interactions are shown surrounded by shaded boxes. The mutants are identified as follows: 5′ loop mutant (CL1), 3′ side single mutant (CL2), and the combined mutant (CL3) containing both mutations. EBS3 mutants are also shown.
Effects of Mutation in RmInt1 Excision Reaction in Vivo
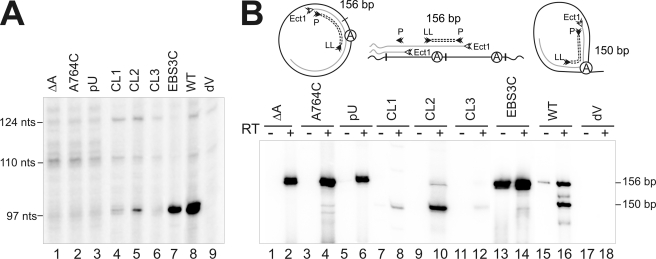
We assessed the efficiency of the intron excision reaction by carrying out primer extension analyses with an oligonucleotide sequence (see supplemental Table 1) complementary to a sequence located 80–97 nt from the 5′ end of the intron (Fig. 3A). Assays with total RNA from S. meliloti RMO17 expressing the RmInt1 intron from pKGEMA4T7 showed a major, strong 97-nt extension product corresponding to excised intron RNA (Fig. 3A, lane 8). This 97-nt band was not detected with RNA from cells harboring pKG4dVT7 (Fig. 3A, lane 9), in which the ribozyme carries mutations affecting the critical conserved AGC-GUU pairing of the intron RNA domain V (GUU→CGA). Similarly, all of the branch site point mutations (Fig. 3A, lanes 1–3) resulted in a lack of detectable excision, even after long periods of exposure, suggesting an inability of the intron to branch efficiently and to form a lariat. These branch site substitutions are known to prevent branching and lead to the relatively inefficient production of linear intron molecules in other introns (6). Nonetheless, no linear intron RNA was detected in RmInt1 branch site mutants by primer extension, 5′-RNA ligase-mediated rapid amplification of cDNA 5′ ends (RLM-RACE) or cRT-PCR. By contrast to branch point mutants, some of the putative branch site receptor mutants gave a band corresponding to the excision product. The CL2 mutant, in which the 3′ side of the loop was affected, displayed ∼8% wild-type splicing activity (Fig. 3A, lane 5), whereas the 5′-loop mutant CL1 (Fig. 3A, lane 4) displayed a stronger splicing defect (∼4% wild-type). The band generated on intron excision was not detected in the combined mutant, CL3 (Fig. 3A, lane 6). These results are consistent with those reported for similar mutations in the coordination loop of ai5γ in vitro (8). However, we cannot rule out the possibility that the branch site receptor mutations have effects on the structure of the ribozyme leading to the splicing defect observed in vivo for RmInt1, rather than specifically affecting the branching reaction.
FIGURE 3.
Effect of branch site, coordination loop, and EBS3 mutations on the splicing reaction in vivo. A, proteinase K-treated total RNA from the S. meliloti RMO17 strain harboring the wild-type intron pKGEMA4T7, the splicing-defective mutant pKG4dVT7, and the corresponding mutants was reverse transcribed with an intron-specific primer (P). The splicing defect of mutants EBS3U and A was similar to that of the EBS3C mutant (not shown). The 97-nt cDNA products obtained from excised intron RNA are indicated, together with the more slowly migrating bands corresponding to unspliced precursor derived transcripts (110 and 124 nt), which appear to be processed at specific 5′U residues as indicated by RACE mapping. B, the products of RmInt1 in vivo were detected by reverse transcription and PCR. Reactions were carried out with (+) and without (−) prior RT of RNA from S. meliloti RMO17 harboring pKGEMA4T7, pKG4dVT7, and domain I and branch-site mutations. The sizes of the identified products are indicated in bp; a schematic representation of the PCR products for the intron lariat, head-to-tail intron dimers, and intron circles is also shown, in which the circled A corresponds to the bulged adenosine in domain VI.
The EBS3-IBS3 interaction stabilizes and aligns the 3′ exon during the second step of splicing. Substitutions affecting EBS3, the binding site for the 3′ exon, have been shown to have minimal effects on the structure of the loop but strongly inhibit the second step of the splicing reaction (23). On the other hand, it has been suggested that the branch site and the 3′ exon bind the coordination loop independently (8). The EBS3C, U, and A mutations (Fig. 3A, lane 7 and data not shown) resulted in significantly lower levels of excision product (∼30–40% wild-type), which nonetheless remained higher than those for the other loop mutants. According to previous observations, our results would involve a different type of impairment of lariat formation in the EBS3 mutant. Although the CL mutants could derivate from structural destabilization that reduces the efficiency of the first step of the splicing reaction, the EBS3 mutants presumably display inhibition of the second step. Branching reaction is highly reversible (24, 25), which requires that the second step of splicing serve as a kinetic trap (25). Thus, the impairment in the second step of splicing in the EBS3 mutant could contribute to the reversibility in the first step giving rise to a reduction of excision product detected by primer extension.
Detection of RmInt1 Products in Vivo by Reverse Transcription and PCR
In a previous work, we demonstrated, by RT-PCR, the existence in vivo of RmInt1 RNA molecules compatible with circular intron forms in which the first G residue of the intron is linked to its last C residue (16). RT-PCR assays (Fig. 3B) were carried out to determine the ability of the mutants to generate these intron forms. Two major RT-PCR products of 156 and 150 nt were observed in cells expressing the wild-type intron from pKGEMA4T7 (Fig. 3B, lane 16), but not in cells expressing the splicing-defective mutant pKG4dVT7 (Fig. 3B, lane 18). The more slowly migrating 156-nt band corresponds to the amplification of templates in which the 5′and 3′ extremities of the intron are joined together, whereas the 150-nt RT-PCR product is derived from a substrate consisting of an intron lariat in which the branch point is the bulged adenosine in domain VI (16). The intron lariat and putative intron circles molecules seem to have different hydrodynamic properties that may be influenced by both the type of folding of the different excised intron molecules and a distinctive interaction of these forms with the IEP (16). Although the degradation rate of both intron molecules is currently unknown, we observed that the amount of putative circles decreased when cells were collected at stationary phase, whereas the levels of lariat were unchanged or slightly raised (data not shown). For this reason, our experiments were carried out at early logarithmic phase. Thus, the conclusions in the current work rest on the assumption that degradation rates are unchanged.
RT-PCR analysis revealed the presence of the larger RT-PCR product in the three branch-site mutants (Fig. 3B, lanes 2, 4, and 6), whereas the product derived from the lariat was barely detectable only in the A764C mutant (lane 4), consistent with the low efficiency of cytosine as the branch point nucleotide demonstrated both in vitro and in vivo in other introns (6, 26, 27). The larger RT-PCR products of ΔA, A764C, and pU mutants were cloned and sequenced. All the clones displayed the expected linkage between the first and last residue of the intron. Therefore, we conclude that the production of RNA molecules with joined 5′ and 3′ intron ends is independent of the branching reaction.
By contrast to what was observed for the branch-site mutations, RT-PCR assays showed the product of the lariat to be present in all coordination loop mutants (Fig. 3B, lanes 8, 10, and 12) at different levels, consistent with the amount of product detected on primer extension analysis. In addition, the product derived from RNA molecules with joined intron extremities was detectable only in the CL2 mutant, suggesting that these intron species may be produced inefficiently by these mutants.
PCR and RT-PCR assays revealed the presence of both DNA (Fig. 3B, lane 13) and RNA (Fig. 3B, lane 14) templates with linked 5′ and 3′ intron extremities in the EBS3C mutant, whereas the product derived from the intron lariat is no longer detectable. All cloned 156 nt RT-PCR products displayed the expected linkage between the first G residue of the intron and its last C residue. These results are particularly striking because the extension product (Fig. 3A, lane 7) is thought to be mostly derived from intron lariat molecules. We have shown previously that primer extension assays using RNA and RNP particles from S. meliloti cells expressing the wild-type intron revealed two extension products that differ by one nucleotide (97–98 nt) relative to the position expected for the 5′ end of the intron, with the shorter product as the major one (16). Interestingly, primer extension performed on a circular intron has been reported to result in two extension products that differ by one nucleotide, with the longer product as the major one (14). Furthermore, reverse transcriptase is known to pause after reading through the 2′–5′ linkage in a nonlariat RNA (28), whereas with lariat RNA, the pause occurs before reading through the 2′–5′ linkage (14). Thus, one possible interpretation of our data is that the extension product observed in the EBS3C mutant could be mostly the larger 98-nt product and that the mutant intron might excised in vivo as putative intron circles.
Quantification of Intron Species with Linked 5′ and 3′ Ends
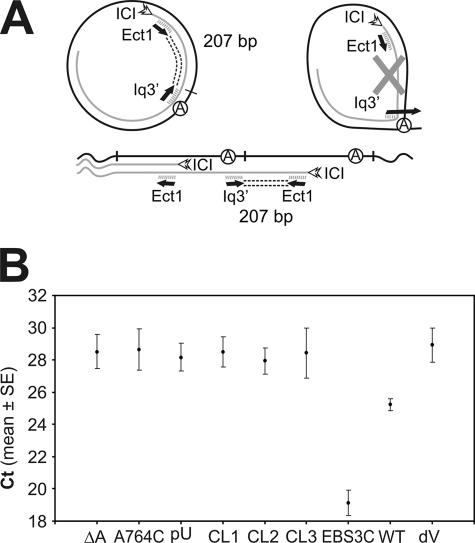
We further investigated the effect of branch site, coordination loop, and EBS3 mutations in the production of RNA molecules with linked 5′ and 3′ intron ends, by carrying out quantitative real-time RT-PCR with the Ect1 and Iq3′ primers (Fig. 4A), on a cDNA template synthesized by the reverse transcription of total RNA with the intron-specific ICI primer. As the 3′ end of the Iq3′ primer is complementary to the last six nucleotides of the intron (tail of the lariat molecule), this amplification generates a 207-bp product from intron RNA templates with linked ends (circular introns or tandem, head-to-tail intron copies) but not from a lariat template (see also supplemental Fig. 1). Our results indicate that 50–100 times more intron RNA molecules with linked intron 5′ and 3′ ends are produced by the EBS3C mutant than by the wild-type intron, whereas the number of such molecules are an order of magnitude lower and close to background levels (dV mutant) for branching and coordination loop mutants (Fig. 4B). Consistent with the results of RT-PCR assays, qPCR in the absence of reverse transcription generated only one-third to one-half as many DNA templates with linked intron extremities as qPCR of equivalent RNA templates in the EBS3C mutant (see supplemental Fig. 2). These results clearly indicate that the replacement with a C residue of EBS3G boosts the production of products compatible with a circular template, or tandem intron copies, whereas branch site and coordination loop mutations behave differently, decreasing the production of these intron species.
FIGURE 4.
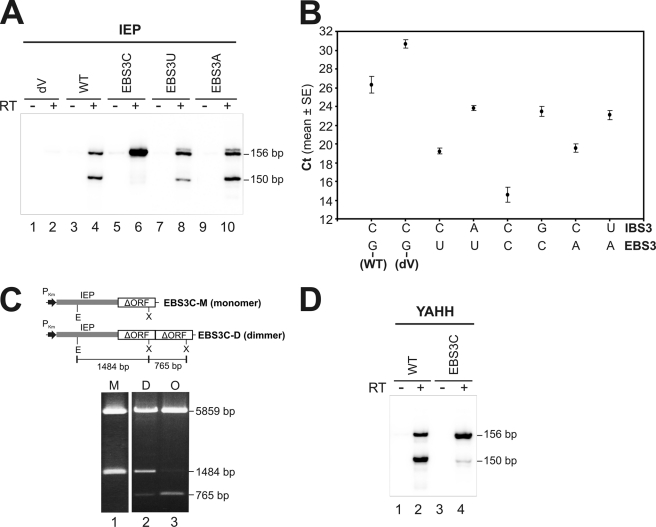
Quantitative real-time RT-PCR data analysis of RmInt1 products in which the 5′ and 3′ extremities are linked. A, schematic diagram of the expected PCR products. cDNA was synthesized from S. meliloti RMO17 total RNA carrying various plasmid constructs, with the ICI oligonucleotide primer. qPCR was performed in the iCycler iQ system (Bio-Rad), with the Ect1 and Iq3′ primers and SYBR Green I. Amplification generates a 207-bp product from intron RNA templates with linked extremities, whether in the form of circular introns or tandem, head-to-tail intron copies, but not from a lariat template. B, amplification levels of the 207-bp product for WT, coordination loop, EBS3, branch site, and domain V mutants. The graph shows the threshold cycle (Ct) for the indicated constructs. The Ct is the point at which the fluorescence intensity of the reaction first exceeds the background level. The data shown are means ± S.E. of at least two replicates in 96-well plates and three independent reactions, each with a different RNA preparation. Three main groups can be distinguished: the WT group, Ct ≈ 25 (∼2.7 × 104 cDNA copies/50 ng total RNA); the putative branch site-docking group, Ct ≈ 28.36 (∼103 cDNA copies/50 ng total RNA), which includes splicing-defective domain I and domain VI mutants, with levels close to the background level displayed by the domain V mutant (Ct ≈ 29); and the EBS3 group, Ct ≈ 19 (∼2.6 × 106 cDNA copies/50 ng total RNA). (qPCR parameters: E > 92% and R2 > 0.991). DNA standard curves were generated for quantification, and melting curve analysis was carried out in all PCR runs, to check that there were no primer dimers (not shown). RNA samples not treated with reverse transcriptase and negative controls containing no template were included to monitor the reactions (not shown).
Nature of Intron Forms in EBS3 Mutants
Our results demonstrate that the replacement of the nucleotide in intron position EBS3G by a C residue increases the abundance of intron products in which the first G residue of the intron is linked to the last C residue. We therefore investigated whether the phenotypes associated with EBS3 substitution were nucleotide-specific. In contrast to the EBS3C mutation, the RT-PCR product derived from lariat intron is clearly detected in both the EBS3U and EBS3A mutants (Fig. 5A, lanes 8 and 10), which suggests a lower abundance of intron RNA molecules with linked 5′ and 3′ ends. That this was the case was confirmed by qPCR assays (Fig. 5B), which show that these intron species are increased by ∼100-fold in the EBS3C mutant, but by 25- and 37-fold in the EBS3U and EBS3A mutants, respectively. As expected for a Watson-Crick pair, when the EBS3-IBS3 base pairing was reestablished by compensatory mutations at IBS3, the production of these intron forms showed a significant reduction. Thus, one possible interpretation of our data were that in addition to putative intron circles, for the EBS3C mutant, these intron products may result from EBS3 (C) being complementary to the first intron nucleotide (G), making it possible for the intron to undergo reverse splicing at the junction with the 5′ exon, resulting in the production of tandem head-to-tail intron copies. We tested this hypothesis, by isolating and analyzing the intron constructs harbored by the EBS3C mutant. Plasmids from four of 13 colonies analyzed showed a restriction profile compatible with head-to-tail intron copies (Fig. 5C), which was further confirmed by DNA sequencing. These findings indicate that the EBS3C mutant is able to reverse splice efficiently into the intron-insertion site in the 5′ exon (DNA target), resulting in the formation of stable tandem intron copies potentially generating DNA and RNA intron molecules with linked 5′ and 3′ ends, intron species that would not be detected by primer extension, but would be detected in RT-PCR assays. Another evidence that supports the existence of head-to-tail intron DNA molecules is that the PCR product obtained in the absence of reverse transcription is no longer detectable by using RNA isolation methods (see “Experimental Procedures”) that efficiently eliminated most of the DNA from the RNA samples (Fig. 5A, lane 5 and compare with Fig. 3B, lane 13).
FIGURE 5.
Effect of the different EBS3 substitutions on the splicing reaction in vivo. A, RT-PCR was performed with (+) and without (−) prior RT of RNA from S. meliloti RMO17 harboring intron constructs bearing various EBS3 substitutions and a wild-type IEP: the EBS3G (WT) was replaced with C, U, and A residues. B, RT-qPCR analysis of RmInt1 molecules with linked 5′ and 3′ ends is shown in different EBS3-IBS3 combined mutants. The EBS3G (WT) was replaced with U, C, and A nucleotides as well as the compensatory mutations were introduced into the IBS3 position to restore the interaction. The threshold cycles were plotted as the mean ± S.E. The background level is displayed by the splicing-defective mutant dV. The different EBS3 mutations exhibit a differential increase in the amount of 5′-3′-linked intron extremities molecules, being the EBSC mutant which highly swells these levels. The rate of these molecules is reduced when a compensatory change in the IBS3 is introduced. C, the EBS3C mutant can generate intron dimers. Head-to-tail intron copies were found when 13 colonies from an EBS3C mutant were analyzed. The EcoRI/XhoI digestions of three clones representative of the three profiles observed are shown: clones with only one copy of the ΔORF intron (M, monomer), others formed from ribozyme dimers (D), and others that may correspond to intron oligomers (O). Schematic diagrams of the various constructs are drawn to scale at the top. PKm, the promoter of the Km resistance gene, is represented as a black arrow; the gray box corresponds to the IEP; white boxes represent the ribozyme; E, EcoRI; and X, XhoI restriction sites. D, total RNA from S. meliloti RMO17 harboring intron constructs with the EBS3G (WT) or the EBS3C substitution and a RT-deficient IEP mutant (YAHH) were assayed by RT-PCR.
We investigated whether the EBS3C mutation also allowed the intron to attack the 5′ splice site (RNA target) efficiently, by introducing this mutation into a construct carrying nucleotide substitutions in the IEP (YAHH mutant) that abolish reverse transcriptase activity and thus intron mobility (22). RT-PCR assays and DNA sequencing revealed that this combined mutant had a phenotype similar to the single mutant EBS3C, with a greater abundance of the 156-nt product with the expected 5′ and 3′ intron junction (Fig. 5D), which was further confirmed by qPCR (data not shown). We therefore conclude that the replacement of EBS3 with a C nucleotide leads the intron to attack both the DNA and RNA sites efficiently, resulting in the production of stable (DNA) or transient (RNA) tandem head-to-tail intron copies, respectively.
Involvement of γ-γ′ Pairing in Intron Forms with Linked 5′ and 3′ Ends
It has been suggested that free 5′ exon attacks the 3′ splice site in intron circle formation, thereby releasing the downstream intron terminus while splicing to the 3′ exon. In addition to EBS3, base pairing (γ-γ′) between the last nucleotide (γ′) of the intron and the γ nucleotide, which is located in the J2/3 segment connecting domains II and III, is required for an efficient second splicing step (29, 30). Toor et al. (31) recently showed that the γ nucleotide was located near the domain V bulge, positioning the 3′ splice site for catalysis. We investigated the nature of intron species with linked 5′ and 3′ intron ends, by first checking the effect of γ and γ′ mutations in the production of these intron forms. The RmInt1 ΔORF-intron and mutant derivatives were analyzed with pICG constructs carrying a Rho-independent transcription terminator complemented with the IEP expressed in trans (21).
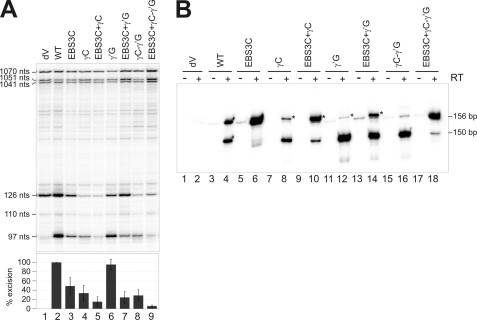
Two mutations were introduced to prevent tertiary Watson-Crick γ-γ′ pairing (Fig. 2A and B): replacement of the γ nucleotide G452 with a C residue in the J2/3 region (hereafter referred to as the γC mutant) and replacement of the γ′ nucleotide C1884 with a G residue (hereafter referred to as the γ′G mutant). A third mutant was also constructed to restore γ-γ′ pairing, but with a C452-G1884 combination. In addition, the EBS3C mutation was introduced into each of these three mutants. As for pKGEMA4T7 constructs, the EBS3C mutation in pICG significantly decreased intron excision (49% wild-type, Fig. 6A) and RT-PCR assays showed the production of larger amounts of the 156-nt product (Fig. 6B, lane 6).
FIGURE 6.
Effect of γ-γ′ mutations on the splicing reaction in vivo. A, total RNA from the S. meliloti RMO17 strain harboring the wild-type intron (pICGterΔORF/pKGIEP), the splicing-defective mutant dV and the corresponding mutants, was reverse-transcribed with an intron-specific primer (P). The cDNA products obtained from excised intron RNA (97 nt) and unspliced precursor derived transcripts (110 and 126 nt) are indicated, together with the more slowly migrating bands corresponding to a characteristic pattern of primary precursor transcripts (1041, 1051, and 1070 nt). Splicing efficiency is plotted as a percentage of wild-type values, with error bars indicating S.D. B, RmInt1-derived products were identified by RT-PCR. PCR was carried out with (+) and without (−) prior RT of RNA from S. meliloti RMO17 harboring the indicated constructs. Total RNA was isolated with the RNeasy Mini Kit (Qiagen), which minimizes the amount of DNA present in the RNA samples. The type of product amplified is indicated (156 and 150 bp, respectively). The products of the mutants marked with an asterisk have an extra cytosine residue at the junction.
The γC mutation was associated with 34% wild-type splicing activity (Fig. 6A, lane 4), probably due to blockage of the second step of splicing. As expected, the EBS3C+γC double mutant (Fig. 6A, lane 5) displayed a stronger splicing defect (15% wild-type) and produced larger amounts of the 156-nt RT-PCR product (Fig. 6B, lane 10). Surprisingly, the γ′G mutation had no significant effect on intron splicing activity (95% wild-type, Fig. 6A, lane 6), and the main signal obtained in RT-PCR assays corresponded to the lariat-derived product (Fig. 6B, lane 12). One possible explanation for these findings is that, unlike the γC mutation, the wild-type γG452 residue in the γ′G mutant may establish a compatible interaction that restores equivalent γ-γ′ pairing at cryptic 3′ splice sites (30). Note that the RmInt1 3′ splice site is located in the midst of a run of three C residues. As expected, the EBS3C+γ′G double mutant displayed a strong splicing defect (24% wild-type, Fig. 6A, lane 7), with an increase in the amount of the 156-nt RT-PCR product, although the lariat-derived product remained the major signal detected by RT-PCR (Fig. 6B, lane 14).
The putative restoration of γ-γ′ pairing with the C452-G1884 combination was not accompanied by a recovery of the splicing efficiency of the intron, which remained at 29% of wild-type values (Fig. 6A, lane 8) or even lower (6% wild-type, lane 9) in the EBS3C+γC-γ′G combined mutant. Thus, the γG452 residue appears to be required for wild-type splicing activity. As observed with the γC and γ′G mutations, RT-PCR assays for the γC-γ′G mutant indicated no increase in the production of the 156-nt product (Fig. 6B, lane 16) but, as expected, the RT-PCR assay (lane 18) showed the levels of intron species with linked 5′ and 3′ extremities be higher after the introduction of the EBS3C mutation.
Evidence of Intron Species with a 2′–5′ Phosphodiester Bond between Last and First Residue of Intron
It has been reported (16, 32, 33) that, in the products amplified from the lariat intron, adenosine is incorporated instead of thymidine when the reverse transcriptase encounters the branched adenosine nucleotide, indicative of a 2′-5′ junction. Similarly, a branched G nucleotide causes reverse transcriptase to incorporate a guanosine residue in place of a cytosine residue (34). As our results showed that the γ′G mutation allows the splicing and production of intron species with linked 5′ and 3′ extremities, we investigated the nature of the junction by sequencing the cloned RT-PCR products. Three of the 38 clones from the γ′G mutant analyzed were indeed derived from these intron forms, whereas the remaining clones corresponded to lariat forms. Two had an extra cytosine residue at the site of the junction, and one was of the expected size but for this last clone, we observed misincorporation of a guanosine residue in place of the cytosine residue when the reverse transcriptase encountered the terminal nucleotide (G) of the intron at the junction. Similarly, eight of 24 clones from the EBS3C+γ′G mutant analyzed were derived from intron species with linked ends. Six had the extra cytosine residue at the junction, and two were of the expected size but, again, for these two clones, we observed misincorporation of a guanosine residue at the terminal nucleotide (G) of the intron. These results suggest that the intron species generated with joint ends have a 2′–5′ phosphodiester linkage, which may correspond to intron RNA circles. By contrast, however, in the EBS3C+γC-γ′G mutant, 11 of 24 cloned RT-PCR products derived from the 156-nt product had the expected junction between the last and first residues of the intron, but a cytosine residue was incorporated by the reverse transcriptase when it encountered the terminal nucleotide of the intron (G). The more plausible explanation of these results is that, in the EBS3C+γC-γ′G mutant the predominant RT-PCR products of 156 nt were derived from tandem, head-to-tail intron templates.
DISCUSSION
We previously reported that RmInt1 is excised in vivo both as an intron lariat and as putative intron circles (16). These conclusions were based on RT-PCR data, but it is not possible, on the basis of these assays alone, to determine whether the templates correspond to genuinely circular intron forms or, alternatively, result from the production of tandem, head-to-tail intron copies. However, the second of these two possibilities was considered unlikely because these intron forms were also generated in the absence of general recombination and by an intron carrying an IEP mutation that abolishes reverse transcriptase activity and thus mobility. We show here that mutations in the predicted coordination loop of RmInt1 lead to intron excision behavior different from that observed for nucleotide substitutions at the branch site adenosine and that intron forms with joined 5′ and 3′ extremities are produced independently of the branching reaction. The replacement of EBS3 with nucleotides preventing the EBS3-IBS3 interaction increases the abundance of intron products in which the first G residue of the intron is linked to the last C residue. These intron forms are likely produced as a result of intron circularization with a 2′–5′ phosphodiester bond at the junction as well as a consequence of intron reverse splicing into either DNA sites (stable intron dimers) or RNA sites (transient intron dimers), generating head-to-tail intron molecules, by pairing EBS3 (C) with the first intron nucleotide (G).
The coordination loop has been identified as a possible receptor for branch site docking, which is required for trans-esterification, and it has been suggested that the conformational state of the intron core is fixed early and persists throughout the entire splicing process (8). If the coordination loop is the binding site for the branch point, we would expect mutations in this loop to result in a phenotype similar to that for nucleotide substitutions at the branch site. However, for RmInt1, branch site substitutions prevent branching, whereas the coordination loop mutants CL1 and CL2 display moderate to strong effects on splicing, but branched templates clearly predominate among splicing products. Thus, we cannot rule out the possibility that the branch site receptor mutations have effects on the structure of the ribozyme leading to the splicing defect observed in vivo for RmInt1. In addition, intron molecules with linked 5′ and 3′ ends are produced in branch site mutants in the absence of lariat formation, suggesting that the generation of such linked products is independent of the branching reaction. Nonetheless, mutations affecting either the branch site or the coordination loop significantly decrease the production of these intron forms (by a factor of 10). It would therefore seem that the correct structure and proper folding of the intron is required for wild-type production of these intron-derived products.
Branch site substitutions are known to prevent branching and to lead to the relatively inefficient production of linear intron molecules in other introns. It has been reported that the linear group II intron ai5γ readily reverses the second step of splicing with high yield and precision in vitro (35). However, we have no evidence that RmInt1 generates excised linear intron molecules in vivo: no linear intron RNA was detected by 5′RLM-RACE, primer extension or cRT-PCR. In addition, none of the RmInt1 branch site and coordination loop mutants showed retrohoming capability (only barely detectable in the A764C mutant that produces some lariat molecules) into DNA recipient target sites (data not shown). It is highly significant that the RT-PCR products derived from intron forms with linked 5′ and 3′ ends generated by γ′G mutants contained an abnormality, in that a G residue was incorporated instead of a C nucleotide when reverse transcriptase encountered the terminal G residue of the intron at the junction. These findings suggest the presence of a 2′-5′ junction and may therefore represent intron circles (14). It therefore seems that these intron forms with linked ends produced by RmInt1 may result as suggested previously (14) from intron molecules that undergo cleavage at the 3′ splice site rather than the first step of the branching pathway, by a trans-splicing reaction triggered by 5′ exon molecules generated by the so-called spliced exon reopening reaction. After release of the 3′ exon, the 2′-OH of the terminal cytosine residue attacks the 5′ splice site, releasing the 5′ exon and an intron circle with a 2′–5′ phosphodiester bond at the junction.
Primer extension analysis revealed that the EBS3C, U, and A mutants had a low splicing efficiency, probably due to blockage of the second step of splicing. Surprisingly, RT-PCR assays revealed the presence of templates with linked 5′ and 3′ intron extremities in the EBS3C mutant, whereas the product derived from the intron lariat is no longer detectable. In addition, RT-qPCR assays revealed that the EBS3C mutant generated ∼100 times as many of these intron forms as the wild-type intron. Furthermore, analysis of the plasmid content of the EBS3C mutant revealed the presence of dimeric and oligomeric head-to-tail species. The simplest interpretation of the data is that these intron forms are derived from intron RNA circles linked by a 2′–5′ phosphodiester bond (detected by primer extension) and head-to-tail intron molecules (revealed by RT-PCR). These findings reveal the extraordinary capability of RmInt1 to spread into both RNA and DNA target sites.
Supplementary Material
Acknowledgments
We thank Isabel Chillón Gázquez for assistance with RT-qPCR analysis and Francisco Martínez-Abarca for critical reading of this manuscript. We thank P. S. Perlman for suggestions to improve this work.
This work was supported by grants from the Spanish Ministerio de Ciencia e Innovación (BIO2008-00740 and CSD 2009-0006 of Programme Consolider-Ingenio 2010), Junta de Andalucía (CVI-01522), and Fondo Europeo de Desarrollo Regional (funding for open access charge from Programme Consolider-Ingenio 2010 from the Spanish Ministerio de Ciencia e Innovación).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1 and Figs. 1 and 2.
- EBS3
- 3′ exon binding site
- qPCR
- quantitative PCR
- RLM-RACE
- RNA ligase-mediated rapid amplification of cDNA 5′ ends
- nt
- nucleotide
- IEP
- intron-encoded protein.
REFERENCES
- 1. Pyle A. M. (2008) Ribozymes and RNA Catalysis, pp. 201–228, RCS Publishing, Cambridge, UK [Google Scholar]
- 2. van der Veen R., Arnberg A. C., van der Horst G., Bonen L., Tabak H. F., Grivell L. A. (1986) Cell 44, 225–234 [DOI] [PubMed] [Google Scholar]
- 3. Peebles C. L., Perlman P. S., Mecklenburg K. L., Petrillo M. L., Tabor J. H., Jarrell K. A., Cheng H. L. (1986) Cell 44, 213–223 [DOI] [PubMed] [Google Scholar]
- 4. Michel F., Ferat J. L. (1995) Annu. Rev. Biochem. 64, 435–461 [DOI] [PubMed] [Google Scholar]
- 5. Pyle A. M. (1996) Nucleic Acids and Molecular Biology, pp. 75–107, Springer-Verlag, Berlin, Germany [Google Scholar]
- 6. Podar M., Chu V. T., Pyle A. M., Perlman P. S. (1998) Nature 391, 915–918 [DOI] [PubMed] [Google Scholar]
- 7. Chu V. T., Adamidi C., Liu Q., Perlman P. S., Pyle A. M. (2001) EMBO J. 20, 6866–6876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hamill S., Pyle A. M. (2006) Mol. Cell 23, 831–840 [DOI] [PubMed] [Google Scholar]
- 9. de Lencastre A., Hamill S., Pyle A. M. (2005) Nat. Struct. Mol. Biol. 12, 626–627 [DOI] [PubMed] [Google Scholar]
- 10. Michel F., Costa M., Westhof E. (2009) Trends Biochem. Sci. 34, 189–199 [DOI] [PubMed] [Google Scholar]
- 11. van der Veen R., Kwakman J. H., Grivell L. A. (1987) EMBO J. 6, 3827–3831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jarrell K. A., Dietrich R. C., Perlman P. S. (1988) Mol. Cell. Biol. 8, 2361–2366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Daniels D. L., Michels W. J., Jr., Pyle A. M. (1996) J. Mol. Biol. 256, 31–49 [DOI] [PubMed] [Google Scholar]
- 14. Murray H. L., Mikheeva S., Coljee V. W., Turczyk B. M., Donahue W. F., Bar-Shalom A., Jarrell K. A. (2001) Mol. Cell 8, 201–211 [DOI] [PubMed] [Google Scholar]
- 15. Li-Pook-Than J., Bonen L. (2006) Nucleic Acids Res. 34, 2782–2790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Molina-Sánchez M. D., Martinez-Abarca F., Toro N. (2006) J. Biol. Chem. 281, 28737–28744 [DOI] [PubMed] [Google Scholar]
- 17. Toro N. (2003) Environ. Microbiol. 5, 143–151 [DOI] [PubMed] [Google Scholar]
- 18. Zimmerly S., Hausner G., Wu Xc. (2001) Nucleic Acids Res. 29, 1238–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Costa M., Michel F., Molina-Sánchez M. D., Martinez-Abarca F., Toro N. (2006) Biochimie 88, 711–717 [DOI] [PubMed] [Google Scholar]
- 20. Nisa-Martínez R., Jiménez-Zurdo J. I., Martínez-Abarca F., Muñoz-Adelantado E., Toro N. (2007) Nucleic Acids Res. 35, 214–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chillón I., Martínez-Abarca F., Toro N. (2011) Nucleic Acids Res. 39, 1095–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Muñoz-Adelantado E., San Filippo J., Martínez-Abarca F., García-Rodríguez F. M., Lambowitz A. M., Toro N. (2003) J. Mol. Biol. 327, 931–943 [DOI] [PubMed] [Google Scholar]
- 23. Costa M., Michel F., Westhof E. (2000) EMBO J. 19, 5007–5018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Augustin S., Müller M. W., Schweyen R. J. (1990) Nature 343, 383–386 [DOI] [PubMed] [Google Scholar]
- 25. Chin K., Pyle A. M. (1995) RNA 1, 391–406 [PMC free article] [PubMed] [Google Scholar]
- 26. Query C. C., Strobel S. A., Sharp P. A. (1996) EMBO J. 15, 1392–1402 [PMC free article] [PubMed] [Google Scholar]
- 27. Liu Q., Green J. B., Khodadadi A., Haeberli P., Beigelman L., Pyle A. M. (1997) J. Mol. Biol. 267, 163–171 [DOI] [PubMed] [Google Scholar]
- 28. Lorsch J. R., Bartel D. P., Szostak J. W. (1995) Nucleic Acids Res. 23, 2811–2814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jacquier A., Michel F. (1990) J. Mol. Biol. 213, 437–447 [DOI] [PubMed] [Google Scholar]
- 30. Jacquier A., Jacquesson-Breuleux N. (1991) J. Mol. Biol. 219, 415–428 [DOI] [PubMed] [Google Scholar]
- 31. Toor N., Keating K. S., Taylor S. D., Pyle A. M. (2008) Science 320, 77–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Vogel J., Hess W. R., Börner T. (1997) Nucleic Acids Res. 25, 2030–2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Vogel J., Börner T. (2002) EMBO J. 21, 3794–3803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nyberg T. M. (2006) In Vivo Studies of Yeast Mitochondrial Intron Splicing: Ectopic Branching and a Screen for Nuclear Encoded Splicing Factors. Doctoral dissertation, University of Texas Southwestern Medical Center, Dallas [Google Scholar]
- 35. Roitzsch M., Pyle A. M. (2009) RNA 15, 473–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.