Abstract
The PEB4 protein is an antigenic virulence factor implicated in host cell adhesion, invasion, and colonization in the food-borne pathogen Campylobacter jejuni. peb4 mutants have defects in outer membrane protein assembly and PEB4 is thought to act as a periplasmic chaperone. The crystallographic structure of PEB4 at 2.2-Å resolution reveals a dimer with distinct SurA-like chaperone and peptidyl-prolyl cis/trans isomerase (PPIase) domains encasing a large central cavity. Unlike SurA, the chaperone domain is formed by interlocking helices from each monomer, creating a domain-swapped architecture. PEB4 stimulated the rate of proline isomerization limited refolding of denatured RNase T1 in a juglone-sensitive manner, consistent with parvulin-like PPIase domains. Refolding and aggregation of denatured rhodanese was significantly retarded in the presence of PEB4 or of an engineered variant specifically lacking the PPIase domain, suggesting the chaperone domain possesses a holdase activity. Using bioinformatics approaches, we identified two other SurA-like proteins (Cj1289 and Cj0694) in C. jejuni. The 2.3-Å structure of Cj1289 does not have the domain-swapped architecture of PEB4 and thus more resembles SurA. Purified Cj1289 also enhanced RNase T1 refolding, although poorly compared with PEB4, but did not retard the refolding of denatured rhodanese. Structurally, Cj1289 is the most similar protein to SurA in C. jejuni, whereas PEB4 has most structural similarity to the Par27 protein of Bordetella pertussis. Our analysis predicts that Cj0694 is equivalent to the membrane-anchored chaperone PpiD. These results provide the first structural insights into the periplasmic assembly of outer membrane proteins in C. jejuni.
Keywords: Chaperone Chaperonin, Crystal Structure, Crystallography, Protein Folding, Protein Structure, Campylobacter, Outer Membrane, Periplasm, Virulence Factor
Introduction
Outer membrane protein assembly in Gram-negative bacteria is a multistep process in which specific periplasmic chaperones bind proteins emerging from the Sec system in an unfolded or partially folded state and present them to a specialized insertion system (the β-barrel assembly machinery complex or BAM)5 in the outer membrane (1, 2). Several periplasmic chaperones have been found to participate in the maturation of OMPs. In Escherichia coli, some OMPs associate with the periplasmic chaperone SurA after they exit the SecYEG translocation complex (3, 4). A “back-up” chaperone, Skp, can also bind these polypeptide substrates in the event that they fail to productively interact with SurA (5). However, in Neisseria meningitidis Skp has a more important role than SurA (6). The chaperone then delivers its cargo to the BAM complex, which completes the insertion and assembly of the OM β-barrel protein.
SurA is the best characterized OMP chaperone. It is a monomer composed of three domains: two parvulin-type peptidyl-prolyl cis/trans isomerase (PPIase) domains and a chaperone domain comprising the N- and C-terminal regions of the protein (7). In vitro proline-limited folding of a protein substrate (RCM-ribonuclease T1) (8) and a synthetic oligopeptide substrate (N-succinyl-Ala-Ala-Pro-Phe p-nitroaniline) (9) showed that PPIase domain I of SurA had no detectable activity, whereas domain II exhibited a relatively low level of PPIase activity comparable with that of intact SurA (8, 9). The chaperone domain exhibits a PPIase-independent “holdase”-like activity; it has been demonstrated in vitro that a SurA variant lacking its parvulin domains was still able to prevent the aggregation of heat-denatured citrate synthase (8). Thus, the role of the active PPIase domain in the chaperone function of SurA is not clear, whereas the inactive PPIase domain has been shown to have a role in the recognition of C-terminal aromatic residues of client proteins (10).
SurA interacts with in vitro synthesized porins 50-fold more efficiently than with similarly sized non-porin proteins (8), suggesting a specific role in the maturation of OMPs. Differential proteomics has shown that mutational loss of SurA affects the abundance of at least eight β-barrel OMPs, namely FadL, LptD, FhuA, OmpX, and FecA as well as the major porins OmpA, OmpF, and LamB (11). Although the negative effect of surA mutation on the three latter proteins had been well documented (5, 9), Vertommen et al. (11) observed that FhuA and LptD are the only two for which the decrease in protein abundance could not be attributed to decreased mRNA levels in the surA mutant strain. They proposed that LptD, and possibly FhuA, were true SurA substrates, and that only a subset of OMPs depends on SurA for folding and insertion in the OM.
The client proteins bound to SurA subsequently interact with the BAM complex, which is composed of a core subunit, BamA (formerly YaeT; a member of the Omp85 protein family) and four lipoproteins BamBCDE (formerly YfgL, NlpB, YfiO, and SmpA, respectively) (1, 2). BamA has an N-terminal region containing repeated polypeptide transport-associated (POTRA) domains and a C-terminal β-barrel domain. It recognizes OMP substrates via an aromatic residue motif at their C termini (12) and deletion of any one of the POTRA domains has partial effects on OMP assembly (13). The three-dimensional structure of the POTRA domains of BamA provides a basis for understanding how the protein interacts with a large number of β-barrel substrates (13, 14). Deletion mutations have demonstrated that POTRA domains 2, 3, and 4 are required to mediate interactions with BamB. In addition, POTRA domain 3 was reported to mediate the augmentation of β-barrel formation in the substrate proteins, whereas domain 5 is crucial in the interactions with other lipoprotein partners (13). The structure of BamB (15–17) suggests that this protein has a scaffolding role within the BAM complex, by optimally orienting the flexible periplasmic domain of BamA for interaction with other BAM proteins and chaperones (15). Other recent structural studies show that BamE interacts with BamD and also binds phosphatidylglycerol, consistent with anchoring to the OM inner leaflet (18).
Despite the importance of the OM in pathogenicity and host-cell interactions, very little is currently known about the biogenesis of OMPs in Gram-negative pathogens other than E. coli and N. meningitidis. Campylobacter jejuni is an ϵ-proteobacterium and is one of the leading causes of acute food-borne gastroenteritis worldwide (19). The bacterium is a commensal in the caecum of poultry, as well as wild bird species (20). Although the majority of human infections result in a severe but self-limiting diarrheal illness, a minority (0.1–1%) can result in serious sequelae such as Guillain-Barré syndrome, a neuromuscular paralysis resulting from an autoimmune reaction against neuronal gangliosides (21). The pathogenic mechanisms of C. jejuni are not completely understood but involve mucosal adherence, host cell invasion, and toxin production (22). Efficient adhesion to host cells is crucial for infection and is mediated by several OMPs (23, 24) including the fibronectin-binding protein CadF, the lipoprotein JlpA, the autotransporter CapA, and the major outer membrane porin (MOMP), PorA.
The abundant periplasmic PEB4 protein has previously been implicated in OMP assembly in C. jejuni (25–27). The cognate gene (cj0596) encodes a predicted periplasmic PPIase and is conserved in all strains that have been genome sequenced to date. Asakura et al. (25) showed that a cj0596 null mutant made in strain NCTC11168 was considerably less adherent to INT407 cells than the wild-type, displayed a reduced level and duration of intestinal colonization of a mouse model of infection, and was deficient in biofilm formation. A cj0596-null mutant made in the highly pathogenic strain 81–176 was also less able to colonize mice but was more motile than the parent strain (26, 27). This mutant actually showed an increased propensity to invade INT407 cells but was not significantly affected in adhesion or intracellular survival. Despite these strain differences, these studies revealed a significant growth defect in the cj0596 mutant and, most significantly, iTRAQ-based proteomic methods revealed decreases in the abundance of several outer membrane and periplasmic proteins in the NCTC11168 cj0596 mutant (25). Mass spectrometric identification of proteins from one- and two-dimensional gels of the strain 81–176 cj0596 mutant also showed a variety of protein expression changes, including a decrease in three outer membrane proteins: Omp50, major OMP, and the fibronectin binding-protein CadF (26, 27), although some other proteins were increased in expression.
Although primary sequence analysis readily detects the presence of a single parvulin-type PPIase domain in PEB4, the homology of the sequence flanking this domain is more difficult to analyze. At the time of initiating this study the top structural prediction hit made by the three-dimensional protein-threading server Phyre (28) was to the E. coli periplasmic chaperone SurA (7). The region of predicted structural similarity with PEB4 included substantial portions of the SurA chaperone domain. This raised the possibility that PEB4 is a functional homologue of SurA in C. jejuni.
Here, we report the 2.2-Å crystal structure of PEB4. This confirms a SurA-like domain organization and fold, but shows that unlike SurA, PEB4 forms a highly intertwined three-dimensional domain-swapped dimer. We confirm that the single parvulin domain in PEB4 is an active PPIase, and show that PEB4 has the properties of a holdase-type chaperone, preventing protein aggregation, consistent with a role in ferrying client OM proteins to the C. jejuni BAM complex. However, we also show that two other periplasmic proteins containing SurA-like domains (Cj1289 and Cj0694) are present in C. jejuni, which may also be involved in OMP biogenesis. Cj1289 was purified and shown to possess PPIase activity. Our structure of Cj1289 determined to 2.3 Å reveals that it is a closer structural homologue of SurA than is PEB4. Although the Cj0694 protein could not be expressed in a soluble form, bioinfomatic evidence suggests it is related to the membrane anchored PpiD chaperone of E. coli (29).
EXPERIMENTAL PROCEDURES
Construction of Overexpression Plasmids
The entire coding region of cj0596 (peb4) including the signal sequence but excluding the stop codon, was amplified from genomic DNA of C. jejuni NCTC11168 by PCR with Pwo DNA polymerase (Roche Applied Science) and oligonucleotides PEB4-NdeI-F and PEB4-XhoI-R (supplemental Table S1). The amplicon was cloned into the NdeI and XhoI restriction sites of pET-21a(+) (Merck Chemicals Ltd., United Kingdom) in-frame with a C-terminal His6 tag to generate pCP0596, expressing native His-tagged PEB4. A PEB4 variant with a PPIase domain deletion lacking residues 123–231 was also constructed (PEB4ΔPPI). This involved sequential cloning of two DNA fragments into pET-21a(+), separately amplified using two pairs of primers: PEB4VN-NdeI-F and PEB4VN-BamHI-R for the N-terminal fragment and PEB4VC-BamHI-F and PEB4VC-XhoI-R for the C-terminal fragment (supplemental Table S1). This plasmid construct was then used as the template for PCR amplification with PEB4-NdeI-F and PEB4-XhoI-R primers (supplemental Table S1) to produce the entire cj0596 coding region lacking the PPIase domain. The final PCR product corresponded to nucleotides 4 to 366 (without the ATG initiation codon) linked to nucleotides 694 to 819 (without the TAA stop codon) by an in-frame BamHI restriction site (producing a glycine-serine linker) that replaced the entire PPIase domain (nucleotides 367 to 693). This was cloned into pET-21a(+), to generate pCP0596V. For cj1289 and cj0694, the entire coding regions excluding the stop codons and N-terminal signal sequences, were amplified by PCR as above, using primer pairs cj1289-NdeI-F/cj1289-XhoI-R and cj0694-NdeI-F/cj0694-XhoI-R (supplemental Table S1). The amplicons were cloned into pET-21a(+), in-frame with the C-terminal His6 tag, to generate pCP1289 and pCP0694 overexpression vectors, respectively. Automated DNA sequencing (Lark Technologies Inc., United Kingdom) confirmed that the sequence of each of the cloned genes was correct. E. coli DH5α was used for all plasmid constructions and screenings, before transformation into E. coli BL21(λDE3) for overexpression.
Purification and Crystallization of PEB4 and Cj1289
PEB4, PEB4ΔPPI, and Cj1289, each carrying a C-terminal His6 tag, were produced by heterologous expression in E. coli BL21(λDE3) cultured in Luria-Bertani (LB) broth supplemented with 100 μg ml−1 of carbenicillin at 37 °C. At an A600 nm of 0.6, 1 mm isopropyl 1-thio-β-d-galactopyranoside was added, and the induced cells were then grown for a further 5 h at 37 °C before harvesting by centrifugation (5,000 × g, 20 min, 4 °C). To obtain selenomethionine-incorporated PEB4 and Cj1289, cells were cultured in M9 medium supplemented with l-selenomethionine and other natural amino acids (30) and the incorporation was verified by mass spectrometry (Astbury Centre for Structural Molecular Biology, University of Leeds, UK). Induction conditions were as above. Cell pellets (up to 15 g wet weight) were suspended in 50 mm Tris-HCl, pH 8.0, 250 mm NaCl (buffer A), disrupted in a French press (1050 p.s.i.), and the soluble fraction was isolated as the supernatant after centrifugation (15,000 × g, 30 min, 4 °C). This was subsequently loaded onto a 5-ml HiTrap Ni-NTA affinity column (GE Healthcare). Protein was eluted with 250 mm imidazole, pH 8.0, in buffer A. The protein solution was diluted with 50 mm MES buffer, pH 6.5 (buffer B), to a final NaCl concentration of 50 mm and was subsequently loaded onto a Resource-S 6-ml cation exchange column (GE Healthcare), previously equilibrated with buffer B and 50 mm NaCl. The PEB4 protein was eluted by applying a linear gradient of 50–1000 mm NaCl, the protein containing fractions were pooled and then further purified on a Superdex 200 10 × 300-mm gel filtration column (GE Healthcare), using 50 mm Tris-HCl, 500 mm NaCl as running buffer. PEB4 eluted from the column as a single peak with an apparent molecular mass ∼87 kDa. To purify the PEB4ΔPPI variant, the same purification strategy was adopted, except that the protein passed through the Resource-S column in the unbound fraction. From gel filtration, the variant protein corresponded to a globular species with an apparent molecular mass of ∼40 kDa, approximating that of a dimer (∼38 kDa). This protein was concentrated to 3.5 mg ml−1 for use in activity assays. Purification of Cj1289 was carried out on 5-ml HiTrap Ni-NTA affinity columns as for PEB4, followed by removal of imidazole with 5-ml HiTrap desalting columns (GE Healthcare). Gel filtration analysis of Cj1289 indicates the presence of a species with apparent molecular mass of ∼40 kDa, intermediate between a monomer and dimer molecular mass (∼60 kDa). Purified proteins were judged to be pure by Coomassie Blue staining on overloaded SDS-PAGE gels and their identities were confirmed by N-terminal sequencing performed by Dr. A. J. G. Moir (Department of Molecular Biology and Biotechnology, University of Sheffield). This analysis confirmed that the signal sequences of PEB4 and PEB4ΔPPI (residues 1–21) were correctly processed and removed by E. coli during overexpression.
For crystallization, the purified PEB4 was concentrated to 30 mg ml−1 in 10 mm Tris-HCl buffer, pH 8.0, plus 50 mm NaCl and Cj1289 was concentrated to 10 mg ml−1 in 20 mm Tris-HCl buffer, pH 8.0. Both proteins were tested for crystallization with a variety of commercial screens. Subsequent optimization for PEB4 resulted in the growth of x-ray diffracting crystals in a condition comprising 0.1 m succinate, pH 3.5, 40% (v/v) Jeffamine at 7 °C. The crystals for native and the selenomethionine derivative of PEB4 were monoclinic and reached an average size of 0.1 × 0.1 × 0.1 mm3 within 4 days. X-ray diffracting crystals for selenomethionine incorporated Cj1289 were obtained in a condition constituting 0.1 m CHES, pH 8.0, 25% (w/v) PEG 8000, 5% (w/v) sucrose. These crystals were orthorhombic and reached an average size of 0.2 × 0.1 × 0.1 mm3 within 14 h at 17 °C.
Data Collection and Structure Determination
Diffraction data for the PEB4 crystals (both native and selenomethionine derivatized) and Cj1289 (selenomethionine derivatized) were measured with synchrotron radiation at the Diamond Light Source (DLS, Oxfordshire, UK). Prior to data collection, crystals of PEB4 were equilibrated against the cryo-protecting solution (comprising 0.1 m succinate, pH 3.5, 45% (v/v) Jeffamine) for 24 h and subsequently flash-cooled in liquid nitrogen. For PEB4 multiple datasets at peak, inflection, and high-energy remote wavelengths were collected on a single selenomethionine derivative crystal on two different beamlines and the respective datasets were merged and scaled together to obtain high redundancy. For Cj1289, selenomethionine crystals were cryo-protected with 0.1 m CHES, pH 6.0, 25% (w/v) PEG 8000, and 30% (w/v) sucrose prior to flash cooling in liquid nitrogen. Multiple datasets were collected at peak, inflection, and high-energy remote wavelengths plus an additional higher resolution set at a lower energy wavelength. The data were processed with Mosflm (31) and merged using Scala (32) as implemented in the Collaborative Computational Project, Number 4 (CCP4) software suite (33).
The native PEB4 crystal and selenomethionine derivative crystals diffracted to 2.2- and 2.5-Å resolutions, respectively, and belonged to space group C2. Analysis of the Matthews's coefficient and inspection of self-rotation Patterson maps indicated the presence of a monomer per asymmetric unit, with a solvent content of 67%. An initial set of phases were obtained from the selenomethionine data by multiwavelength anomalous diffraction using Solve (34) followed by density modification of the resultant map using Resolve (35). The electron density map for the native PEB4 was further modified with Pirate (36), using the 2.2-Å resolution native data and the phase information from the selenomethionine derivative provided by Hendrickson Lattmann coefficients obtained from Solve/Resolve. Subsequent automated model building was carried out using Buccaneer (37). Several further iterations of manual building using the program Coot (38) were alternated with maximum-likelihood refinement using the program Refmac5 (39). Water molecules were added using ARP/wARP (40, 41) during the final refinement cycles. The final model was complete apart from the C-terminal His6 tag and the signal sequence (residues 1–21), which had been processed and removed in the E. coli expression host to give the mature protein prior to crystallization.
The Cj1289 selenomethionine derivative crystals diffracted up to 2.3-Å resolution and belonged to space group P212121. Matthews coefficient calculations suggested the presence of two monomers per asymmetric unit, with a solvent content of 49%. An initial set of phases was obtained from the selenomethionine data to 2.8 Å by multiwavelength anomalous diffraction using Solve (34) followed by density modification of the resultant map using Resolve (35). Initially, the datasets for all the wavelengths were combined, scaled together, and maps were calculated using phase information from the selenomethionine derivative provided by Hendrickson Lattmann coefficients obtained from Solve/Resolve, which were subjected to density modification with Pirate (36), followed by automated model building using Buccaneer (37). Several further iterations of manual building using the program Coot (38) were alternated with maximum-likelihood refinement using the refinement program option (phenix.refine) within the Phenix suite (42). Initial stages of refinement involved the use of a simulated annealing protocol using default parameters, followed by TLS refinement using the recommendations obtained from the TLSMD server (43), considering chain A as TLS group 1 (residues 21–271) and chain B subdivided in two domains comprising TLS group 2 (21–186) and TLS group 3 (187–271). The model was eventually refined against a separate 2.3-Å selenomethionine dataset retaining the same set of reflections for calculation of the free R. Water molecules were added in the final stages of refinement. The final model was complete apart from residues 102–104 in protein chain A, the C-terminal His6 tags in both chains and the leader sequences (residues 1–20), which had been deleted from the expression construct. The model quality was validated using Coot and MolProbity (44).
A summary of the relevant data statistics is shown in Table 1, with a more detailed description in supplemental Table S2 (PEB4) and supplemental Table S3 (Cj1289). Structure factors and coordinates have been deposited at the PDB with accession codes 3RFW (PEB4) and 3RGC (Cj1289). All figures were generated using PyMol (pymol.org) and topology diagrams prepared using Topdraw (45).
TABLE 1.
Summary structural statistical data for PEB4 and Cj1289
Full data are given under supplemental Table S2 (PEB4) and supplemental Table S3 (Cj1289).
| PEB4 | Cj1289 | |
|---|---|---|
| Data collection | ||
| Space group | C2 | P212121 |
| Unit cell (Å) | a = 81.4 Å, b = 91.6 Å, c = 61.6 Å | a = 49.6 Å, b = 94.6 Å, c = 124.6 Å |
| α = γ = 90°, β = 102.3° | α = β = γ = 90° | |
| Resolution range (Å) | 39.8–2.20 | 62.3–2.30 |
| No. of measured reflections | 251,133 | 189,232 |
| No. of unique reflections | 22,272 | 26,871 |
| Completeness (%)a | 99.5 (100.0) | 100.0 (100.0) |
| Rpima,b | 0.023 (0.138) | 0.052 (0.379) |
| Mn〈I/sd〉a | 20.3 (5.7) | 14.3 (3.2) |
| Refinement | ||
| R/Rfreec,d | 0.234/0.276 | 0.225/0.280 |
| Overall B-factor (Å2) | 48.2 | 56.5 |
| R.m.s. deviation in bond distances (Å) | 0.011 | 0.008 |
| R.m.s. deviation in bond angles (°) | 1.1 | 1.1 |
| Ramachandran | ||
| Most favored (%) | 97.2 | 96.0 |
| Additionally allowed (%) | 2.8 | 4.0 |
a Data in parentheses correspond to the highest resolution shell.
b Rpim = Σhkl[1/(N-1)]1/2 Σi|Ii(hkl) −I(hkl)|/ΣhklΣi Ii(hkl).
c R-factor = Σ|Fobs − Fcalc|/ΣFobs.
d Rfree calculated as in footnote c but using 5% of experimental data excluded from refinement for validation.
Calculation of Hydrophobicity of Surface Residues
The surface exposed area of all individual atoms was calculated using AREAIMOL from the CCP4 suite. Fractions of surface area contributed by polar and non-polar atoms were calculated using an in-house Python script, which implements a method similar to that of Miller et al. (46). In this method all nitrogen, oxygen, and cysteine sulfur atoms were counted as polar, and all other atoms were counted as non-polar. No hydrogens were included in the calculations. Following the analysis of Fukuchi and Nishikawa (47) non-polar residues were defined as Ile, Leu, Met, Phe, Trp, Tyr, and Val. The inner surfaces were defined as: Chaperone, residues 40, 41, 43, 44, 46, 64, 67, 68, 71,85, 89, 92, 93, 96, 97, 99, 100, 103, 104, 107, 120, 128, 129, 130, 131, 235, 239, 240, 243, 244, 246, 247, 250, 253, and 257; and PPIase, residues 133, 135, 138, 140, 175, 176, 177, 178, 179, 182, 185, 186, 187, 188, 189, 190, 192, 193, 194, 195, 198, 217, 218, 219, 222, and 227.
PPIase Activity Measurements by NMR Exchange Spectroscopy
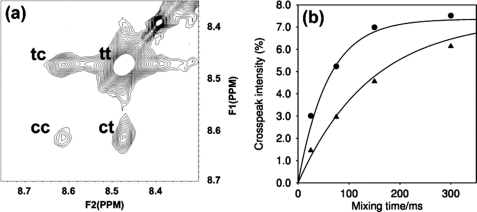
NOESY spectra were acquired at 298 K on a 600-MHz Bruker Avance Spectrometer, equipped with a cryoprobe, using the Bruker pulse sequence noesygpph19. The acquisition times were 500 and 142 ms in the direct and indirect dimensions, respectively. The spectra were run folded with an indirect spectral width of 3 ppm, with 16 scans per increment, giving a total time for each spectrum of ∼2 h. The solid curves plotted in Fig. 3b are 100 × fc × ft × (1-exp(−kextmix)), where fc and ft are the fractional populations of prolyl-peptide bonds that are cis or trans, respectively; kex is the exchange rate constant; and tmix is the NOESY mixing time. The peptide substrate used was N-succinyl-Ala-Ala-Pro-Phe-p-nitroanilide (Sigma). Samples were prepared by dilution of a stock solution of a 20:1 molar ratio of peptide/PEB4 solution, to ensure a constant ratio of peptide to PEB4.
FIGURE 3.
PPIase activity of PEB4 measured with N-succinyl-Ala-Ala-Pro-Phe-p-nitroanilide. a, portion of 75 ms mixing time NOESY spectrum of 3 mm peptide, 0.15 mm PEB4, showing the peaks for the amide proton of Phe4. The diagonal peak, arising from molecules that are in the cis(trans) state both before and after the mixing time, is labeled cc(tt). The cross-peak arising from molecules that are in the cis(trans) state before the mixing time and the trans(cis) state after the mixing time is labeled ct(tc). b, variation with mixing time of the cross-peak intensity for Phe4 (expressed as the mean of the intensity of the ct and tc cross-peaks relative to the total intensity of all four peaks). Data are shown for 1 mm peptide, 0.05 mm PEB4 (triangles) and 3 mm peptide, 0.15 mm PEB4 (circles). The curves are for a cis:trans ratio of 8:92, with exchange rate constants of 7 s−1 (1 mm peptide) and 17 s−1 (3 mm peptide), corresponding to rate constants for the PEB4 molecules of 140 and 340 s−1, respectively (using the ratio of peptide:PEB4 of 20:1).
PPIase-enhanced Refolding of Reduced and Carboxymethylated RNase T1
Proline isomerization limited protein folding was assayed with wild-type RNase T1 from Aspergillus oryzae (Sigma) as previously described (48). Disulfide reduced and S-carboxymethylated (RCM)-RNase T1 was prepared according to Mücke and Schmid (49). The refolding of wild-type RCM-RNase T1 is rate-limited by prolyl isomerization even when the disulfide bonds are broken (50). Refolding was monitored by intrinsic tryptophan fluorescence and initiated by a 30-fold dilution of the unfolded protein (stored in the absence of NaCl) to a final concentration of 0.5 μm RCM-RNase T1 in buffer containing 0.1 m Tris-HCl, pH 8.0, and 2 m NaCl at 15 °C. Changes in the steady-state tryptophan fluorescence were measured at 320 nm (10 nm slit-width) with excitation at 268 nm (2.5 nm slit width) using a Varian Cary Eclipse spectrofluorimeter over a 1-h period. PEB4, PEB4ΔPPI, or Cj1289 were added to various final concentrations prior to the dilution. The effect of PPIase inhibitors cyclosporin A and juglone (5-hydroxy-1,4-naphthoquinone) were determined at 5 and 7 μm final concentrations, respectively, by incubation of the chaperone with the inhibitors for 1 h at 10 °C prior to the determination of PPIase activity. Under the conditions of the assay, refolding of RCM-RNase T1 was dominated by a slow reaction (τ = 1700 s, 88% amplitude), whereas the fast reaction (τ = 9 s) had a minor amplitude of only 2% (49). Therefore, the fast reaction was mostly complete within the dead time of mixing of 2 to 3 s.
Rhodanese Refolding Assay
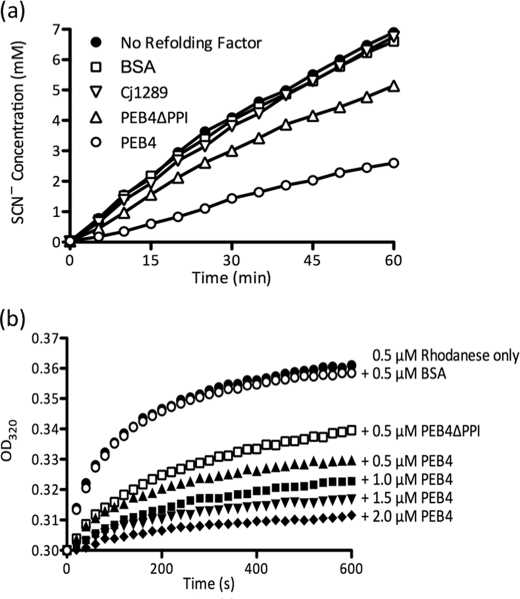
Chaperone activity was assessed by the rate of re-folding of chemically denatured bovine mitochondrial rhodanese (thiosulfate:cyanide sulfurtransferase; EC 2.8.1.1; Sigma) as measured by the formation of thiocyanate as the red iron complex, (FeSCN)2+, after addition of ferric nitrate. Following the method of Horowitz and Westley (51), 38 μm rhodanese was first denatured for 2 h at 25 °C in 50 mm Tris-HCl, pH 7.8, containing 6 m guanidine-HCl and 10 mm DTT. Renaturation was initiated by a 76-fold dilution at 37 °C in 50 mm Tris-HCl, pH 7.8, containing 50 mm Na2S2O3, 10 mm KCN, and 10 mm DTT (refolding solution). 300-μl aliquots of the refolding mixture were withdrawn at various time points and added to 200 μl of 38% (v/v) formaldehyde to stop all enzymatic activity. After completion of the time course, samples were microcentrifuged for 5 min to pellet the precipitates. The supernatants were subsequently mixed with 500 μl of ferric nitrate solution containing 165 mm Fe(NO3)3 dissolved in 8.67% (v/v) HNO3 and absorbance was determined at 460 nm. Where indicated, PEB4, PEB4ΔPPI, Cj1289, or bovine serum albumin (BSA) were present in the refolding solution prior to the addition of the unfolded rhodanese to give 0.5 μm final concentration.
Rhodanese Aggregation Assay
Rhodanese was also used as a model protein to investigate the effect of PEB4 and PEB4ΔPPI on protein aggregation during renaturation. The formation of protein aggregates was detected by light scattering, as measured by the absorbance increase at 320 nm in a Shimadzu UV-2401PC spectrophotometer. Unfolding and refolding of rhodanese was carried out as previously described (52). Rhodanese (30 μm) was first denatured for 2 h at 25 °C in 100 mm sodium phosphate buffer, pH 7.4, containing 6 m guanidine-HCl and 10 mm DTT. Renaturation was initiated by a 60-fold dilution (final rhodanese concentration 0.5 μm) at 25 °C in 100 mm sodium phosphate buffer, pH 7.4, with a 2-s dead time of mixing, in the absence or presence of PEB4 (0.5–2.5 μm), PEB4ΔPPI (0.5 μm), or BSA (0.5 μm; negative control).
RESULTS
The C. jejuni Genome Encodes Three Periplasmic Proteins Related to SurA
To test whether C. jejuni possesses any members of the SurA superfamily that might be involved in the OMP assembly pathway in addition to PEB4, we scanned the Pfam SurA_N HMM (PF09312.4; Pfam release 24.0) against the C. jejuni NCTC11168 predicted translated ORFs (1643 sequences) using HMMSEARCH from the HMMER3 package (53). The only hit above the default inclusion threshold of E = 0.01 was the protein Cj1289 (E = 7.5e-12, score = 42.0), which is annotated as a putative periplasmic protein. Only with a relaxed inclusion threshold did PEB4 (Cj0596) appear on the output list and was ranked sixth (E = 0.34, score = 7.7), whereas the Trigger Factor was ranked 27th (E = 4.5, score = 4.0). We used Phyre to further analyze all of the top 30 sequences in the list of hits. From the analysis, Cj0694 (ranked 4th in the list, E = 0.17, score = 8.6, annotated as a putative periplasmic protein) was also identified as a SurA homologue. The only other possible hit suggested by Phyre was Cj1495 (ranked 10th in the list). However, for Cj1495, Phyre does not rank SurA as the top hit, and the secondary structure alignment is not convincing. The full complement of SurA homologues detected in C. jejuni NCTC11168 is therefore Cj1289, Cj0596 (PEB4), and Cj0694. The presence of two of these significantly below the typical HMMER3 inclusion threshold demonstrates the challenge of categorically identifying all SurA homologues in the protein repertoire of an organism. In BLAST searches we could find no homologues of the alternative chaperone Skp.
PEB4 Forms a Domain-swapped Dimer
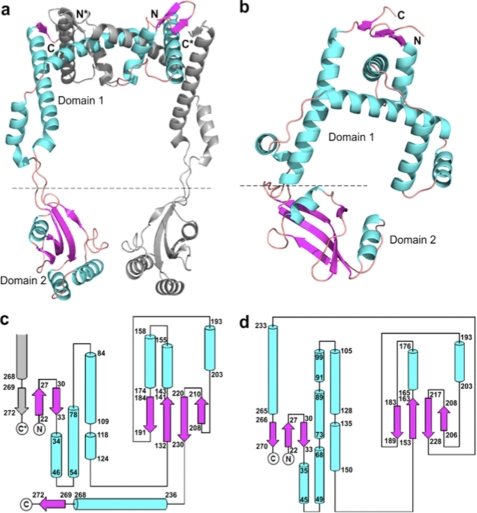
The crystal structure of PEB4 was elucidated to a resolution of 2.2 Å. A PEB4 monomer was formed from two domains: a predominantly helical domain 1, composed of N- and C-terminal regions of the protein (residues 22–127 and 236–273) and domain 2 (residues 132–231), which has a classical parvulin PPIase-fold. These two domains are joined by an extended linker region (residues 128–131 and 232–235) (Fig. 1). The C-terminal His6 tag along with a short linker is not discernible in the density maps, indicating its highly flexible nature. Gel filtration during purification indicated a species in solution that was intermediate between a dimer and trimer (∼90 kDa). Examination of the crystal packing reveals a probable biological dimer of PEB4 formed by extensive interactions between two crystal symmetry-related monomers (Fig. 1). This dimer has an elongated structure and might be expected to run with an anomalously large molecular weight as estimated by gel filtration. Most of the buried surface area on formation of the dimer is between domains 1 from each chain, with a small contact area between domains 2. The overall surface area of the monomer is ∼17,000 Å2 and that of dimer is ∼30,000 Å2 with a total buried fraction of ∼4,000 Å2, as calculated using the PISA server (54).
FIGURE 1.
The structures of PEB4 and Cj1289. a, a dimer of PEB4 as observed in the asymmetric unit of the crystal depicted in ribbon form with α-helices and β-strands shown as coils and arrows, respectively. One monomer is colored gray and the second monomer with helices in cyan and strands in magenta. The domains are indicated (domain 1 is the chaperone domain and domain 2 is the PPIase) and termini from the monomers are differentiated with *. Formation of an inter-monomer 3-stranded β-sheet can be clearly seen in the top right-hand corner and emphasizes the domain swapped architecture. b, a monomer of Cj1289 depicted in the same manner and with the same color scheme as in a. The equivalent 3-stranded β-sheet is clearly intra-monomer. c and d, topology diagrams of PEB4 and Cj1289, respectively. The α-helices and β-strands are shown as cylinders and arrows and the color scheme is maintained from a. Residue numbers marking the extent of the secondary structure elements are given. The PPIase domain 2 is shown on the right of each diagram. In c the formation of the inter-monomer β-sheet is indicated by the presence of an extra, gray colored strand from the other monomer in the dimer.
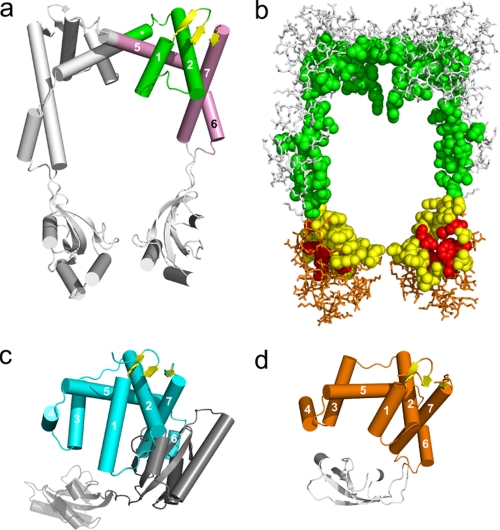
As suspected from bioinformatics searches, the folds of the domains in PEB4 and SurA are similar. However, the extent of the similarity of domain 1 in PEB4 and the chaperone domain of SurA is only evident when the structure of PEB4 is interpreted as a domain-swapped dimer. Thus, a unit of structure containing elements from both chains of the dimer can be identified (Fig. 2a), which has a very similar fold to the chaperone domain of SurA (Fig. 2c). These two units of structure overlay with an r.m.s. deviation of 3.2 Å for 72 Cα atoms. The extensive interface in PEB4 between domains 1 from each monomer involves a “swapping” or “inter-twining” of α-helices (Figs. 1 and 2). One consequence is that a small 3-stranded anti-parallel β-sheet is formed from the first two β-strands in one monomer and the terminal β-strand of the second monomer. The equivalent small sheet in SurA, in contrast, is formed by intra-monomer contacts.
FIGURE 2.
a, PEB4 in schematic representation with the helices (represented as cylinders) of the chaperone domain numbered sequentially and in the same orientation as in Fig. 1a. One unit equivalent to the SurA chaperone domain is highlighted in color. The unit comprises elements from the two chains. The helices and loops are colored green or pink according to chain identity. Note that the sheet colored yellow contains elements from both chains in PEB4. The equivalent sheet is also colored yellow in panels c and d. The helices are numbered according to their equivalent in c and d. b, space-filled representation of the residues contributing to the inner surface of the central cavity of PEB4 (orientated as in a), and wireframe representation of the remaining residues. Residues from the chaperone domain are colored green and white. Residues from the PPIase domain are colored red (active site), yellow (other inner surface residues), and orange (remaining residues). c, schematic representation of SurA. Helices and loops of the chaperone domain are in cyan. The three-stranded sheet (yellow) comprises elements from the extreme N terminus and the extreme C terminus of the monomer. The PPIase domains 1 (inactive) and 2 (active) are shown in dark gray and light gray, respectively. The chaperone domain is oriented for optimal comparison with the representation of PEB4 in panel a. d, the SurA-chaperone domain-like unit of Cj1289 (helices and loops in orange), with the single PPIase domain also shown in gray.
When the PPIase domain of PEB4 alone was submitted to the DALI server (55), a good alignment was obtained to several PPIases, major differences being observed in the loop regions. The closest match was found to be to the PPIase domain of PrsA (r.m.s. deviation 1.6 Å for 89 Cα positions, PDB code 1ZK6), a foldase from Bacillus subtilis, with a sequence identity in the PPIase domain of 46% (the structure of full-length PrsA has not been determined). Unlike PEB4, E. coli SurA has two PPIase domains of which the first is inactive (7). The PEB4 PPIase domain shares a high structural similarity with both PPIase domains of SurA (r.m.s. deviation 1.6 Å for 91 and 92 Cα positions, respectively; PDB code 1M5Y), but the sequence identity is higher to the second (active) domain (27 versus 35%). In both PEB4 and SurA structures, the PPIase domain(s) is inserted immediately before the long C-terminal helix and strand, which fold back to complete the chaperone domain 1.
The overall crystal structure of the PEB4 dimer is reminiscent of a pair of headphones with a substantial central cavity (∼55 × ∼50 Å). However, whereas the highly intertwined domain-swapped dimer is likely to be very stable, the relative positions of the PPIase domains to the chaperone domain may be quite variable in solution, as judged by an elevated average B-factor (66 Å2) for residues in the PPIase domain relative to that in the chaperone domain (44 Å2).
The Inner Surface of the PEB4 Chaperone Domain Is Markedly Hydrophobic
Initial examination of the surface of PEB4 suggested to us that the interior surface of the central cavity was substantially more hydrophobic than the exterior surface of the protein. To analyze the surface composition in greater detail we first selected residues belonging to the inner surface of the central cavity (space-filled residues in Fig. 2b). We further subdivided these areas into chaperone domain (green residues in Fig. 2b) and PPIase domain (yellow and red residues in Fig. 2b). We then calculated (see “Experimental Procedures”) the fraction of surface-exposed residues that were non-polar (supplemental Fig. S1) and the corresponding values for the remaining surface residues of the two domains (which we define as the exterior surface). As a second measure (see “Experimental Procedures”) the fractions of surface area contributed by polar and non-polar atoms to these surfaces was also calculated (supplemental Fig. S1).
By both measures the inner surface is markedly more hydrophobic than the outer surface. In typical mesophiles, only 17% of the surface residues of soluble proteins are non-polar (47). In PEB4, the total inner surface, and in particular the chaperone domain is notably more hydrophobic, with ∼50% of surface residues being non-polar. In contrast, the outer surfaces show values much more similar to typical mesophiles, with a strong preponderance of polar residues (supplemental Fig. S1). The atom-based approach counts substantial fractions of classically polar residues such as arginine as non-polar. For a typical mesophile, the ratio of polar to non-polar surface atoms in soluble proteins is therefore rather more even with 47% of surface atoms non-polar (56). By this measure also, the inner surfaces have a significantly elevated proportion of non-polar atoms, whereas the outer surfaces show values very close to those expected for typical mesophiles (supplemental Fig. S1).
By homology with other PPIases, the active site of the PEB4 PPIase domain incorporates residues His138, Leu186, Met194, Phe198, Phe219, and His222 (corresponding to residues His289, Leu341, Phe349, Phe353, Phe373, and His376 of SurA). These residues are colored red in Fig. 2b. In the crystallized conformation, the active site is oriented toward the central cavity, and is therefore well positioned to access proline residues in an unfolded or partially folded substrate sequestered within the cavity.
PPIase Activity of PEB4 with Peptide and Protein Substrates
To gain evidence for a potential dual PPIase and chaperone role for PEB4, we conducted a series of experiments to determine the PPIase activity of the protein and investigate the nature of its chaperone activity. The classical PPIase assay where a chromogenic proline-containing peptide is cleaved by chymotrypsin in a spectrophotometric assay for cis-trans isomerization has previously been employed with PEB4 (26), but in our hands gave unreliable results due to digestion of PEB4 by the protease. We have used two-dimensional 1H NMR exchange spectroscopy to measure the isomerization kinetics based on the resonances of protons in the peptide amino acids adjacent to the proline residue in N-succinyl-Ala-Ala-Pro-Phe-p-nitroanilide, in the presence and absence of purified PEB4 (Fig. 3). The rates of exchange (per PEB4 molecule) of 140 and 340 s−1, for substrate concentrations of 1 and 3 mm, respectively, indicate that the catalytic ability of PEB4 is comparable with that of cyclophilin, which has kcat ∼ 600 s−1 (57).
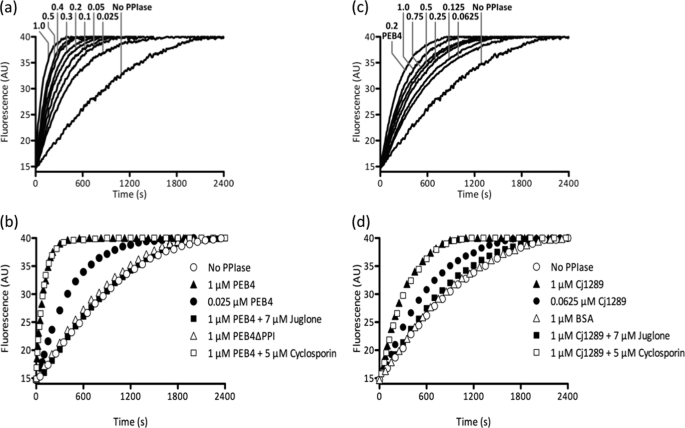
The PPIase activity of PEB4 with a protein substrate was demonstrated by monitoring the acceleration of the proline isomerization limited refolding of reduced and carboxymethylated ribonuclease T1, as reported by tryptophan fluorescence (Fig. 4, a and b). This revealed a marked PEB4 concentration-dependent enhancement of the rate of refolding (Fig. 4a) that was specificially inhibited by the parvulin family inhibitor juglone, but not by the cyclophilin family inhibitor cyclosporin (Fig. 4b). A variant of PEB4 lacking the PPIase domain was engineered using the structural information obtained above. Residues 123–231 of the native protein were deleted and replaced by a Gly-Ser linker. This PEB4ΔPPI variant was stable and could be purified by a modification of the standard procedure used for PEB4 (see “Experimental Procedures”). It was completely inactive in accelerating the refolding of ribonuclease T1 (Fig. 4b).
FIGURE 4.
Comparison of the PPIase activity of PEB4, PEB4ΔPPI, and Cj1289 proteins as measured by ribonuclease refolding. At time 0, refolding of RCM-RNase T1 was initiated by a 30-fold dilution to give a final concentration of 0.5 μm in the presence of 2 m NaCl. The fluorescence change was measured at 15 °C and the data shown are representative of three independent experiments. a, the refolding of RCM-RNase T1 is systematically accelerated by increasing concentrations of PEB4. The numbers on the graph are the final concentrations of PEB4 in μm. b, deletion of the PPIase domain of PEB4 (PEB4ΔPPI) abolishes the rate enhancement observed with full-length PEB4 protein. Preincubation of PEB4 with cyclosporin A, a cyclophilin inhibitor, had no effect on its ability to assist RNase T1 refolding, whereas preincubation with juglone, a parvulin inhibitor, completely eliminated the acceleration of refolding. c, as in panel a but using increasing concentrations of Cj1289 (numbers are final concentration in μm) in comparison to 0.2 μm PEB4. d, juglone inhibits the PPIase activity of Cj1289, whereas cyclosporin A is without effect. BSA is used here as an additional control to show that the rate enhancement is Cj1289 specific.
PEB4 and PEB4ΔPPI Inhibit the Refolding of Denatured Rhodanese
To test the ability of PEB4 to act as a chaperone, we used assays based on refolding of the model protein rhodanese (a sulfurtransferase). In the presence of PEB4, there was a reduced rate of thiocyanate formation compared with the control, indicating a reduced yield of active folded rhodanese (Fig. 5a). Similarly, in a protein aggregation assay (Fig. 5b), increasing PEB4 concentrations progressively inhibited aggregation of denatured rhodanese, as measured by light scattering kinetics, whereas BSA was without effect. These effects were mediated largely by the chaperone domain of PEB4, as the purified variant protein (PEB4ΔPPI), specifically lacking the PPIase domain, still showed substantial inhibition of rhodanese refolding (Fig. 5a) and aggregation (Fig. 5b). Therefore, it is clear that PEB4 is a holdase-type chaperone that can slow protein folding and aggregation, a role consistent with binding and transporting client proteins maintained in only a partially folded state to the BAM complex for insertion in the OM.
FIGURE 5.
Chaperone activity of PEB4, PEB4ΔPPI, and Cj1289. a, PEB4 inhibits the refolding of chemically denatured rhodanese, whereas Cj1289 does not. At time 0, refolding of denatured rhodanese was initiated by dilution into 50 mm Tris-HCl, pH 7.8, containing 50 mm Na2S2O3, 10 mm KCN, and 10 mm dithiothreitol and the restoration of sulfurtransferase activity (accumulation of thiocyanate) monitored as described under “Experimental Procedures.” PEB4, PEB4ΔPPI, Cj1289, or BSA were present in the refolding solution prior to the addition of the denatured rhodanese to give 0.5 μm final concentration, (equimolar with rhodanese). b, effect of PEB4 on rhodanese aggregation. The data points show the increase in optical density at 320 nm accompanying aggregation of denatured rhodanese after a 60-fold dilution into buffer at time 0. PEB4 and the PEB4ΔPPI deletion variant significantly reduced the formation of rhodanese aggregates, whereas the presence of BSA at an equimolar concentration had no effect.
Cj1289 Is Structurally More Similar to SurA Than Is PEB4
Cj1289 was readily expressed in a soluble form, purified and crystallized. Gel filtration analysis during purification suggested that the protein exists as an elongated monomer (∼40 kDa) in solution. Examination of the crystal packing reveals one of the two monomers in the asymmetric unit is somewhat more globular in nature than the other. This is in agreement with measurements of surface area; the PISA (30) server indicates the overall surface areas for the two monomers differ by ∼1000 Å2 (14,837 versus 15,813 Å2). The crystal structure was elucidated to a resolution of 2.3 Å (Fig. 1). Like PEB4, a monomer of Cj1289 consists of two structural domains. Domain 1 (residues 22–146 and 233–270, Figs. 1 and 2) has a fold similar to the SurA chaperone domain. Domain 2 (residues 152–228) has a parvulin-type PPIase fold. These two domains are joined by a linker region (residues 147–151 and 229–232). The highly flexible C-terminal His6 tag along with a short linker is not discernible in the density maps for either monomer A or B, nor are residues 102–104 (inclusive) in monomer A. Like E. coli SurA, but unlike PEB4, the N and C termini of the same monomer of Cj1289 form a small, 3-stranded anti-parallel β-sheet. Thus, unlike PEB4 there is no domain swapping or inter-twining of monomers observed in the case of Cj1289.
PPIase and Chaperone Activities of Cj1289
In the RNase T1 refolding assay, purified Cj1289 catalyzed an enhancement of the rate of refolding in a concentration-dependent manner (Fig. 4c) showing that the PPIase domain of the protein is active. However, compared with PEB4, Cj1289 appeared to exhibit lower catalytic activity as a PPIase; the rate constant at 1 μm Cj1289 in the assay was ∼4-fold lower than with 1 μm PEB4 under the same conditions (Fig. 4, b and d). The parvulin-specific inhibitor juglone completely inhibited the Cj1289 catalyzed reaction, whereas cyclosporin was without effect (Fig. 4d), the same pattern as seen with PEB4 and consistent with the parvulin-like fold of the Cj1289 PPIase domain. However, unlike PEB4, purified Cj1289 did not inhibit the refolding of denatured rhodanese (Fig. 5a). We were also unable to demonstrate any Cj1289-dependent inhibition of aggregation of this protein (data not shown). These data are surprising given the structural similarity to SurA, and suggest that the chaperone domain may be specific for particular client proteins.
Bioinformatic Analysis Suggests That Cj0694 Is an Orthologue of PpiD
In a BLAST search, the closest corresponding E. coli protein to Cj0694 is PpiD (bit score = 86 using BLOSUM62 for an alignment of 399 residues of Cj0694; next highest bit score is 33 for alignment to SurA across 236 residues of Cj0694). Inspection of the predicted secondary structure from Phyre also suggests a much closer relationship to PpiD than to SurA (data not shown). PpiD is anchored on the periplasmic face of the inner membrane via an N-terminal transmembrane domain, and is believed to function as a folding chaperone for proteins that are exported via the Sec machinery (58). Cj0694 is also likely to be membrane anchored in C. jejuni, as SignalP does not predict the presence of a signal peptidase I cleavage site in the N-terminal region, unlike both Cj1289 and PEB4, which have typical cleaved signal sequences. Overexpression of Cj0694 resulted in the formation of only insoluble protein, even after removal of the predicted N-terminal membrane-spanning region, thus precluding structural comparisons with PEB4 and Cj1289.
DISCUSSION
In this work we have begun to elucidate the basis for OMP assembly in C. jejuni by determination of the structure and activity of two periplasmic chaperones that have folds similar to the well characterized E. coli SurA protein. C. jejuni does not appear to contain an Skp homologue, and PEB4 is currently the only protein implicated in OMP assembly, based on changes in OMP abundance in peb4 mutants (25–27). The 2.2-Å structure determined here reveals a monomer composed of independently folded chaperone and PPIase domains, connected by a short linker region. The chaperone domain is clearly related to that in the monomeric SurA but is distinct in the way in which helices from two monomers closely interlock to form a biological dimer. The result is to form a larger substrate-binding cradle than that proposed for SurA (59). The central cavity of PEB4, bounded by domains 1 and 2, may be analogous to the “cargo bay” of Trigger Factor (60). We have highlighted the increased hydrophobicity of the inner surface of the central cavity and the likely location of the PPIase active site, which are both consistent with the binding of unfolded or partially folded protein cargoes in this internal region of PEB4. A possible consequence of the postulated mobility of the PPIase domains could be to contract the central cavity around a bound substrate to either align the PPIase domain for enhanced catalytic activity or to enhance binding.
The formation of a domain-swapped dimer in PEB4 is most similar to that seen in the partial structure of Par27 (61), a periplasmic chaperone from Bordetella pertussis. Par27 is involved in assembly of the filamentous hemagglutinin, a secreted virulence factor (62). As assessed from the published data, the overall topology of the chaperone domain is very similar to that seen in PEB4, but the electron density of the PPIase domains in the Par27 dimer could not be seen in the crystal structure (61), implying a high degree of flexibility. However, at the time of publication of our manuscript, coordinates for the Par27 structure were not available and we were thus unable to perform a full three-dimensional structure comparison.
Although the biological role of the single PPIase domain of PEB4 is unknown, it is catalytically active based on the high rates of exchange in two-dimensional 1H NMR experiments with a peptide substrate and the PPIase activity of PEB4 in the ribonuclease T1 assay. The latter revealed a marked PEB4-dependent acceleration of the rate of refolding that was completely inhibited by the parvulin family inhibitor juglone, but not by cyclosporin A. SurA, unlike PEB4, has two parvulin-like PPIase domains, one of which is catalytically inactive (9) but packs closely against the chaperone domain, whereas the active PPIase forms a more loosely tethered satellite domain (63). There is considerable evidence that the first (inactive) PPIase domain is involved in client protein recognition. SurA binds proteins bearing the C-terminal Ar-Ar or Ar-X-Ar motif (where Ar is an aromatic residue, and X is any other residue) that is characteristic of many OMPs (64–66). Even short peptides (<11 amino acids) interact with SurA as long as they contain this signature motif (10). The inactive parvulin-like domain confers substrate specificity by interacting with the aromatic residues of the tested peptides (63). C. jejuni OMPs also commonly have a C-terminal aromatic residue motif. However, the PEB4 PPIase domain is most similar to the second (active) PPIase domain of SurA in terms of sequence similarity and remoteness to the chaperone domain. Thus the mechanism of substrate recognition may be substantially different.
The nature of the chaperone activity of PEB4 was demonstrated in refolding studies with the classical model protein rhodanese. We found that PEB4 reduced the yield of active protein in the refolding assay. Increasing PEB4 concentrations strongly inhibited aggregation of renaturing rhodanese, as measured by light scattering kinetics. Taken together, these data indicate that PEB4 is a holdase-type chaperone that can inhibit protein folding and aggregation by sequestration of unfolded protein. The same role has been demonstrated for a number of other chaperones involved in OMP assembly, notably SurA itself (8) and Par27 (62). A variant protein (PEB4ΔPPI), consisting only of the chaperone domain, was constructed to test the hypothesis that the chaperone activity may be independent of the PPIase domain. As expected, this protein was totally inactive in the RNase T1 PPIase assay but still exhibited inhibition of refolding and reduced aggregation of rhodanese, although it was less effective than the full-length PEB4. This would indicate that the PPIase domain does make a contribution to the overall chaperone activity.
Our results show that PEB4 is not the only SurA-like chaperone present in C. jejuni; bioinformatics identified two additional homologues. Cj1289 was the top hit in our searches, whereas PEB4 was only detected at a substantially lower similarity level. This suggested that Cj1289 was likely to be structurally more similar to SurA, which was confirmed by the 2.3-Å crystal structure obtained in this work. There is no domain swapping in the chaperone domain of Cj1289, and the protein behaves as a monomer in solution. As in PEB4, there is a single parvulin-like PPIase domain rather than two as in SurA. The PPIase domain of Cj1289 is active in enhancing the rate of refolding of RNase T1, but with a lower catalytic activity compared with PEB4. Given the overall similarity to both SurA and PEB4, it is surprising that we could not demonstrate chaperone activity in the rhodanese refolding and aggregation assays. It is thus possible that Cj1289 exhibits stronger substrate specificity related to its physiological role in the C. jejuni periplasm.
The third periplasmic SurA-like chaperone we discovered in this work is Cj0694. BLAST searches indicate weak sequence similarity to the PpiD protein of E. coli. PpiD was first identified as a putative OMP chaperone by the fact that its overexpression can rescue bacteria in which surA is deleted (29). More recent evidence refutes this, and shows that PpiD is not specifically involved in the maturation of OMPs (67). It also has a catalytically inactive parvulin domain (68). PpiD is not free in the periplasm but membrane anchored by an N-terminal uncleaved signal sequence (58, 67). From the retardation in a ppiD mutant of the release of OmpA into the periplasm through the Sec system (58) a role for PpiD in immediately interacting with a variety of proteins exiting from the SecYEG translocon was suggested; it may thus act as the periplasmic equivalent of Trigger Factor (67). Unfortunately, recombinant Cj0694 could not be obtained in a soluble form for structural studies. However, the sequence similarity to PpiD coupled with the predicted N-terminal membrane spanning region lacking an obvious signal peptidase cleavage site, argues for a similar role for this protein in C. jejuni as in E. coli.
The current model of outer membrane protein assembly in which specific periplasmic chaperones bind OMPs emerging from the Sec system in a partially folded state and present these to the BAM complex in the outer membrane, has come largely from studies on only E. coli and N. meningitidis (1, 2, 69), although there are significant differences in OMP assembly between these species. In E. coli only BamA and BamD are essential for viability (70). In C. jejuni, an obvious homologue of BamA can be identified by homology (Cj0129), and a probable BamD is encoded by cj1074c. However, our extensive bioinformatics searches have revealed no homologues of BamB, BamC, or BamE, suggesting either that BamAD is the minimal complex required for OM assembly, or that C. jejuni employs additional novel proteins in its Bam complex. In addition to distinct chaperones, it is thus evident that fundamental and poorly understood variations exist in the OMP assembly pathway in different bacterial groups. The recent successful reconstitution of the BAM complex (69) provides an in vitro system with which to elucidate the different roles of chaperones like PEB4 and Cj1289 that may have distinct physiological functions.
Supplementary Material
Acknowledgments
We acknowledge the support of Dr. S. Sedelnikova and Dr. P. Baker with protein purification and x-ray data collection, and Andrea Hounslow for NMR data collection.
This work was supported in part by a grant from the United Kingdom Biotechnology and Biological Sciences Research Council.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1 and Tables S1–S3.
The atomic coordinates and structure factors (codes 3RFW and 3RGC) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- BAM
- β-barrel assembly machinery
- OMP
- outer membrane protein
- PPIase
- peptidyl-prolyl cis/trans isomerase
- POTRA
- polypeptide-transport-associated
- RCM
- reduced and S-carboxymethylated
- r.m.s.
- root mean square
- CHES
- 2-(cyclohexylamino)ethanesulfonic acid
- TLS
- Translation/Libration/Screw
- PDB
- Protein Data Bank.
REFERENCES
- 1. Bos M. P., Robert V., Tommassen J. (2007) Annu. Rev. Microbiol. 61, 191–214 [DOI] [PubMed] [Google Scholar]
- 2. Knowles T. J., Scott-Tucker A., Overduin M., Henderson I. R. (2009) Nat. Rev. Microbiol. 7, 206–214 [DOI] [PubMed] [Google Scholar]
- 3. Mogensen J. E., Otzen D. E. (2005) Mol. Microbiol. 57, 326–346 [DOI] [PubMed] [Google Scholar]
- 4. Tamm L. K., Hong H., Liang B. (2004) Biochim. Biophys. Acta 1666, 250–263 [DOI] [PubMed] [Google Scholar]
- 5. Sklar J. G., Wu T., Kahne D., Silhavy T. J. (2007) Genes Dev. 21, 2473–2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Volokhina E. B., Grijpstra J., Stork M., Schilders I., Tommassen J., Bos M. P. (2011) J. Bacteriol. 193, 1612–1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bitto E., McKay D. B. (2002) Structure 10, 1489–1498 [DOI] [PubMed] [Google Scholar]
- 8. Behrens S., Maier R., de Cock H., Schmid F. X., Gross C. A. (2001) EMBO J. 20, 285–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rouvière P. E., Gross C. A. (1996) Genes Dev. 10, 3170–3182 [DOI] [PubMed] [Google Scholar]
- 10. Stymest K. H., Klappa P. (2008) FEBS J. 275, 3470–3479 [DOI] [PubMed] [Google Scholar]
- 11. Vertommen D., Ruiz N., Leverrier P., Silhavy T. J., Collet J. F. (2009) Proteomics 9, 2432–2443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Robert V., Volokhina E. B., Senf F., Bos M. P., Van Gelder P., Tommassen J. (2006) PLoS Biol. 4, e377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kim S., Malinverni J. C., Sliz P., Silhavy T. J., Harrison S. C., Kahne D. (2007) Science 317, 961–964 [DOI] [PubMed] [Google Scholar]
- 14. Misra R. (2007) ACS Chem. Biol. 2, 649–651 18041813 [Google Scholar]
- 15. Noinaj N., Fairman J. W., Buchanan S. K. (2011) J. Mol. Biol. 407, 248–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Heuck A., Schleiffer A., Clausen T. (2011) J. Mol. Biol. 406, 659–666 [DOI] [PubMed] [Google Scholar]
- 17. Kim K. H., Paetzel M. (2011) J. Mol. Biol. 406, 667–678 [DOI] [PubMed] [Google Scholar]
- 18. Knowles T. J., Browning D. F., Jeeves M., Maderbocus R., Rajesh S., Sridhar P., Manoli E., Emery D., Sommer U., Spencer A., Leyton D. L., Squire D., Chaudhuri R. R., Viant M. R., Cunningham A. F., Henderson I. R., Overduin M. (2011) EMBO Rep. 12, 123–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Suzuki H., Yamamoto S. (2009) J. Vet. Med. Sci. 71, 255–261 [DOI] [PubMed] [Google Scholar]
- 20. Janssen R., Krogfelt K. A., Cawthraw S. A., van Pelt W., Wagenaar J. A., Owen R. J. (2008) Clin. Microbiol. Rev. 21, 505–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jacobs B. C., Koga M., van Rijs W., Geleijns K., van Doorn P. A., Willison H. J., Yuki N. (2008) J. Neuroimmunol. 194, 181–190 [DOI] [PubMed] [Google Scholar]
- 22. Hu L., Tall B. D., Curtis S. K., Kopecko D. J. (2008) Infect. Immun. 76, 5294–5304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Flanagan R. C., Neal-McKinney J. M., Dhillon A. S., Miller W. G., Konkel M. E. (2009) Infect. Immun. 77, 2399–2407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Konkel M. E., Larson C. L., Flanagan R. C. (2010) J. Bacteriol. 192, 68–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Asakura H., Yamasaki M., Yamamoto S., Igimi S. (2007) FEMS Microbiol. Lett. 275, 278–285 [DOI] [PubMed] [Google Scholar]
- 26. Rathbun K. M., Hall J. E., Thompson S. A. (2009) BMC Microbiol. 9, 160–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rathbun K. M., Thompson S. A. (2009) FEMS Microbiol. Lett. 300, 188–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kelley L. A., Sternberg M. J. (2009) Nat. Protoc. 4, 363–371 [DOI] [PubMed] [Google Scholar]
- 29. Dartigalongue C., Raina S. (1998) EMBO J. 17, 3968–3980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Doublié S. (1997) Methods in Enzymology, Macromolecular Crystallography, Part A, Academic Press, San Diego [Google Scholar]
- 31. Leslie A. G. (1992) Joint CCP4 + ESF-EAMCB Newsletter on Protein Crystallography 26, 27–33 [Google Scholar]
- 32. Evans P. (2006) Acta Crystallogr. D 62, 72–82 [DOI] [PubMed] [Google Scholar]
- 33. Potterton E., Briggs P., Turkenburg M., Dodson E. (2003) Acta Crystallogr. D 59, 1131–1137 [DOI] [PubMed] [Google Scholar]
- 34. Terwilliger T. C., Berendzen J. (1999) Acta Crystallogr. D 55, 849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Terwilliger T. C. (2000) Acta Crystallogr. D 56, 965–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cowtan K. (2000) Acta Crystallogr. D 56, 1612–1621 [DOI] [PubMed] [Google Scholar]
- 37. Cowtan K. (2006) Acta Crystallogr. D 62, 1002–1011 [DOI] [PubMed] [Google Scholar]
- 38. Emsley P., Cowtan K. (2004) Acta Crystallogr. D 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 39. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 40. Morris R. J., Perrakis A., Lamzin V. S. (2003) Methods Enzymol. 374, 229–244 [DOI] [PubMed] [Google Scholar]
- 41. Morris R. J., Zwart P. H., Cohen S., Fernandez F. J., Kakaris M., Kirillova O., Vonrhein C., Perrakis A., Lamzin V. S. (2004) J. Synchrotron. Radiat. 11, 56–59 [DOI] [PubMed] [Google Scholar]
- 42. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) Acta Crystallogr. D 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Painter J., Merritt E. A. (2006) J. Appl. Cryst. 39, 109–111 [Google Scholar]
- 44. Chen V. B., Arendall W. B., 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C. (2010) Acta Crystallogr. D 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bond C. S. (2003) Bioinformatics 19, 311–312 [DOI] [PubMed] [Google Scholar]
- 46. Miller S., Janin J., Lesk A. M., Chothia C. (1987) J. Mol. Biol. 196, 641–656 [DOI] [PubMed] [Google Scholar]
- 47. Fukuchi S., Nishikawa K. (2001) J. Mol. Biol. 309, 835–843 [DOI] [PubMed] [Google Scholar]
- 48. Scholz C., Stoller G., Zarnt T., Fischer G., Schmid F. X. (1997) EMBO J. 16, 54–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mücke M., Schmid F. X. (1994) Biochemistry 33, 14608–14619 [DOI] [PubMed] [Google Scholar]
- 50. Mücke M., Schmid F. X. (1992) Biochemistry 31, 7848–7854 [DOI] [PubMed] [Google Scholar]
- 51. Horowitz P., Westley J. (1970) J. Biol. Chem. 245, 986–990 [PubMed] [Google Scholar]
- 52. Ideno A., Yoshida T., Furutani M., Maruyama T. (2000) Eur. J. Biochem. 267, 3139–3149 [DOI] [PubMed] [Google Scholar]
- 53. Eddy S. R. (2008) PLoS Comput. Biol. 4, e1000069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Krissinel E., Henrick K. (2007) J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 55. Holm L., Rosenström P. (2010) Nucleic Acids Res. 38, W545–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Britton K. L., Stillman T. J., Yip K. S., Forterre P., Engel P. C., Rice D. W. (1998) J. Biol. Chem. 273, 9023–9030 [DOI] [PubMed] [Google Scholar]
- 57. Kern D., Kern G., Scherer G., Fischer G., Drakenberg T. (1995) Biochemistry 34, 13594–13602 [DOI] [PubMed] [Google Scholar]
- 58. Antonoaea R., Fürst M., Nishiyama K., Müller M. (2008) Biochemistry 47, 5649–5656 [DOI] [PubMed] [Google Scholar]
- 59. Stirling P. C., Bakhoum S. F., Feigl A. B., Leroux M. R. (2006) Nat. Struct. Mol. Biol. 13, 865–870 [DOI] [PubMed] [Google Scholar]
- 60. Martinez-Hackert E., Hendrickson W. A. (2009) Cell 138, 923–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Clantin B., Leyrat C., Wohlkönig A., Hodak H., Ribeiro Ede A., Jr., Martinez N., Baud C., Smet-Nocca C., Villeret V., Jacob-Dubuisson F., Jamin M. (2010) J. Struct. Biol. 169, 253–265 [DOI] [PubMed] [Google Scholar]
- 62. Hodak H., Wohlkönig A., Smet-Nocca C., Drobecq H., Wieruszeski J. M., Sénéchal M., Landrieu I., Locht C., Jamin M., Jacob-Dubuisson F. (2008) J. Mol. Biol. 376, 414–426 [DOI] [PubMed] [Google Scholar]
- 63. Xu X., Wang S., Hu Y. X., McKay D. B. (2007) J. Mol. Biol. 373, 367–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Bitto E., McKay D. B. (2003) J. Biol. Chem. 278, 49316–49322 [DOI] [PubMed] [Google Scholar]
- 65. Bitto E., McKay D. B. (2004) FEBS Lett. 568, 94–98 [DOI] [PubMed] [Google Scholar]
- 66. Hennecke G., Nolte J., Volkmer-Engert R., Schneider-Mergener J., Behrens S. (2005) J. Biol. Chem. 280, 23540–23548 [DOI] [PubMed] [Google Scholar]
- 67. Matern Y., Barion B., Behrens-Kneip S. (2010) BMC Microbiol. 10, 251–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Weininger U., Jakob R. P., Kovermann M., Balbach J., Schmid F. X. (2010) Protein Sci. 19, 6–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Hagan C. L., Kim S., Kahne D. (2010) Science 328, 890–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Onufryk C., Crouch M. L., Fang F. C., Gross C. A. (2005) J. Bacteriol. 187, 4552–4561 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.