Abstract
Acquired chemoresistance not only blunts anticancer therapy but may also promote cancer cell migration and metastasis. Our previous studies have revealed that acquired tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) resistance in lung cancer cells is associated with Akt-mediated stabilization of cellular caspase 8 and Fas-associated death domain (FADD)-like apoptosis regulator-like inhibitory protein (c-FLIP) and myeloid cell leukemia 1 (Mcl-1). In this report, we show that cells with acquired TRAIL resistance have significantly increased capacities in migration and invasion. By gene expression screening, tissue transglutaminase (TGM2) was identified as one of the genes with the highest expression increase in TRAIL-resistant cells. Suppressing TGM2 dramatically alleviated TRAIL resistance and cell migration, suggesting that TGM2 contributes to these two phenotypes in TRAIL-resistant cells. TGM2-mediated TRAIL resistance is likely through c-FLIP because TGM2 suppression significantly reduced c-FLIP but not Mcl-1 expression. The expression of matrix metalloproteinase 9 (MMP-9) was suppressed when TGM2 was inhibited, suggesting that TGM2 potentiates cell migration through up-regulating MMP-9 expression. We found that EGF receptor (EGFR) was highly active in the TRAIL-resistant cells, and suppression of EGFR dramatically reduced TGM2 expression. We further determined JNK and ERK, but not Akt and NF-κB, are responsible for EGFR-mediated TGM2 expression. These results identify a novel pathway that involves EGFR, MAPK (JNK and ERK), and TGM2 for acquired TRAIL resistance and cell migration in lung cancer cells. Because TGM2 couples TRAIL resistance and cell migration, it could be a molecular target for circumventing acquired chemoresistance and metastasis in lung cancer.
Keywords: Akt PKB, Cell Migration, Drug Resistance, ERK, JNK, EGFR, Lung Cancer, TRAIL, Tissue Transglutaminase
Introduction
Anticancer chemotherapy is one of the main approaches for treating patients with late stage lung cancer. Commonly used anticancer drugs kill cancer cells mainly through induction of apoptosis. However, cancer cells readily escape this cytotoxic mechanism of therapeutics. Many cancer cells are primarily resistant to apoptosis, which is usually due to down-regulation of the apoptosis pathways and/or activation of cell survival signals through genetic and epigenetic aberrations acquired during transformation (1, 2). In addition, cancer cells acquire apoptosis resistance during chemotherapy (secondary or acquired apoptosis resistance) (3–5). Acquired apoptosis resistance has been an increasing concern because it has emerged as an important mechanism underlying acquired chemoresistance (6, 7). Indeed, acquired apoptosis resistance is detrimental because it not only dampens the anticancer activity of the drugs but also promotes cancer progression and metastasis (8). For example, when acquired resistance to TNF-related apoptosis-inducing ligand (TRAIL)-induced apoptosis arises, instead of killing the cancer cells, TRAIL3 promotes proliferation and metastasis in the apoptosis-resistant cancer cells, converting TRAIL from a cancer killer to a cancer promoter during therapy (9, 10). Therefore, it is crucial to understand the mechanism of acquired apoptosis resistance and its associated cancer cell metastasis to retain the cancer-killing activity while circumventing the cancer promoting potential of chemotherapeutics such as TRAIL.
Due to its selective cytotoxicity in transformed cells, TRAIL is regarded as the most promising anticancer agent in the TNF superfamily of cytokines (11). TRAIL as well as TRAIL receptor agonist antibodies are currently tested as anticancer agents in clinical trials for treating solid tumors, including lung cancer (12, 13). TRAIL kills cancer cells mainly through activating the extrinsic apoptosis pathway via ligation to its functional receptors, death receptor 4 (DR4)/TRAIL-R1 and DR5/TRAIL-R2 (11). TRAIL-induced apoptosis is executed through recruitment of caspase-8 to the death-inducing signal complex and subsequent activation of downstream effectors caspase-3 and -7 (14). The apoptotic signal can be amplified through caspase-8-mediated cleavages of the BH3-only Bcl-2 family member BID that activates the mitochondrial apoptosis pathway (14). However, there are mechanisms that keep the TRAIL-induced apoptosis pathway in check, which are adapted by cancer cells to escape from the cytotoxicity of TRAIL. For example, the activation of the transcription factor nuclear factor-κB (NF-κB) by DR4 or DR5 signaling prevents the initiation of apoptosis (15, 16). Decoy receptor 1 (DcR1)/TRAIL-R3 and DcR2/TRAIL-R4 and osteoprotegerin compete with the functional receptors to blunt TRAIL-induced apoptosis (17).
The molecular basis for acquired TRAIL resistance has not been well elucidated (4, 5, 18). We have recently established acquired TRAIL resistance in lung cancer cell lines by continuously exposing the TRAIL-sensitive lung cancer cells to nontoxic doses and gradually increasing the concentrations of TRAIL (19, 20). With these cells, we have demonstrated that the acquired TRAIL resistance was associated with Akt-mediated stabilization of c-FLIP and Mcl-1 and overexpression of cyclooxygenase 2 (COX-2) (19, 20).
Although apoptosis resistance-associated metastasis has been widely observed, the underlying mechanisms for which, particularly for TRAIL resistance-associated cancer cell migration and invasion, are poorly studied (10, 21). Although cancer metastasis consists of multiple steps involving invading the extracellular matrix and migrating from the primary site to distal sites, migration and invasion represent the important early processes of this malignant behavior of cancer (22, 23). Understanding how acquired TRAIL resistance-associated cancer cell migration and invasion holds a key for prevention of this acquired detrimental effect of TRAIL in cancer therapy.
In this report, we determined that the lung cancer cells with acquired TRAIL resistance have significantly increased migration and invasion capacities. By gene expression screening, tissue transglutaminase (tTG, also called TGM2) was identified as one of the genes with the highest expression increase in TRAIL-resistant cells. Further experiments demonstrate that TGM2 plays an important role in both TRAIL resistance and cell migration. The results reveal a novel pathway consisting of EGFR, MAPK (JNK and ERK), and TGM2 that promotes TRAIL resistance through c-FLIP and migration/invasion through MMP-9, and suggest TGM2 as a molecular target for circumventing acquired chemoresistance and metastasis because this protein couples TRAIL resistance and cell migration in lung cancer.
MATERIALS AND METHODS
Reagents
Glutathione S-transferase (GST)-TRAIL was prepared as described previously (15, 24). Cystamine was from Sigma-Aldrich. EGFR inhibitor III (a selective and irreversible inhibitor that blocks EGFR autophosphorylation), U0126, SP600125, SB203580, IKK inhibitor II, and LY294002 were purchased from Calbiochem. Small interfering RNA (siRNA; SiGenome SMARTpool) for TGM2, MMP-9, EGFR, and negative control siRNA were purchased from Dharmacon. The TGM2 siRNAs targeting the 3′-UTR of the TGM2 mRNA were synthesized by Integrated DNA Technologies. The targeted sequences are as follows: TGM2 siRNA-2, 5′-CTCCTCTCTCTAAGCCTCAGTCTCC-3′; TGM2 siRNA-4, 5′-GGACAGAAGGTGGTCACAGTCATGG-3′. Matrigel was from BD Biosciences. The following antibodies were used for Western blot: anti-TGM2 (Santa Cruz Biotechnology), anti-β-tubulin and β-actin (Sigma-Aldrich); anti-MMP-1, MMP-2, and MMP-9 (Calbiochem); anti-c-FLIP and Mcl-1 (Alexis Laboratories); anti-phospho-EGFR (Y1086, Abcam), anti-EGFR, anti-ERK, anti-Akt, and anti-phospho-Akt (Ser-473) (Cell Signaling Technology); anti-phospho-ERK and phospho-JNK (BIOSOURCE); and anti-JNK1 (BD Biosciences). The TGM2-expressing vector pCDNA-TGM2 was provided generously by Dr. Gail V. W. Johnson from the University of Rochester (25).
Cell Culture
The human lung cancer cell lines A549, H460, and H1568 were obtained from American Type Culture Collection (Manassas, VA) and grown in RPMI 1640 medium supplemented with 10% FBS, 1 mm glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin. A549-TRAIL-resistant (TR), H460-TR, and H1568-TR cells were established as described previously (19, 20) and were maintained in the same growth medium supplemented with TRAIL. The parental wild-type cell lines were designated as A549-WT, H460-WT, and H1568-WT, respectively. For ectopic expression of TGM2, A549-WT and -TR cells were transfected with pCDNA-TGM2, stable transfectant clones were selected and maintained in medium containing G418 (200 μg/ml).
RNA Interference
A549- or H1568-TR cells were seeded in a six-well plate the day before transfection at 30–50% confluency. siRNA was transfected with INTERFERinTM siRNA transfection reagent (Polyplus-transfection). Forty-eight hours after transfection, TGM2, MMP-9, and EGFR protein levels were measured by Western blot.
Migration Wound Healing Assay
A549- or H1568-WT TR cells, cystamine-treated or -untreated TR cells, and NC or TGM2 or MMP-9 siRNA-transfected TR cells were grown to 90% confluency in RPMI 1640 medium with 10% FBS, and then they were incubated with medium containing 2% FBS overnight. The cultured cells were scratched using a 200-μl pipette tip and rinsed with PBS twice to remove free floating cells and debris. After being incubated with medium containing 10% FBS, the cells were visualized by light microscopy at 0 and 16 h after scratch. The experiments were repeated three times.
Migration Transwell Assay
To assess cell migration in vitro, A549 or H1568-WT TR cells, cystamine-treated or -untreated TR cells, and NC or TGM2 or MMP-9 siRNA-transfected TR cells (2 × 104 in 100 μl serum-free medium) were placed in the top chamber of transwell migration chambers (pore size, 5 μm; Corning Co.). The lower chamber was filled with 600 μl of medium with 2% FBS. After 6 h, cells that had not migrated to the lower chamber were wiped away from the upper surface of the transwell membrane with a cotton swab. Migrated cells on the lower membrane surface were fixed, stained, photographed, and counted. The experiment was repeated at least three times. The migration index was calculated by taking the migrated cell number of the control as 1.
Invasion Transwell Assay
In vitro invasion assays were done in Matrigel-coated transwells. A549 or H1568-WT TR cells, cystamine-treated or -untreated TR cells, and NC or TGM2 or MMP-9 siRNA-transfected TR cells (5 × 104 in 200 μl serum-free medium) were put in the top chamber, whereas the lower chamber was filled with 600 μl of medium with 10% FBS as chemoattractant. After 24 h, cells that had not invaded to the lower chamber were wiped away from the upper surface of the transwell membrane with a cotton swab. Invaded cells on the lower membrane surface were fixed, stained, photographed, and counted. The invasion index was calculated by taking the invaded cell number of the control sample as 1.
RT-PCR
Total RNA was extracted with the RNeasy kit (Qiagen). Two microgram of RNA from each sample was used as a template for cDNA synthesis with a reverse transcription kit (Promega). An equal volume of cDNA product was used in the PCR. The primers were used as follows: TGM2, 5′-TCCTCTCTGGGCCTTTGTTTCCTT-3′ (forward primer) and 5′-TATGGCTTAAGGCTTCGTGGAGCA-3′ (reverse primer); β-actin, 5′-CCAGCCTTCCTTCCTGGGCAT-3′ (forward primer) and 5′-AGGAGCAATGATCTTGATCTTCATT-3′ (reverse primer). The reaction condition was as follows: 95 °C for 2 min, 94 °C for 45 s, 55 °C for 45 s, and 72 °C for 45 s and after indicated cycles, 72 °C for 6 min. For TGM2, the PCR cycles were 30, whereas for β-actin, the cycles were 22. PCR products were resolved in 2% agarose gels with 0.5 μg/ml ethidium bromide, visualized, and photographed.
Western Blot
Total cell protein was extracted in M2 buffer (20 mm Tris-HCl, pH 7.6, 0.5% Nonidet P-40, 250 mm NaCl, 3 mm EDTA, 3 mm EGTA, 2 mm dithiothreitol, 0.5 mm phenylmethylsulfonyl fluoride, 20 mm glycerophosphate, 1 mm sodium vanadate, and 1 μg/ml leupeptin). Equal amounts of cell proteins were resolved in 12% SDS-polyacrylamide gels and then transferred to PVDF membranes. The proteins were visualized by enhanced chemiluminescence reagent according to the manufacturer's instructions (GE Healthcare). The intensity of the individual bands was quantified by densitometry (NIH ImageJ, version 1.62) and normalized to the corresponding input control (β-actin or β-tubulin) bands. Fold changes were calculated with the control taken as 1.
Cytotoxicity Assay
Cytotoxicity was determined using a lactate dehydrogenase release-base cytotoxicity detection kit (Promega). Cells were seeded in 48-well plates at 70 to 80% confluency, cultured overnight, and then treated as indicated in the figure legends. Lactate dehydrogenase release was determined, and cell death was calculated as described previously (19, 26). The experiments were repeated three times, and representative results are shown in the figures.
Statistics
Data were expressed as mean ± S.D. Statistical significance was examined by one-way analysis of variance. In all analyses, p < 0.05 was considered statistically significant.
RESULTS
Lung Cancer Cells with Acquired TRAIL Resistance Have Elevated Migration and Invasion Capacities
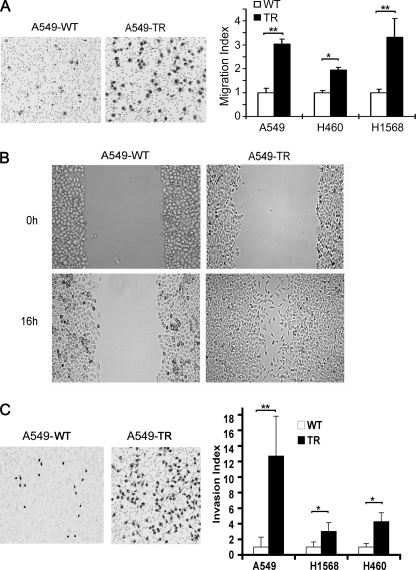
Cancer cells with chemoresistance may have increased migration and metastasis potential (8, 9). Our previous studies have established lung cancer cells with acquired TRAIL resistance by treating the cells with chronic exposure to increasing concentrations of TRAIL and have determined a pathway involving the cell survival kinase Akt, caspase inhibitor c-FLIP, the prosurvival Bcl-2 family member Mcl-1 and COX-2 that contribute to the cell's resistance to TRAIL-induced apoptosis (19, 20). To investigate whether the cells with acquired TRAIL resistance (TR) have increased migration potential, a migration transwell assay was performed. Compared with A549-WT, A549-TR cells exhibited a 3.0-fold increase of cells migrating through the membrane (Fig. 1A, left). To substantiate this observation, a wound healing migration assay was employed, which consistently showed that A549-TR cells had higher migration potential than A549-WT cells (Fig. 1B). Similarly, compared with their respective WT cells, H460-TR and H1568-TR cells showed a 1.9- and 3.3-fold increases in migration, respectively (Fig. 1A, right). Because tumor cell invasiveness relies heavily on cell migration (27), we further examined whether the TR cells have elevated invasion potential with an in vitro Matrigel invasion transwell assay. Significantly more A549-TR (12.7-fold) than A549-WT cells (1-fold) invaded the Matrigel (Fig. 1C, left). Notably, there was no difference in cell proliferation between A549-WT and A549-TR cells (data not shown), suggesting that the increased cell migration and invasion ability in A549-TR cells was unlikely due to change of cell proliferation rates. Similarly, H1568-TR and H460-TR cells had 3.0- and 4.1-fold higher invasion compared with their respective WT cells, respectively (Fig. 1C, right). These results suggest that the lung cancer cells with acquired TRAIL resistance have elevated capacities in migration and invasion.
FIGURE 1.
Increased migration and invasion capacities in the cells with acquired TRAIL resistance. A, migration assay. WT and TR of A549, H460, and H1568 cells (2 × 104 in 100 μl of serum-free medium) were seeded in the top chamber of transwell chambers for migration for 6 h (see “Materials and Methods”). Representative images of migrated A549-WT and -TR cells are shown (left). Quantification of cell migration of each cell line is shown (right). Data shown are mean ± S.D. *, p < 0.05; **, p < 0.01. B, wound healing assay. Cell cultures in confluence were scratched and photographed immediately after scratching the wound (0 h) or 16 h post scratching. Representative images are shown. C, invasion assay. A549-, H460-, and H1568-WT or TR cells (5 × 104 in 200 μl serum-free medium) were seeded in the top chamber that contained a layer of Matrigel, whereas the lower chamber contained medium with 10% FBS as chemoattractant. After incubation for 24 h, invaded cells on the lower membrane surface were detected. Representative images of migrated A549-WT and -TR cells are shown (left). Quantification of cell migration of each line is shown (right). Data shown are mean ± S.D. *, p < 0.05; **, p < 0.01.
TGM2 Expression Is Significantly Increased in TR Cells
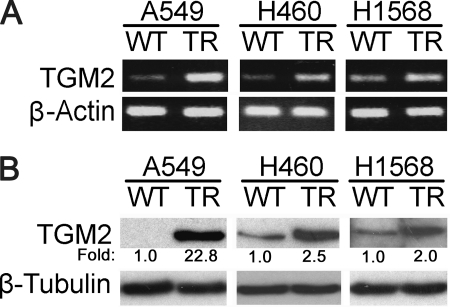
To explore the molecular basis of increased cell migration and invasion in the TRAIL-resistant cells, a gene expression microarray assay was employed for comparing the H460-TR to H460-WT cells. TGM2 was identified to be one of the genes that had the most significant expression increase (19-fold) in H460-TR cells (data not shown). To validate the result, RT-PCR and Western blot were used to detect TGM2 mRNA and TGM2 protein expression, respectively. In agreement with the gene expression microarray results, TGM2 mRNA and protein levels were remarkably higher in A549-, H460-, and H1568-TR cells than their respective WT cells (Fig. 2, A and B). Because TGM2 was reportedly involved in regulation of apoptosis and cell migration (28), we focused on this multifunctional factor in this study.
FIGURE 2.
Increased TGM2 expression in A549-TR, H460-TR, and H1568-TR cells. A, equal amounts of total RNA from the indicated cells were detected for TGM2 mRNA expression by RT-PCR. β-Actin was detected as an input control. B, equal amounts of cell extract from the indicated cells were used for detection of TGM2 protein expression by Western blot. β-Tubulin was detected as an input control. Fold changes were calculated with the control taken as 1.
TGM2 Is Involved in Migration and Invasion in TR Cells
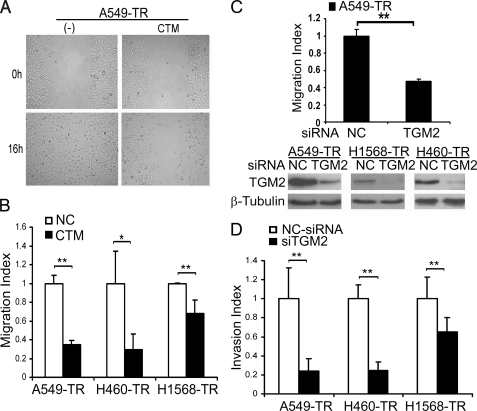
We then investigated the involvement of TGM2 in the migration and invasion of the TR cell. The TGM2 inhibitor cystamine was first used to suppress TGM2 activity. Cystamine markedly suppressed A549-TR cell migration, which was detected with both the wound healing and transwell migration assays (Fig. 3, A and B). A similar inhibitory effect of cystamine on migration in H1568-TR and H460 cells was also detected (Fig. 3B). To substantiate this result, TGM2 siRNA was used to deplete TGM2 from the TR cells (Fig. 3C and supplemental Fig. S1). Consistently, knockdown of TGM2 expression effectively blocked A549-TR cell migration (Fig. 3C and supplemental Fig. S1). A similar inhibitory effect of TGM2 expression suppression on A549-TR, H460, and H1568-TR cell invasion was observed (Fig. 3D). Notably, the inhibitory effect of TGM2 suppression with either cystamine or siRNA on cell migration and invasion was much stronger in A549-TR than in H1568-TR cells (Fig. 3, B and D), which was well correlated with the extent of TGM2 expression increase (Fig. 2). Furthermore, ectopic overexpression of TGM2 in A549-WT cells significantly increased cell migration and invasion (supplemental Fig. S2). More importantly, the migration suppression by TGM2 knockdown was effectively alleviated when TGM2 expression in A549-TR cells was restored by ectopic expression with a TGM2-expressing vector that escaped the TGM2 siRNA targeting (supplemental Fig. S3). These results strongly suggest that TGM2 plays an important role in the increased migration and invasion in the TR cells.
FIGURE 3.
TGM2 contributes to increased migration and invasion in the cells with acquired TRAIL resistance. A, A549-TR cells treated or untreated with cystamine (CTM; 1 mm) for 24 h before wound healing assay as described in the legend to Fig. 1. Representative images are shown. B, A549- or H1568-TR cells were treated with cystamine (1 mm) for 24 h before the migration assay as described in the legend to Fig. 1. Data shown are the mean ± S.D. **, p < 0.01. C, A549-TR cells transfected with negative control siRNA (NC-siRNA) or TGM2 siRNA (pool). The migration was performed 48 h post transfection (top). Data shown are the mean ± S.D. **, p < 0.01. Knockdown of TGM2 expression was confirmed by Western blot (bottom). D, invasion assay. The cells were transfected with indicated siRNA as described in C, and cell invasion was detected as described in the legend to Fig. 1. Data shown are mean ± S.D. *, p < 0.05.
TGM2-mediated MMP-9 Expression Contributes To Cell Migration and Invasion
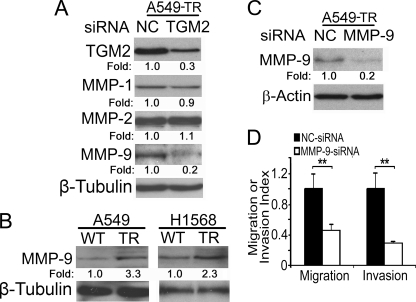
One of the early steps in cancer cell metastasis is to penetrate the extracellular matrix, a process involving matrix metalloproteinases (MMPs). Previous reports suggested that MMP-1, MMP-2, and MMP-9 are involved in human lung cancer cell metastasis (29). We then explored which MMP is regulated by TGM2 in TR cells. When TGM2 expression was knocked down, the expression of MMP-9 but not MMP-1 and -2 was substantially suppressed (Fig. 4A). Indeed, MMP-9 protein expression was markedly higher in A549-TR and H1568-TR cells compared with their respective WT cells (Fig. 4B). Knockdown of MMP-9 expression in A549-TR cells dramatically inhibited migration and invasion (Fig. 4, C and D, and supplemental Fig. S4). These results suggest that TGM2 regulates TR cell migration and invasion at least partly through MMP-9.
FIGURE 4.
MMP-9 is involved in TGM2-mediated migration and invasion in the TRAIL-resistant cells. A, A549-TR cells were transfected with negative control siRNA or TGM2-siRNA and incubated for 48 h. Equal amounts of cell extracts were detected for TGM2, MMP-1, MMP-2, and MMP-9 protein expression by Western blot. β-Tubulin was detected as an input control. B, equal amounts of cell extracts from the indicated cells were detected for MMP-9 protein expression by Western blot. β-Tubulin was detected as an input control. C, A549-TR cells were transfected with NC siRNA and MMP-9 siRNA. Equal amounts of cell extracts were detected for MMP-9 protein expression by Western blot. β-Actin was detected as a loading control. Fold changes were calculated with the control taken as 1. D, migration and invasion assays were performed as described under “Materials and Methods.” Data shown are mean ± S.D. **, p < 0.01.
Suppressing TGM2 Attenuates TRAIL Resistance by Down-regulating c-FLIPL Expression
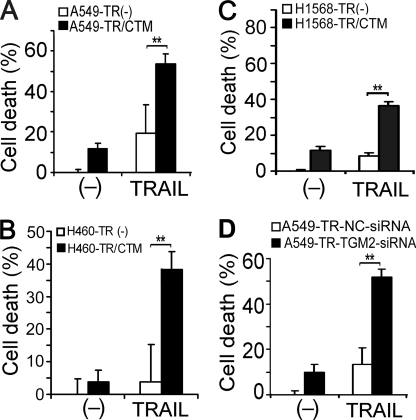
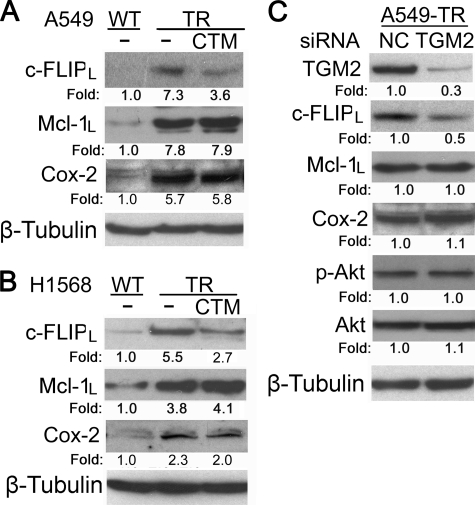
To investigate the role and mechanism by which TGM2 contributes to TRAIL resistance, the effects of suppressing TGM2 activity with cystamine or TGM2 expression with siRNA on TRAIL-induced cytotoxicity were examined. Cystamine substantially sensitized A549-TR, H460-TR, and H1568-TR cells to TRAIL-induced cytotoxicity (Fig. 5, A and B). Likewise, TGM2 siRNA significantly potentiated TRAIL-induced death in A549-TR cells (Fig. 5C and supplemental Fig. S5). Similar effects were seen in H1568-TR cells (data not shown). More importantly, the sensitization of the cytotoxicity of TRAIL by TGM2 knockdown in A549-TR cells was effectively attenuated when TGM2 expression was restored by ectopic expression with a TGM2-expressing vector that has escaped the TGM2 siRNA targeting (supplemental Fig. S6). Furthermore, artificially overexpressing TGM2 in A549-WT cells significantly suppressed TRAIL-induced cytotoxicity (supplemental Fig. S7). These results imply that TGM2 overexpression plays a substantial role in acquired TRAIL resistance. Our previous studies found that Akt-mediated overexpression of c-FLIPL, Mcl-1L, and COX-2 plays an important role in acquired TRAIL resistance (19, 20). Thus, we examined whether TGM2 regulates c-FLIPL, Mcl-1L or COX-2 expression in TR cells. Cystamine treatment substantially reduced the expression levels of c-FLIPL but not Mcl-1L and COX-2 in both A549-TR and H1568-TR cells (Fig. 6, A and B). Consistently, knockdown of TGM2 expression in A549-TR cells resulted in a similar reduction of c-FLIPL but not Mcl-1L or COX-2 expression (Fig. 6C). These results suggest that c-FLIPL is one of the downstream effectors of TGM2 in mediating acquired TRAIL resistance. Although the expression and activity of Akt were barely affected by TGM2 siRNA (Fig. 6C), it appears that TGM2 regulates c-FLIPL expression through a novel pathway, which is Akt-independent.
FIGURE 5.
TGM2 contributes to acquired TRAIL resistance. A–C, the indicated cells were pretreated with cystamine (CTM; 1 mm) for 1 h or were left untreated, followed by exposure to TRAIL (150 ng/ml for A549 and H1568, 100 ng/ml for H460) for 30 h. Cell death was detected by lactate dehydrogenase leakage assay (cytotoxicity assay). Data shown are mean ± S.D. **, p < 0.01. D, A549-TR cells were transfected with 20 nm of TGM 2 siRNA or NC siRNA. Forty-eight hours after transfection, the cells were treated with TRAIL (150 ng/ml) for 30 h or were left untreated. Cell death was detected as described in A.
FIGURE 6.
TGM2 suppression reduced expression of c-FLIP-1L but not Mcl-1L and COX-2. A and B, the indicated cells were treated with cystamine (CTM; 1 mm) overnight or were left untreated. c-FLIP-1L, Mcl-1L, and COX-2 were detected by Western blot. β-Tubulin was detected as an input control. C, A549-TR cells were transfected with TGM 2 siRNA (pool) or NC siRNA. Forty-eight hours after transfection, cell lysates were prepared, and the indicated proteins were detected by Western blot. β-Tubulin was detected as an input control. Fold changes were calculated with the control taken as 1.
EGFR Activation Contributes to TGM2 Overexpression through the ERK/JNK Pathway in TR Cells
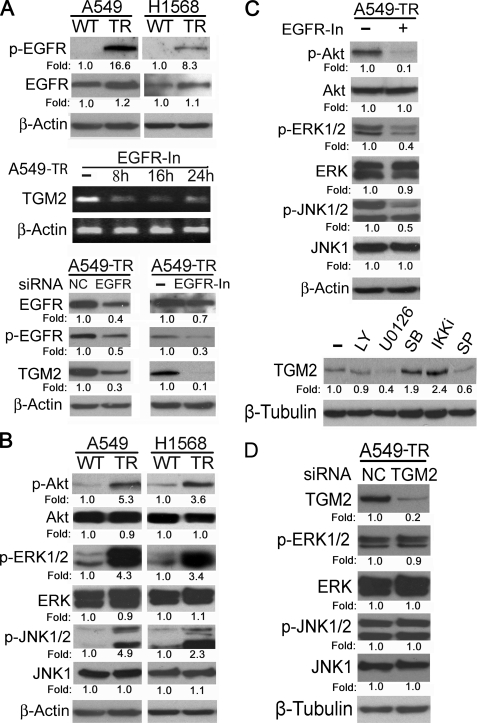
EGFR-mediated cell signaling is involved in cancer development and drug resistance in a variety of human tumors (30, 31). Because EGFR signaling was reported to up-regulate TGM2 expression (32, 33), we examined the expression and activity of EGFR in the TR cells. Indeed, the active form (phosphorylated) of EGFR was markedly elevated, whereas the total EGFR protein was slightly increased, in A549-TR and H1568-TR cells (Fig. 7A, top). Blocking EGFR activity with the EGFR inhibitor greatly reduced TGM2 mRNA and protein expression (Fig. 7A). To further validate the role of EGFR in regulating TGM2 protein expression, RNA interference was employed to knock down EGFR expression in A549-TR cells. TGM2 protein level was down-regulated when EGFR expression and activity were suppressed with EGFR siRNA (Fig. 7A, bottom). These results clearly suggest that the activation of EGFR is responsible for increased TGM2 expression in the TR cells.
FIGURE 7.
EGFR-mediated activation of ERK and JNK contributes to TGM2 overexpression in cells with acquired TRAIL resistance. A, equal amounts of cell extracts from the indicated cells were detected for p-EGFR and total EGFR expression by Western blot. β-Actin was detected as an input control (top). Equal amounts of total RNA from A549-TR cells treated with the EGFR inhibitor (10 μm) for the indicated times were detected for TGM2 mRNA expression by RT-PCR. β-Actin was detected as an input control (middle). Equal amounts of cell extract from A549-TR, transfected with the indicated siRNA, or treated with the EGFR inhibitor (10 μm) were detected for the indicated proteins by Western blot. β-Actin was detected as an input control. C, A549-TR cells were treated with the EGFR inhibitor (10 μm) overnight or were left untreated. The indicated proteins were detected by Western blot. β-Actin was detected as an input control (top). A549-TR cells were treated with 10 μm of LY294002, U0126, SB203580, IKK inhibitor II (IKKi), or SP600125 (SP) overnight or were left untreated, and TGM2 protein expression was detected by Western blot. β-Tubulin was detected as an input control. D, equal amounts of cell extract from A549-TR, transfected by TGM2-siRNA or NC-siRNA were detected for the indicated proteins by Western blot. β-Tubulin was detected as an input control. Fold changes were calculated with the control taken as 1.
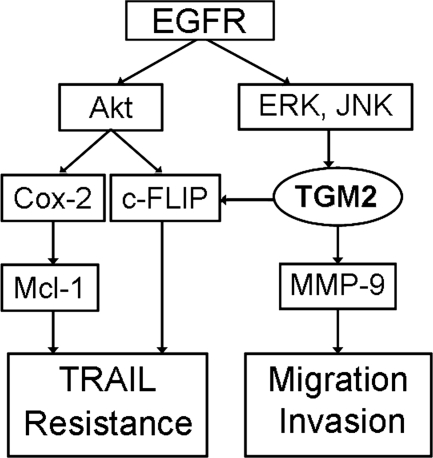
We further delineated the signaling pathway that mediates the EGFR-dependent activation of TGM2. In TR cells, the main signaling pathways activated by EGFR, Akt, JNK, and ERK were activated in A549-TR and H1568-TR cells (Fig. 7B). Suppressing EGFR dramatically reduced the activity of these pathways, suggesting the increased EGFR activity is the main upstream event of these pathways in the TR cells (Fig. 7B). To examine the role of these pathways in TGM2 expression, inhibitors for each pathway, PI3K/Akt (LY294002), MEK/ERK (U0126), and JNK (SP600125) were used to treat the cells. The inhibitors for MEK/ERK and JNK but not Akt significantly suppressed TGM2 expression in A549-TR cells. As controls, the p38 inhibitor SB203580 and NF-κB inhibitor IKKin exerted no inhibitory effect on TGM2 expression. All the inhibitors were effective in suppressing their respective pathways (supplemental Fig. S8). A moderate stimulatory effect of the p38 and NF-κB inhibitors was noted. The roles and mechanisms for the negative effect on TGM2 expression of these two pathways deserve further investigation. Nevertheless, our results suggest that the two MAPKs, JNK and ERK, are the major downstream kinases for EGFR-mediated TGM2 expression. Conversely, suppression of TGM2 had no effect on ERK and JNK activity (Fig. 7D), further placing TGM2 downstream of JNK and ERK. Collectively, these results establish a novel pathway that involves EGFR, MAPK (ERK and JNK), and TGM2 for acquired TRAIL resistance and cell migration in lung cancer cells, and TGM2 is important for mediating both TRAIL resistance and cell migration through c-FLIP and MMP-9, respectively (Fig. 8).
FIGURE 8.
A model of acquired TRAIL resistance and migration/invasion. EGFR-mediated TGM2 overexpression couples acquired TRAIL resistance and migration involving c-FLIP and MMP-9, respectively. The EGFR-mediated Akt activation is also involved in c-FLIP- and COX-2-mediated TRAIL resistance (see “Discussion”).
DISCUSSION
In this study, we determined that a novel pathway consisting of EGFR, MAPK (JNK and ERK), and TGM2 is responsible for acquired TRAIL resistance-associated migration and invasion in lung cancer cells. Specifically, the expression of TGM2 was up-regulated by EGFR-mediated MAP kinases JNK and ERK, and TGM2 plays a dual role in both TRAIL resistance and cell migration through up-regulation of c-FLIP and MMP-9, respectively. Suppression of TGM2 was able to simultaneously alleviate TRAIL resistance and suppress cell migration and invasion. Therefore, TGM2 could be a molecular target for circumventing acquired TRAIL resistance-associated metastasis.
Our previous studies have established a pathway consisting of Akt, c-FLIP, Mcl-1, and COX-2 that is responsible for acquired TRAIL resistance in lung cancer cells (19). Akt-mediated COX-2 activation triggers Mcl-1 but not c-FLIP expression (20). In this report, we identify a new branch of cell signaling that up-regulates c-FLIP expression through TGM2. The observation is consistent with recent reports showing that TGM2 conveys cancer cell resistance to TRAIL-induced apoptosis (34, 35). Interestingly, this new pathway is driven by EGFR-mediated MAPKs (JNK and ERK) but not Akt. The results add more complexity to acquired TRAIL resistance and suggest that overexpression of c-FLIP in the TRAIL-resistant cells is achieved in at least two distinct pathways: that through Akt and MAPK/TGM2, respectively (Ref. 19 and this study). Blockage of both pathways may be required to sufficiently reduce c-FLIP expression to prevent or alleviate acquired TRAIL resistance in lung cancer. It was reported that TGM2 suppresses DR5 expression (34). However, we did not detect changes in DR5 expression in our TRAIL-resistant cells (data not shown).
As one of the major obstacles that hinder cancer therapy, acquired chemoresistance not only neutralizes anticancer activity of the drugs but also promotes tumor progression (8). Although chemoresistance-associated migration/invasion has been observed, the underlying mechanism of which has not been well studied. By gene expression screening, we identified TGM2 as one protein factor coupling TRAIL resistance and migration/invasion. Consistent with previous reports (34, 35), suppressing TGM2 activity with cystamine or expression with siRNA dramatically alleviated TRAIL resistance and cell migration. Interestingly, TGM2 suppression greatly reduced expression of MMP-9, a matrix metalloproteinase that is involved in cancer cell migration/invasion. Suppression of MMP-9 expression with siRNA substantially inhibited cell migration/invasion, suggesting that MMP-9 is one of the major MMPs that contribute to acquired TRAIL resistance-associated migration/invasion. Although regulated by TGM2 in certain cancer cell types, other MMPs such as MMP-2 are unlikely involved in migration in the lung cancer cells with acquired TRAIL resistance (36). This observation suggests that the mechanisms of TGM2 in cancer cell migration/invasion are cell type-specific or that the functioning mechanism of TGM2 in acquired TRAIL resistance-associated migration/invasion is distinct to that in cancer cells with primary apoptosis resistance. Indeed, it was reported that in primary TRAIL-resistant cholangiocarcinoma cells, NF-κB pathway was utilized for TRAIL-triggered cancer cell migration, invasion, and metastasis (10, 21). However, NF-κB is apparently not involved in the TGM2-mediated MMP9 expression and migration/invasion in our TRAIL-resistant cells. Although it remains to be determined how TGM2 promotes MMP9 expression in the context of acquired TRAIL resistance in lung cancer cells, our results establish a novel signaling cascade containing TGM2 and MMP-9 for acquired TRAIL resistance-associated migration/invasion. Targeting the key factors such as TGM2 in this pathway may prevent or attenuate invasion or metastasis during chemotherapy with TRAIL.
As the most diverse and ubiquitous member of the transglutaminase family of enzymes that catalyze posttranslational modification of proteins, TGM2 has been found to play roles in a variety of diseases, including cancer (37). It has been shown that TGM2 promotes cell growth, survival, and motility or invasion (38, 39). Although increased TGM2 expression in cancer cells has been linked to increased drug resistance, metastasis, and poor patient survival (36, 37, 40), to our knowledge, this is the first report showing that TGM2 is overexpressed when acquired TRAIL resistance arises with the association of increased cell migration and invasion. Although the regulation of TGM2 expression is complex, EGFR-mediated cell signaling is implicated in up-regulation of TGM2 expression in cancer cells (32). In this study, we reveal that EGFR-mediated activation of JNK and ERK increases TGM2 expression, which contributes to acquired TRAIL resistance and cell migration. Because EGFR is the main determinant of Akt activity that plays an important role in apoptosis resistance in the cells with acquired TRAIL resistance (19, 20), findings in this study place EGFR at the upstream of the two signaling cascades that involve Akt and TGM2, respectively (Fig. 8). Future studies are needed to elucidate the underlying mechanism by which EGFR is activated when lung cancer cells are chronically exposed to TRAIL. Nevertheless, our results for the first time demonstrate that EGFR activation is required for establishing cancer cell resistance to TRAIL and subsequently enhanced cancer cell migration and invasion.
Collectively, results from this study establish a new signaling pathway consisting of EGFR, ERK or JNK, and TGM2 for acquired TRAIL resistance and cell migration in lung cancer cells. Because TGM2 couples acquired TRAIL resistance and cell migration, it could be a molecular target for circumventing acquired chemoresistance and metastasis in lung cancer.
Supplementary Material
Acknowledgments
We thank Dr. Gail V.W. Johnson from the University of Rochester for providing the TGM2-expressing plasmid and Mabel Padilla for help in manuscript preparation.
This work was supported in part by National Institutes of Health NIEHS Grant R01ES017328 and Department of Energy Low Dose Radiation Research Program DE-SC0001173.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S8.
- TRAIL
- TNF-related apoptosis-inducing ligand
- EGFR
- EGF receptor
- MMP-9
- matrix metalloproteinase 9
- TR
- TRAIL-resistant
- COX-2
- cyclooxygenase 2
- Mcl-1
- myeloid cell leukemia 1
- TGM2
- tissue transglutaminase
- DR
- death receptor
- NC
- negative control.
REFERENCES
- 1. Fulda S. (2009) Int. J. Cancer 124, 511–515 [DOI] [PubMed] [Google Scholar]
- 2. Zhang L., Fang B. (2005) Cancer Gene Ther. 12, 228–237 [DOI] [PubMed] [Google Scholar]
- 3. Wilson T. R., Johnston P. G., Longley D. B. (2009) Curr. Cancer Drug Targets 9, 307–319 [DOI] [PubMed] [Google Scholar]
- 4. Song J. J., An J. Y., Kwon Y. T., Lee Y. J. (2007) J. Biol. Chem. 282, 319–328 [DOI] [PubMed] [Google Scholar]
- 5. Wenger T., Mattern J., Penzel R., Gassler N., Haas T. L., Sprick M. R., Walczak H., Krammer P. H., Debatin K. M., Herr I. (2006) Cell Death Differ. 13, 1740–1751 [DOI] [PubMed] [Google Scholar]
- 6. Ghavami S., Hashemi M., Ande S. R., Yeganeh B., Xiao W., Eshraghi M., Bus C. J., Kadkhoda K., Wiechec E., Halayko A. J., Los M. (2009) J. Med. Genet. 46, 497–510 [DOI] [PubMed] [Google Scholar]
- 7. Lønning P. E. (2010) Mol. Oncol. 4, 284–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Roché H., Vahdat L. T. (2011) Ann. Oncol. 22, 1000–1010 [DOI] [PubMed] [Google Scholar]
- 9. Malhi H., Gores G. J. (2006) Oncogene 25, 7333–7335 [DOI] [PubMed] [Google Scholar]
- 10. Ishimura N., Isomoto H., Bronk S. F., Gores G. J. (2006) Am. J. Physiol. Gastrointest. Liver. Physiol. 290, G129–136 [DOI] [PubMed] [Google Scholar]
- 11. Abdulghani J., El-Deiry W. S. (2010) Expert. Opin. Ther. Targets 14, 1091–1108 [DOI] [PubMed] [Google Scholar]
- 12. Stegehuis J. H., de Wilt L. H., de Vries E. G., Groen H. J., de Jong S., Kruyt F. A. (2010) Drug Resist. Updat. 13, 2–15 [DOI] [PubMed] [Google Scholar]
- 13. Leong S., Cohen R. B., Gustafson D. L., Langer C. J., Camidge D. R., Padavic K., Gore L., Smith M., Chow L. Q., von Mehren M., O'Bryant C., Hariharan S., Diab S., Fox N. L., Miceli R., Eckhardt S. G. (2009) J. Clin. Oncol. 27, 4413–4421 [DOI] [PubMed] [Google Scholar]
- 14. Wang X. (2001) Genes Dev. 15, 2922–2933 [PubMed] [Google Scholar]
- 15. Lin Y., Devin A., Cook A., Keane M. M., Kelliher M., Lipkowitz S., Liu Z. G. (2000) Mol. Cell. Biol. 20, 6638–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wajant H. (2004) Vitam. Horm. 67, 101–132 [DOI] [PubMed] [Google Scholar]
- 17. Wajant H., Gerspach J., Pfizenmaier K. (2005) Cytokine Growth Factor Rev. 16, 55–76 [DOI] [PubMed] [Google Scholar]
- 18. Lane D., Côté M., Grondin R., Couture M. C., Piché A. (2006) Mol. Cancer Ther. 5, 509–521 [DOI] [PubMed] [Google Scholar]
- 19. Wang X., Chen W., Zeng W., Bai L., Tesfaigzi Y., Belinsky S. A., Lin Y. (2008) Mol. Cancer Ther. 7, 1156–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen W., Bai L., Wang X., Xu S., Belinsky S. A., Lin Y. (2010) Mol. Pharmacol. 77, 416–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fingas C. D., Blechacz B. R., Smoot R. L., Guicciardi M. E., Mott J., Bronk S. F., Werneburg N. W., Sirica A. E., Gores G. J. (2010) Hepatology 52, 550–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Grivennikov S. I., Greten F. R., Karin M. (2010) Cell 140, 883–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lin Y., Shi R., Wang X., Shen H. M. (2008) Curr. Cancer Drug Targets 8, 634–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chen W., Wang X., Zhuang J., Zhang L., Lin Y. (2007) Carcinogenesis 28, 2114–2121 [DOI] [PubMed] [Google Scholar]
- 25. Tucholski J., Johnson G. V. (2003) J. Biol. Chem. 278, 26838–26843 [DOI] [PubMed] [Google Scholar]
- 26. Ju W., Wang X., Shi H., Chen W., Belinsky S. A., Lin Y. (2007) Mol. Pharmacol. 71, 1381–1388 [DOI] [PubMed] [Google Scholar]
- 27. Sahai E. (2007) Nat. Rev. Cancer 7, 737–749 [DOI] [PubMed] [Google Scholar]
- 28. Mehta K., Kumar A., Kim H. I. (2010) Biochem. Pharmacol. 80, 1921–1929 [DOI] [PubMed] [Google Scholar]
- 29. Ohbayashi H. (2002) Curr. Protein Pept. Sci. 3, 409–421 [DOI] [PubMed] [Google Scholar]
- 30. Ho C., Laskin J. (2009) Expert Opin. Investig. Drugs 18, 1133–1145 [DOI] [PubMed] [Google Scholar]
- 31. Zandi R., Larsen A. B., Andersen P., Stockhausen M. T., Poulsen H. S. (2007) Cell Signal 19, 2013–2023 [DOI] [PubMed] [Google Scholar]
- 32. Li B., Antonyak M. A., Druso J. E., Cheng L., Nikitin A. Y., Cerione R. A. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 1408–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Antonyak M. A., Li B., Regan A. D., Feng Q., Dusaban S. S., Cerione R. A. (2009) J. Biol. Chem. 284, 17914–17925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Frese-Schaper M., Schardt J. A., Sakai T., Carboni G. L., Schmid R. A., Frese S. (2010) FEBS Lett. 584, 2867–2871 [DOI] [PubMed] [Google Scholar]
- 35. Jang J. H., Park J. S., Lee T. J., Kwon T. K. (2010) Cancer Lett. 287, 224–230 [DOI] [PubMed] [Google Scholar]
- 36. Satpathy M., Shao M., Emerson R., Donner D. B., Matei D. (2009) J. Biol. Chem. 284, 15390–15399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lorand L., Graham R. M. (2003) Nat. Rev. Mol. Cell Biol. 4, 140–156 [DOI] [PubMed] [Google Scholar]
- 38. Verma A., Wang H., Manavathi B., Fok J. Y., Mann A. P., Kumar R., Mehta K. (2006) Cancer Res. 66, 10525–10533 [DOI] [PubMed] [Google Scholar]
- 39. Park K. S., Kim H. K., Lee J. H., Choi Y. B., Park S. Y., Yang S. H., Kim S. Y., Hong K. M. (2010) J. Cancer Res. Clin. Oncol. 136, 493–502 [DOI] [PubMed] [Google Scholar]
- 40. Miyoshi N., Ishii H., Mimori K., Tanaka F., Hitora T., Tei M., Sekimoto M., Doki Y., Mori M. (2010) Ann. Surg. Oncol. 17, 967–972 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.