Abstract
The transfer of ubiquitin (Ub) to a substrate protein requires a cascade of E1 activating, E2 conjugating, and E3 ligating enzymes. E3 Ub ligases containing U-box and RING domains bind both E2∼Ub conjugates and substrates to facilitate transfer of the Ub molecule. Although the overall mode of action of E3 ligases is well established, many of the mechanistic details that determine the outcome of ubiquitination are poorly understood. CHIP (carboxyl terminus of Hsc70-interacting protein) is a U-box E3 ligase that serves as a co-chaperone to heat shock proteins and is critical for the regulation of unfolded proteins in the cytosol. We have performed a systematic analysis of the interactions of CHIP with E2 conjugating enzymes and found that only a subset bind and function. Moreover, some E2 enzymes function in pairs to create products that neither create individually. Characterization of the products of these reactions showed that different E2 enzymes produce different ubiquitination products, i.e. that E2 determines the outcome of Ub transfer. Site-directed mutagenesis on the E2 enzymes Ube2D1 and Ube2L3 (UbcH5a and UbcH7) established that an SPA motif in loop 7 of E2 is required for binding to CHIP but is not sufficient for activation of the E2∼Ub conjugate and consequent ubiquitination activity. These data support the proposal that the E2 SPA motif provides specificity for binding to CHIP, whereas activation of the E2∼Ub conjugate is derived from other molecular determinants.
Keywords: E3 Ubiquitin Ligase, Enzyme Mechanisms, NMR, Protein-Protein Interactions, Ubiquitin-conjugating Enzyme (Ubc), Ubiquitination
Introduction
Ubiquitination is a key post-translational modification most commonly associated with protein turnover via degradation in the 26 S proteasome but is also involved in the regulation of gene expression, protein activation, and localization (1, 2). To modify a substrate, ubiquitin (Ub)2 is covalently attached to a lysine through its C-terminal glycine by the sequential action of a cascade of three enzymes. The E1 ubiquitin-activating enzyme uses ATP to generate a thioester bond to Ub and then hands off the Ub to an E2 ubiquitin-conjugating enzyme via thioester bond transfer, a process known as charging the E2 enzyme. In the final step, the E3 ubiquitin ligase catalyzes ligation of Ub to the substrate. U-box- and RING-type E3 ligases act as scaffolds to bring together the E2∼Ub conjugate and the substrate. These E3 enzymes also play a critically important role in activating the E2∼Ub conjugate, which is otherwise unable to efficiently transfer Ub to a substrate. Creation of polyubiquitin chains is thought to occur by multiple cycles of E2 charging and cycling through the E3-substrate complex. Recent evidence has shown that polyubiquitination may result from the action of combinations of E2 and E3 enzymes with separate initiation and elongation functions (3–8). The location, length, and linkage of ubiquitination are all important parts of the signal resulting from ubiquitin attachment.
Consistent with the diversity of functional roles, there are numerous potential products that result from protein ubiquitination. Although the basic mechanisms of ubiquitination have been established, many questions remain about the factors that determine ubiquitination products. Part of the answer derives from the diversity of E2 and E3 enzymes; humans have 38 E2 enzymes and >650 U-box/RING E3 ligases (1). Two important issues yet to be fully resolved are the molecular determinants for E2 enzyme function with a given E3 ligase and the structural basis for activation of an E2∼Ub conjugate by interaction with the E3 ligase. These questions are particularly intriguing because there is a great deal of structural similarity among E2 enzymes and among the U-box and RING E2-binding domains of E3 ligases. E2 enzymes all contain a conserved catalytic core domain that interacts with both E1 and E3 (9), and all U-box and RING domains are structurally conserved (10, 11). Furthermore, the accumulated structures of RING and U-box domains and their complexes with E2 enzymes reveal a conserved E2-binding interface, and there appears to be little structural change induced upon binding for either the U-box/RING domain or the E2 catalytic core (12).
Although the E2 enzymes are essential, little is known about how they affect ubiquitination outcomes. Most studies of ubiquitination activity have used a single E2, typically Ube2D1 (UbcH5a), or a mixture of E2 enzymes extracted from cells. A few recent investigations have tested differences in ubiquitination outcome for several specific E2 enzymes, but large-scale screening is rare. The identity of the E2 enzyme in a complex appears to determine the type of ubiquitination produced by U-box and RING domain-containing E3 ligases (4, 5, 13–15). The existence of trends for classes of E3 ligases is only beginning to emerge. A recent yeast two-hybrid screen of RING domain E3 enzymes found that most bind a small number of E2 enzymes and that there are a few E2 enzymes that bind almost every E3 enzyme (16). Thus, although some data are currently available, more systematic screening of E2 interactions for a range of E3 ligases is needed to detect common traits of the E2 enzymes and what role the E3 enzyme plays in the specificity of the interaction.
CHIP (carboxyl terminus of Hsc70-interacting protein) is the best studied of the U-box E3 ligases due to its high abundance and critical position as a co-chaperone of Hsc70, Hsp70, and Hsp90 (17). CHIP has been shown to be involved in regulating the concentration, oligomerization, and toxicity of several heat shock protein substrates, including Tau, cystic fibrosis transmembrane conductance regulator (CFTR), α-synuclein, and ataxin-3 (18–21). The structure of homodimeric CHIP is known (22). It has been established that the isolated U-box domain binds the E2 enzyme, and two structures of CHIPU bound to an E2 enzyme have been reported (22, 23). Here, we report investigations into the binding and ubiquitination activity of CHIP with a set of human E2 conjugating enzymes. These data, combined with an analysis of the type of ubiquitination product generated by CHIP and specific E2 enzymes, were evaluated in the light of results reported previously for other E3 ligases and provide insights into the molecular basis for the E2 selectivity and function of CHIP.
EXPERIMENTAL PROCEDURES
DNA Constructs
A plasmid expressing Hsp70 obtained from Jeff Brodsky (University of Pittsburgh) (24) was subcloned into an in-house pBG102 vector containing an N-terminal His6 tag. The CHIP U-box domain (CHIPU; amino acids 218–303) was selected from a series of C-terminal fragments screened for stability (see below). E2 mutants were produced following the Stratagene QuikChange protocol.
Protein Purification
Wheat E1 (Uba1) was expressed in Escherichia coli BL21(DE3) cells induced with 1 mm isopropyl β-d-thiogalactopyranoside at 37 °C for 3 h. Cells were lysed in buffer containing 50 mm Tris-HCl (pH 8), 1 mm EDTA, 5 mm DTT, and 1 mm PMSF. After the addition of 5 mm MgCl2 and ATP, the E1 protein was purified on an Affi-Gel 10 resin loaded with purified ubiquitin and dialyzed into 50 mm HEPES (pH 7.5) and 1 mm DTT.
All other recombinant proteins were overexpressed in E. coli BL21(DE3) cells grown in LB or M9 minimal medium containing 15NH4Cl and [13C]glucose as needed and then induced with 0.25 mm isopropyl β-d-thiogalactopyranoside at 16 °C for 16–20 h. Cells were lysed in 20 mm Tris (pH 7.5), 300 mm NaCl, and 1 mm PMSF and purified by nickel resin eluting with 200 mm imidazole in the above buffer. E2 enzymes were further purified by cation exchange using 20 mm MES (pH 6.0) and a 50–500 mm NaCl gradient. His-tagged Hsp70 and CHIP were cleaved with H3C protease and purified again by nickel affinity chromatography and then size exclusion chromatography (S75) in 100 mm NaCl and 20 mm HEPES (pH 7.5). His-Ub was cleaved with thrombin and purified by nickel affinity chromatography, followed by benzamidine-Sepharose.
NMR Spectroscopy
All data were recorded at 600 MHz (1H) and 20 °C in buffer containing 20 mm HEPES (pH 7.5), 50 mm NaCl, and 1 mm DTT. All NMR data were processed with NMRPipe (25) and analyzed with SPARKY (26). Backbone resonances of 15N-labeled CHIPU were assigned using a combination of standard three-dimensional experiments, including HNCACB, HN(CO)CACB, HNCA, and HN(CO)CA. Resonance assignments have been deposited at the Biological Magnetic Resonance Bank with accession number 17411.
Titrations used 300 μm 15N-labeled CHIPU and began with a 2:1 complex of E2-CHIPU. Intermediate ratios of 0.125, 0.25, 0.5, and 1 were produced by adding the 2:1 complex to a solution of CHIPU alone. The normalized change in chemical shift was calculated as ((ΔH)2 + (ΔN/5)2)1/2. A significant difference was defined as >1 S.D. past the mean. Residues perturbed by binding were defined as those with significant changes in chemical shift and/or loss of intensity and line broadening throughout the observable portion of the titration for that resonance. Only residues that were consistently observed as perturbed throughout a titration series were included in the final analysis in supplemental Fig. 2.
In Vitro Ubiquitination Assays
The protocol was based on a previously published procedure (27). CHIP autoubiquitination proceeded with 75 nm E1, 0.6 μm E2, 0.5 μm CHIP, and 10 μm Ub incubated in 100 mm NaCl, 40 mm Tris (pH 7.5), 5 mm MgCl2, 1 mm DTT, and 5 mm ATP at 30 °C for 1 h unless indicated otherwise. E2 charging was probed by incubating 150 nm E1, 2 μm E2, and 10 μm Ub in the above buffer without reducing agent for 30 min. Each sample was split, and reducing or nonreducing sample buffer was added. Ubiquitination of Hsp70 by CHIP was similar to autoubiquitination reactions with 50 nm CHIP and 40 μm Ub and the addition of 1.5 μm Hsp70. All reactions were separated by SDS-PAGE and either stained with Coomassie Blue or transferred to PVDF membrane for immunoblotting with either mouse anti-Ub (Abcam) or rabbit anti-CHIP (Calbiochem) antibody. Human E1 ubiquitin-activating enzyme was purchased from Enzo Life Sciences and used for all assays with Ube2K.
RESULTS
Any E3 ubiquitin ligase has the potential to function with multiple E2 conjugating enzymes. CHIP in vitro ubiquitination activity has previously been assayed for eight E2 conjugating enzymes, with only five having activity (23, 28–30). (To avoid confusion, the standard Ube2x nomenclature is used in this study (supplemental Table 1).) Of these E2 enzymes, only Ube2D1 (UbcH5a) and Ube2N (Ubc13) have been specifically shown to interact with CHIPU (22, 23). We initially attempted to screen E2 binding to CHIPU using a yeast-two hybrid approach, but the U-box bait constructs all gave false positives during initial testing. We then turned to a combination of binding studies by NMR and in vitro ubiquitination assays to examine the molecular determinants of the CHIP E2 selectivity.
Structures of CHIPU in complex with Ube2D1 and Ube2N have been reported, and the role of CHIP interactions with the SPA motif in loop 7 of the E2 catalytic core domain was noted (23). Our experiments were therefore directed toward investigating the importance of the E2 SPA motif for binding and function with CHIP. The subset of human E2 enzymes tested in this work included all those with the loop 7 SPA motif, others with conservative mutations in this motif, and a few additional E2 enzymes selected for comparison with previously reported studies of E2 selectivity and function.
CHIPU Binds a Subset of E2 Conjugating Enzymes
To accurately characterize the binding of an E2 conjugating enzyme to CHIPU, we turned to the well established heteronuclear NMR approach (5, 31, 32). Four constructs of C-terminal CHIPU were produced: 212–303, 218–303, 223–303, and 226–303. A 15N-1H heteronuclear single-quantum coherence (HSQC) NMR spectrum was collected for each protein at pH 7.0 and 293 K. The two shortest constructs (223–303 and 226–303) appeared to be aggregated or unfolded under these conditions. CHIP-(212–303) and CHIP-(218–303) had nearly identical spectra; therefore, conditions were optimized for the shorter construct, CHIP-(218–303) (CHIPU), which was used for all further experiments for this domain.
The 15N-1H HSQC NMR spectrum of CHIPU shows dispersed signals indicative of a well folded globular domain (Fig. 1 and Fig. 2, black). CHIPU is a homodimer, and only one set of NMR signals was observed, indicating that the two subunits are identical. To perform a detailed analysis of the E2-binding site on CHIP, backbone 1H, 13C, and 15N resonance assignments were made using standard multidimensional triple-resonance experiments (see “Experimental Procedures”). The physical interactions of CHIPU with E2 conjugating enzymes were then monitored by following the titration of 15N-labeled CHIPU with unlabeled E2 by 15N-1H HSQC NMR spectra. Over the course of the titrations, only one set of signals for the CHIPU dimer was observed, indicating fast exchange on the NMR time scale between free and E2-bound CHIPU.
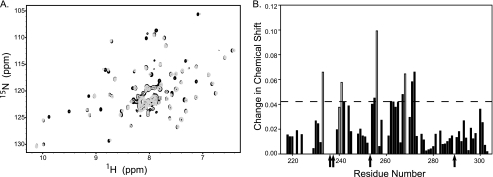
FIGURE 1.
Interaction of Ube2D1 with CHIPU. A, 800-MHz 15N-1H transverse relaxation-optimized spectroscopy-HSQC spectra for 2H,15N-labeled CHIPU collected in the absence (black) and presence (gray) of a 2-fold excess of Ube2D1. E2 binding to CHIPU resulted in resonance perturbations on the fast-to-intermediate time scale. B, analysis of resonance perturbations for the spectra in A. The change in chemical shift is plotted for each observed residue, with the dashed line showing the cutoff used for determining a significant change in chemical shift. The open bars represent resonances that were significantly broadened but still observable following the addition of Ube2D1. Residues that were broadened beyond detection are indicated by arrows.
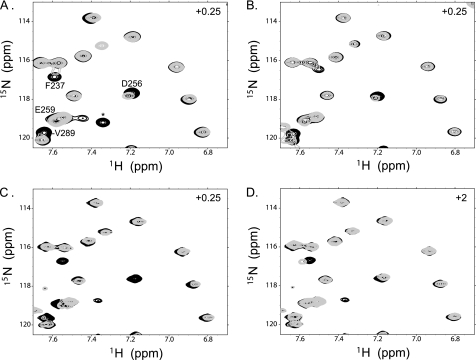
FIGURE 2.
E2-CHIPU binding is specific. Shown here is a region of 15N-1H HSQC NMR spectra for 15N-labeled CHIPU complexes with Ube2D1 (UbcH5a) (A), Ube2E3 (UbcM2) (B), Ube2W (C), and Ube2G2 (D). Free CHIPU is shown in black, with spectra for 0.25 molar eq of E2 added in A–C and 2.0 molar eq in D in gray. Key residues involved in binding are labeled to demonstrate that resonances that are specifically perturbed in A–C are not perturbed in D despite the higher concentration of E2 added. The peak labeled with an asterisk visible in A, C, and D is from an impurity.
The NMR titrations revealed two categories of E2 enzyme binding to CHIPU, one involving specific and the other involving nonspecific interactions. Titration of a nonspecific E2 enzyme resulted in only very small perturbations of CHIPU signals, even under conditions of 2-fold excess E2 (Fig. 2D), indicating very weak binding. In contrast, for an E2 enzyme that bound specifically, perturbations of the signals were observed at substoichiometric ratios (Fig. 2, A–C), and most signals were lost at a 1:1 molar ratio of E2 to CHIPU. This general loss of signals is attributed to the formation of a 2:2 E2-CHIPU complex with a total molecular mass near 60 kDa, which results in a substantial increase in the rate of spin relaxation.
To test our hypothesis about the loss of signals upon formation of the complex, a transverse relaxation-optimized spectroscopy-HSQC spectrum was acquired for uniformly deuterated CHIPU with a 2-fold molar excess of Ube2D1 (Fig. 1A). The transverse relaxation-optimized spectroscopy experiment is much less sensitive to relaxation effects than the standard HSQC experiment, so this spectrum should contain nearly all signals. Indeed, when comparing the spectra acquired for the free protein and the complex (Fig. 1A), we observed the majority of the signals and could determine chemical shift perturbations due to the binding of Ube2D1. Perturbations of signals ranged from differences in chemical shifts to increased line width/decreased intensity to complete loss of signals, as summarized in Fig. 1B. The observation of perturbations of only a subset of resonances of CHIPU confirms the formation of a specific complex. The observation of only one set of signals for the CHIP dimer supports the 2:2 stoichiometry and that the homodimer remains symmetric in the complex with E2. Together, these results confirm that the general loss of signal intensity in the HSQC of non-deuterated CHIPU is due to the large size of the complex.
The results observed for all E2 enzymes tested for binding to CHIPU were analyzed in the manner described for Ube2D1 and are provided in supplemental Fig. 1. Of the E2 enzymes tested in this series, eight (Ube2D1, Ube2D2, Ube2D3, Ube2E1, Ube2E2, Ube2E3, Ube2N, and Ube2W) were found to specifically bind CHIPU, and three (Ube2G2, Ube2K, and Ube2L3) did not. As noted above, binding to CHIP has been shown previously for Ube2D1 and Ube2N (22, 23). Fig. 2 (A–C) shows spectral overlays for NMR titrations with three different specific E2 enzymes (Ube2D1, Ube2E3, and Ube2W), which reveal similar patterns of chemical shift and line width perturbations of CHIPU at low ratios of E2 (0.25:1). In contrast, the spectrum of CHIPU with an excess of the nonspecific E2 Ube2G2 is nearly identical to the spectrum of CHIPU alone (Fig. 2D).
Our detailed analysis of the NMR signal perturbations arising upon titration with each E2 enzyme (supplemental Fig. 1) revealed that the specific E2 enzymes have largely similar effects on CHIPU. The slight differences between them are presumably due to variability in the sequence. The perturbed residues common to all specific E2 enzymes include Gly-233, Phe-237, Glu-238, Arg-241, Asp-253, Asp-256, Glu-259, Asp-268, and Arg-272, all mapping to the E2 interaction surface observed in the crystal structure of the complex of CHIPU and Ube2N (22). Based on the available structural models, Lys-234–Ser-236 should also be involved in binding to E2 enzymes; however, we were unable to observe signals corresponding to these residues in either the free or E2-bound state of CHIPU.
We observed small chemical shift perturbations for nine residues of CHIPU during titration with nonspecific E2 enzymes (including Phe-237, Glu-238, Arg-241, Asp-253, and Arg-272) that were also perturbed by specific E2 binding. Notably, Asp-256, Glu-259, or Asp-268, which were strongly perturbed during specific binding, were not perturbed by the nonspecific E2 enzymes. Our analysis suggests that nonspecific E2 enzymes have weak contacts on one side of the E2-binding surface and involve E2 residues in loop 7. Overall, E2 enzymes that interact nonspecifically with CHIPU bind much more weakly and in a different manner compared with the E2 enzymes that bind specifically to CHIPU.
Autoubiquitination of CHIP Is E2-dependent
To further characterize E2-CHIPU interactions, we analyzed patterns in CHIP in vitro autoubiquitination assays for a set of 14 human E2 conjugating enzymes selected as described above (Fig. 3). The results of the assays were classified into four different outcomes: 1) no products or only E2 autoubiquitination, 2) monoubiquitination of CHIP with no Ub chains, 3) polyubiquitination of CHIP, and 4) formation of free polyubiquitin chains.
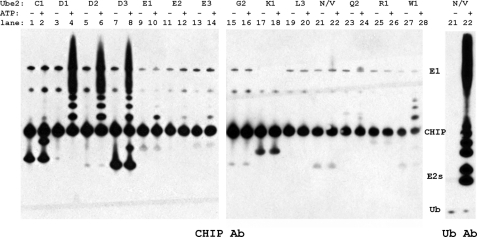
FIGURE 3.
CHIP functions in vitro with multiple E2 enzymes. In vitro autoubiquitination reactions of CHIP for a panel of human E2 enzymes were incubated at 30 °C for 1 h with and without ATP. Samples were analyzed by SDS-PAGE and immunoblotted with anti-CHIP and anti-Ub antibodies as indicated. Only the reaction with Ube2N/Uev1a is shown for the Ub blot to demonstrate its unique production of free polyubiquitin chains.
The E2 enzymes that showed no ubiquitination function with CHIP included Ube2G2, Ube2K, Ube2L3, Ube2Q2, and Ube2R1. To ensure that these E2 enzymes were active under the in vitro assay conditions, the ability of the E1 activating enzyme to charge E2 with Ub was assayed by monitoring the formation of the DTT-sensitive E2∼Ub conjugate. Although these five E2 enzymes did not function with CHIP, they all formed the E2∼Ub conjugate (supplemental Fig. 3, left panel). Of these five E2 enzymes, Ube2G2, Ube2K, and Ube2L3 were shown not to bind CHIPU in our NMR titrations. Although they were not tested, we presume that Ube2Q2 and Ube2R1 also do not bind CHIP.
Eight E2 enzymes (Ube2D1–3, Ube2E1–3, Ube2N/V1a, and Ube2W) functioned with CHIP, and all of them have been shown to bind specifically to the U-box domain. The heterocomplex E2 Ube2N/Uev1a (Ubc13/Uev1a) is a special case and created only free polyubiquitin chains (Fig. 3, lanes 21 and 22). This phenomenon has been attributed to the binding of Ub by the Uev subunit, resulting in a unique orientation of the donor and acceptor Ub molecules, which specifically directs formation of Lys-63-linked Ub chains (33). Notably, four of the E2 enzymes (Ube2W and Ube2E1–3) each attached only one to three ubiquitin molecules to CHIP (Fig. 3, lanes 9–14, 27, and 28). The lack of Ub chain formation by these E2 enzymes is consistent with reports of monoubiquitination at multiple sites on CHIP (4, 34). Previous specific studies of CHIP ubiquitination with the Ube2E1–3 enzymes are inconsistent (see supplemental Table 1): an earlier study reported no function with Ube2E1 (UbcH6) (28), but a more recent study observed some catalytic function with Ube2E2 (23). Here, we have clearly shown that CHIP autoubiquitination by this family of E2 enzymes results in monoubiquitination of CHIP.
The well studied and promiscuous Ube2D1–3 (UbcH5a–c) family of E2 enzymes was observed to polyubiquitinate CHIP (Fig. 3, lanes 3–8). Their ability to add ubiquitin both to a substrate (monoubiquitination) and to ubiquitinated proteins (chain extension) has been attributed to the presence of a backside noncovalent ubiquitin-binding site (35, 36). Other possible properties of these enzymes, such as oligomerization (e.g. Ref. 33), have been proposed as the basis for their efficient polyubiquitination activity but have not been tested extensively.
Polyubiquitination via the Coupled Action of Two E2 Enzymes
Recent reports have revealed that certain E2 conjugating enzymes function with an E3 ligase only after the substrate is already at least monoubiquitinated. For example, Ube2K was active in autoubiquitination of BRCA1-BARD1 only in the presence of another E2 that first produces monoubiquitinated protein (5). To determine whether CHIP functions similarly, in vitro autoubiquitination assays were performed using mixtures of E2 enzymes in a single reaction. In one set of experiments, all tested E2 enzymes that did not function with CHIP on their own (Ube2C1, Ube2G2, Ube2K, Ube2L3, Ube2Q2, and Ube2R1) were tested in combination with the most efficient monoubiquitinating E2 found, Ube2W. No polyubiquitination of CHIP was observed for any of these E2 enzymes, confirming that they do not function even with monoubiquitinated CHIP.
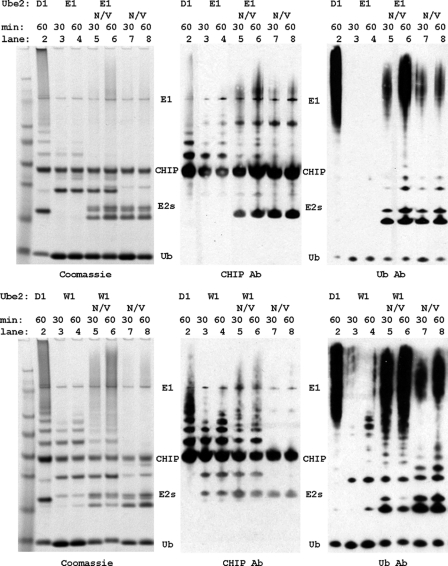
To determine whether monoubiquitination can be extended by CHIP functioning with a different E2 enzyme, in vitro ubiquitination assays were performed using combinations of the monoubiquitinating E2 enzyme Ube2E1 (UbcH6) or Ube2W with Ube2N/Uev1a, which only produces free Ub chains on its own. We observed that when Ube2E1 or Ube2W was complemented with Ube2N/Uev1a, CHIP was polyubiquitinated (Fig. 4). The addition of Ube2N/Uev1a to the reaction (lanes 5 and 6) caused a steep reduction in the amount of monoubiquitinated CHIP compared with the products of Ube2E1 or Ube2W alone (lanes 3 and 4), which we attribute to the monoubiquitinated product serving as substrate for chain extension by Ube2N/Uev1a. Compared with Ube2N/Uev1a alone (lanes 7 and 8), there was a dramatic increase in polyubiquitination and formation of high molecular mass species (lanes 5 and 6). We also found that Ube2N/Uev1a had a greater efficiency for building Ub chains on a previously monoubiquitinated substrate relative to building chains on free Ub. Thus, both Ube2E1 and Ube2W are able to function together with Ube2N/Uev1a to create new products (polyubiquitinated CHIP) that neither E2 could produce alone.
FIGURE 4.
In vitro polyubiquitination of CHIP by Ube2N/Uev1a acting in concert with Ube2E1 (upper panels) and Ube2W (lower panels). CHIP in vitro autoubiquitination reactions were incubated at 30 °C for the times indicated. The products were visualized by Coomassie Blue staining or immunoblotting using anti-CHIP and anti-Ub antibodies as indicated.
Our experiments also revealed interesting differences in E2 activity rates. Figs. 3 and 4 clearly show that Ube2W was much more efficient than Ube2E1–3 at producing monoubiquitinated CHIP (Fig. 3, lanes 9–14 and 28). The same relative rates were observed with E2 mixtures, where Ube2W together with Ube2N/Uev1a produced more of the high molecular mass ubiquitin species than Ube2E1 did with Ube2N/Uev1a. This observation is attributed to monoubiquitination of CHIP being the rate-limiting step for polyubiquitination under these conditions. We also note that the polyubiquitinated species produced by combining E2 enzymes were smaller than those produced by Ube2D1 alone and rarely included products so large that they were retained at the top of the gel (Fig. 4, lanes 2 and 6). This may indicate a difference in the mechanism of ubiquitin chain extension and regulation of chain building for the Ube2D family versus Ube2N/Uev1a.
CHIP Ubiquitination of the Substrate Hsp70
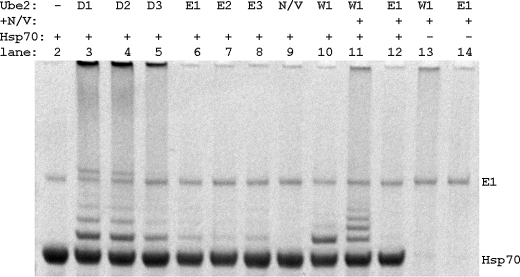
We next asked if CHIP autoubiquitination is representative of ubiquitination of CHIP substrates. The cellular functions of CHIP include the ubiquitination of heat shock protein-bound substrates (30). A role in the regulation of heat shock proteins as well has been proposed, and it has been established that polyubiquitination of Hsp70 by CHIP leads to its degradation by the 26 S proteasome (37). Using in vitro assays, we examined the E2 dependence of Hsp70 ubiquitination by CHIP (Fig. 5). These data show that Hsp70 was efficiently ubiquitinated by CHIP and additionally that the pattern of E2 dependence of Hsp70 ubiquitination by CHIP was the same as that of CHIP autoubiquitination. In particular, we found that 1) Ube2N/Uev1a did not directly ubiquitinate Hsp70, 2) Ube2D1–3 polyubiquitinated CHIP, and 3) Ube2E1–3 and Ube2W monoubiquitinated CHIP. We also found that the combination of Ube2W and Ube2N/Uev1a led to greater ubiquitination of Hsp70 than either E2 alone. Among all these similarities, we did observe one difference: the combination of Ube2E1 and Ube2N/Uev1a resulted in no or very weak Hsp70 polyubiquitination. We note that the activity of Ube2E1 alone in CHIP substrate ubiquitination appeared to be weaker than that in CHIP autoubiquitination; however, the origin of weaker activity in these assays remains unknown. Overall, the ensemble of these data confirms that CHIP substrate ubiquitination is similar to autoubiquitination. Moreover, both substrate and autoubiquitination assays indicated that the outcome of the ubiquitination reaction depends on the identity of the E2 conjugating enzyme.
FIGURE 5.
Ubiquitination of the CHIP substrate Hsp70 is E2-dependent. In vitro ubiquitination reactions of Hsp70 using CHIP as the E3 ligase and different E2 enzymes were incubated for 1 h at 30 °C. Reaction products were separated by SDS-PAGE and visualized by Coomassie Blue staining.
The E2 SPA Motif Is Required but Not Sufficient for CHIP Binding and Function
Eight human E2 conjugating enzymes were found to bind and function in vitro with CHIP. Thus, in comparison with other E3 enzymes that have been studied, CHIP appears to have a limited number of functional E2 partners. In an effort to understand the structural basis for this E2 selectivity, the results from the surveys of E2 binding and ubiquitination activity were used to direct sequence and structural analyses of the factors that differentiate E2 enzymes that are active versus inactive with CHIP.
The catalytic core is structurally conserved in all E2 enzymes, and all utilize the same general interaction surface to bind to E3 ligases. Despite these similarities, E2 enzymes have only limited sequence homology, and many residues at the E3-binding interface vary. A previous study proposed that the E2 SPA motif in loop 7 is required for CHIP recognition and binding (23), and indeed, all eight E2 enzymes for CHIP contain this motif. Although residues outside of E2 loop 7 are also involved in binding, those in helix 1 are conserved across nearly all E2 enzymes, and those in loop 4 are either completely conserved or vary widely among the E2 enzymes that bind CHIPU. Therefore, these other residues are unlikely to be involved in the selectivity for binding to CHIPU and distinguishing the subset of E2 enzymes that function with CHIP.
The E2 SPA motif has a central location at the CHIP-binding interface. We first tested the requirements of CHIP for these exact residues by designing a series of three mutants of Ube2D1 (UbcH5a) based on sequences found in E2 enzymes that do not function with CHIP: Ube2D1 S94K (KPA), S94A/P95A (AAA), and S94A (APA). We note that the KPA and AAA mutants correspond to the naturally occurring sequences in Ube2L3 and Ube2K, respectively.
To assess the effect of these Ube2D1 mutations, each was tested for in vitro ubiquitination activity with CHIP. First, a ubiquitin charging assay was performed, and all of the mutants were shown to be charged with Ub by E1 (supplemental Fig. 3, right panel). These observations demonstrated that the E2∼Ub intermediate could be readily formed and that the mutations did not interfere with E1-E2 activity. However, the CHIP autoubiquitination assay revealed that none of the mutants exhibited activity (Fig. 6E).
FIGURE 6.
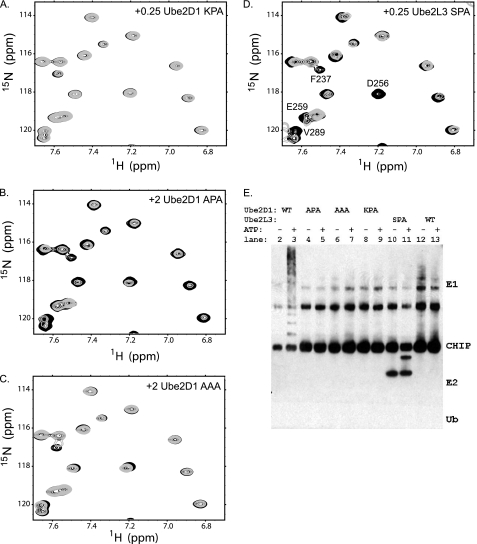
The E2 SPA motif is required for binding and function with CHIP. Shown is a region of 15N-1H HSQC NMR spectra for 15N-labeled CHIPU complexes with Ube2D1 KPA (A), Ube2D1 APA (B), Ube2D1 AAA (C), and Ube2L3 SPA (D). Free CHIPU is shown in black, with spectra for 0.25 molar eq of E2 added in A and D and 2.0 molar eq in B and C in gray. Residues are labeled in D for comparison with Fig. 2. E, CHIP in vitro autoubiquitination assays with all four E2 mutants and their respective WT proteins. The reactions were incubated for 1 h at 30 °C, separated by SDS-PAGE, and visualized by anti-CHIP antibody Western blotting.
All three mutants were then tested for binding to CHIPU using NMR as described above for WT E2 enzymes (Fig. 6, A–C). We found that none of the Ube2D1 mutants interacted as strongly with 15N-labeled CHIPU as the WT, with no observable chemical shift perturbations at a 0.25 molar ratio of E2 to CHIPU. However, a weak interaction was observed between Ube2D1 KPA and CHIPU for a 2-fold excess of E2, stronger than the nonspecific binding seen for Ube2L3, which naturally contains KPA. Nevertheless, this weak interaction is apparently insufficient to enable activation of E2∼Ub because this mutant showed no appreciable activity in the in vitro CHIP ubiquitination assay.
To further test the importance of the SPA motif, we asked if the SPA motif alone is sufficient for E2 binding and function with CHIP. Our approach involved selecting an E2 enzyme that is somewhat similar to the functional E2 enzymes but lacks the SPA motif and engineering an SPA motif into this E2. Ube2L3 (UbcH7) was selected for this experiment, as it showed no ubiquitination activity or specific binding with CHIP. The Ube2L3 SPA mutant was prepared by mutating Lys-96 of the natural KPA motif to serine. As opposed to the Ube2D1 mutants, Ube2L3 SPA did show chemical shift perturbations consistent with binding to CHIPU and nearly identical to the pattern of binding seen for WT Ube2D1 (compare Figs. 2A and 6D). Thus, the SPA motif on an E2 enzyme is required and appears to be sufficient for binding to CHIPU. As Ube2L3 SPA was able to bind CHIPU with a similar affinity to active E2 enzymes for CHIP, we expected that this E2 might also show ubiquitination activity. However, no activity was seen in the in vitro CHIP autoubiquitination assay (Fig. 6E), consistent with a previous report (23). Thus, although the SPA motif may be critical, the presence of the motif alone and the resultant binding to CHIP are not sufficient to support activation of the E2∼Ub conjugate and function in CHIP ubiquitination.
DISCUSSION
There is a growing recognition that ubiquitination of a substrate is not strictly the product of the action of a single E2 conjugating enzyme functioning with a specific E3 ligating enzyme. Most E3 ligases are found to function with several E2 enzymes and produce multiple types of ubiquitination signals. The basis for selectivity of E2-E3 interactions and the mechanism for producing different ubiquitin signals are currently not well understood. Through systematic analysis of binding and in vitro ubiquitination by the E3 ligase CHIP, we have discovered that CHIP functions with only a few E2 enzymes and that the product formed in ubiquitination reactions with CHIP depends on which E2 is involved.
NMR chemical shift perturbations showed that the subset of active E2 enzymes bound CHIP with similar affinity and at the same CHIP-binding surface. The data are also consistent with the previously determined Kd of 1–6 μm for Ubc13 (Ube2N) binding to CHIP (22). Most of the residues in CHIPU identified as affected by E2 binding lie on a single surface of the protein. One additional residue consistently perturbed by E2 binding, Val-289, lies distant from the E2-binding site. This resonance consistently showed broadening and loss of signal intensity upon titration with E2 enzymes, even with low amounts of E2 in solution. Val-289 is on the opposite side of CHIP relative to the E2 interaction surface and on a surface involved in dimerization of the U-box (see supplemental Fig. 2). One explanation of the observed perturbation is that binding of E2 enzymes could stabilize the dimeric state of CHIPU. The chemical shift perturbations observed for Val-289 may reflect an effect on exchange between asymmetric states upon binding an E2, which could arise from partial E2 loading of the CHIP dimer or from conformational exchange paralleling that observed in the intact protein (22, 36).
E3 U-box and RING domains are structurally conserved and, based on the available structures, generally have a conserved E2-binding interface. Therefore, variations in the E3 sequence for residues at the interface are likely to be key to specificity in E2 recognition. A large screen of E2-binding partners for many RING domain E3 ligases revealed that most E3 enzymes bind to only a subset of E2 enzymes and that many E3 enzymes bind to a specific group of eight E2 enzymes (16). In addition, Klevit and co-workers (5) surveyed all human E2 enzymes for binding to BRCA1-BARD1 (BCBD) and found 10 E2 enzymes that bind the BRCA1 RING domain. Here, we have shown that CHIP binds a smaller subset of E2 enzymes than BCBD and only those in the common group identified by Markson et al. (16) in the large screen. Although most of the interacting E2 enzymes are the same for BCBD and CHIP, neither Ube2K nor Ube2L3 binds to CHIP, yet both bind to BCBD. Further investigations into the origin of these differences should provide insights in the basis for the differential E2 selectivity of E3 ligases.
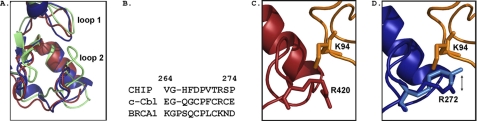
The series of wild-type and mutant E2 enzymes we have tested confirms that CHIP requires the E2 SPA motif in loop 7 for recognition and binding. This explains why Ube2L3 and Ube2K did not bind because they contain KPA and AAA, respectively, at this site. However, this does not explain why BCBD is not as selective as CHIP. We compared the sequences and structures of CHIPU with those of the BRCA1 and c-Cbl RING domains using the available structures (22, 23, 38–40). Our comparative analysis showed several differences at the E2-E3 interface, notably that the second loop (CHIP Val-264–Pro-274) is longer in BRCA1 (Lys-56–Asp-67) (Fig. 7, A and B). This loop in CHIP contains a bulky phenylalanine residue (Phe-267) that results in a smaller pocket at the bottom of the E2 interface of CHIP and therefore is not expected to accommodate larger residues in E2 loop 7. This difference likely contributes to why CHIP binds and functions with fewer E2 enzymes compared with BRCA1. The differences observed between the structures of CHIPU and BRCA1 RING domains suggest that the two E3 ligases should bind E2 conjugating enzymes differently, leading to greater differences than what we have observed. However, the static snapshots of the E2-E3 complexes do not reflect the intrinsic malleability of protein interfaces that may make the E3 ligases more similar than anticipated on this basis.
FIGURE 7.
Comparison of the E2 interface for U-box and RING domains. A, overlay of CHIPU (blue; Protein Data Bank code 2OXQ) with the c-Cbl RING domain (red; code 1FBV) and the BRCA1 RING domain (green; code 1JM7). The coordinates were superimposed on each other by minimizing the root mean square deviation of the backbone C-α atoms. B, U-box and RING loop 2 sequences. C, structure of the E2-E3 complex showing that c-Cbl (red; code 1FBV) can accommodate the large residue Lys-94 in Ube2L3 (orange; code 1FBV) by rotation of Arg-420. D, model of Ube2L3-CHIP based on the alignment with c-Cbl shown in A. The equivalent residue in CHIP, Arg-272 (light blue, code 2C2L; dark blue, code 2OXQ), has a range of positions and may be able to rotate in the same manner as c-Cbl to accommodate Lys-94.
Based on the structure of an E2-CHIPU complex, it has been previously stated that a large side chain at position 94 of E2, such as lysine, could not be accommodated by CHIP (22). Surprisingly, we found that the Ube2D1 S94K mutant does bind and indeed binds more strongly than Ube2L3 (UbcH7), which naturally has KPA at loop 7. Although neither of these enzymes was functionally active with CHIP, the structure of the c-Cbl-Ube2L3 complex shows how the bulky Lys-94 residue could be accommodated by rotation of CHIP Arg-272 (c-Cbl Arg-420) down and away from E2 (Fig. 7, C and D). This position of CHIP Arg-272 is nearly identical to what was seen in the structure of the complex with Ube2D1 (23), but as the Ube2D1 S94K mutant has only very weak binding, we must assume there are other differences affecting the interaction.
Most current theories regarding the ubiquitin transfer mechanism require binding of E2 to E3 for both localization of Ub to the substrate and activation of the E2∼Ub conjugate for Ub release. Although the E2∼Ub conjugate is not highly stable, E3 binding significantly increases the rate of Ub release. Our studies demonstrate that CHIP stimulates ubiquitination with most, but not all, E2 enzymes that bind to the U-box. In particular, although Ube2L3 SPA binds to CHIPU, it is not functionally active in ubiquitination. This observation implies that binding and activation have separable molecular determinants that involve different residues of the E2 enzyme. We note that BRCA1 binds WT Ube2L3 (UbcH7) but also exhibits no ubiquitination activity. Huang et al. (41) studied the reaction of Ube2L3 with c-Cbl and found that the E2∼Ub conjugate is more stable and binds more tightly to E3 compared with Ube2D1. It is conceivable that the Ube2L3 protein is generally less active or not activated by binding to U-box and RING E3 ligases, but more in-depth knowledge of the mechanism for E2∼Ub activation is required before any specific conclusions can be drawn.
There are several reports of E2 enzymes that function in tandem to polyubiquitinate a substrate. Two examples involve a single E3 ligase that utilizes two different E2 enzymes for substrate polyubiquitination (8, 14). For both the anaphase-promoting complex and SCFβ-TRCP2, initiation of ubiquitination can be catalyzed by the Ube2D family (Ubc4/UbcH5). The resulting monoubiquitinated species then becomes a substrate for polyubiquitination by Cdc34 or Ubc1, respectively. Our observation of monoubiquitination (initiation) of CHIP and CHIP substrate Hsp70 by either Ube2E1 or Ube2W and ubiquitin chain elongation by Ube2N/Uev1a further supports recent proposals that initiation and elongation of ubiquitin chains can occur through the action of different E2 enzymes (3–7). Our data specifically show high molecular mass ubiquitinated species formed by the coordinated action of Ube2E1 or Ube2W with Ube2N/Uev1a in CHIP autoubiquitination and in ubiquitination of the CHIP substrate Hsp70.
With the need for rapid cycling of different E2-E3 complexes and selectivity in forming active complexes, the ubiquitination system requires a careful balance of specificity and plasticity. Although there are 38 proposed E2 enzymes in humans and despite their high structural conservation, most E3 ligases function only with a few E2 enzymes. In this work, we have shown that CHIP functions with only a subset of E2 conjugating enzymes. We note that this group of eight functional E2 enzymes matches a set of E2 enzymes found to share selectivity for E3 enzymes in a screen of RING domain E3 ligases (16). All of these E2 enzymes share the SPA motif in loop 7, which implies that this may be a common recognition motif for other E3 ligases that bind this subset of E2 enzymes.
Each E2 conjugating enzyme that functions with CHIP results in ubiquitination products consistent with those seen for the E3 ligases BRCA1-BARD1, TRAF6, and SCFβ-TRCP2, i.e. Ube2D1 always results in polyubiquitination, etc. (4, 5, 8). This supports the idea that the outcome or type of ubiquitination (monoubiquitination, polyubiquitination, chain type) is specified by the identity of the E2 enzyme, once activated by binding to an E3 U-box or RING domain. We have shown that the set of eight E2 enzymes that bind to and function with CHIP produces a range of ubiquitination events (Fig. 3) and also that some produce unique outcomes when combined in vitro (Fig. 4). This set of E2 enzymes appears to be able to produce all possible types of the ubiquitin post-translational modification, and a recent broad screen showed that this same set interacts with a large number of E3 ligases (16). Thus, it is conceivable that these E2 enzymes are a core group for most types of ubiquitination activity, whereas other E2 enzymes have evolved specialized and localized functions.
Supplementary Material
Acknowledgments
We thank Rachel Klevit (University of Washington) for the plasmid expressing wheat Uba1 (E1); Cam Patterson (University of North Carolina at Chapel Hill) for the plasmid expressing CHIP; and Yoana Dimitrova, Kyle Nordquist, Cam Patterson, and Rachel Klevit for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grant R01GM075156 (to W. J. C.) and Grants T32CA009582 and F32GM087050 (to S. E. S.). This work was also supported by the China Scholarship Council (to Y. Y.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3, Table 1, and additional references.
- Ub
- ubiquitin
- CHIPU
- CHIP U-box domain
- HSQC
- heteronuclear single-quantum coherence
- BCBD
- BRCA1-BARD1.
REFERENCES
- 1. Deshaies R. J., Joazeiro C. A. (2009) Annu. Rev. Biochem. 78, 399–434 [DOI] [PubMed] [Google Scholar]
- 2. Hochstrasser M. (2006) Cell 124, 27–34 [DOI] [PubMed] [Google Scholar]
- 3. Petroski M. D., Zhou X., Dong G., Daniel-Issakani S., Payan D. G., Huang J. (2007) J. Biol. Chem. 282, 29936–29945 [DOI] [PubMed] [Google Scholar]
- 4. Windheim M., Peggie M., Cohen P. (2008) Biochem. J. 409, 723–729 [DOI] [PubMed] [Google Scholar]
- 5. Christensen D. E., Brzovic P. S., Klevit R. E. (2007) Nat. Struct. Mol. Biol. 14, 941–948 [DOI] [PubMed] [Google Scholar]
- 6. Hoege C., Pfander B., Moldovan G. L., Pyrowolakis G., Jentsch S. (2002) Nature 419, 135–141 [DOI] [PubMed] [Google Scholar]
- 7. Ye Y., Rape M. (2009) Nat. Rev. Mol. Cell Biol. 10, 755–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wu K., Kovacev J., Pan Z. Q. (2010) Mol. Cell 37, 784–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Eletr Z. M., Huang D. T., Duda D. M., Schulman B. A., Kuhlman B. (2005) Nat. Struct. Mol. Biol. 12, 933–934 [DOI] [PubMed] [Google Scholar]
- 10. Hatakeyama S., Yada M., Matsumoto M., Ishida N., Nakayama K. I. (2001) J. Biol. Chem. 276, 33111–33120 [DOI] [PubMed] [Google Scholar]
- 11. Ohi M. D., Vander Kooi C. W., Rosenberg J. A., Chazin W. J., Gould K. L. (2003) Nat. Struct. Biol. 10, 250–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Capili A. D., Lima C. D. (2007) Curr. Opin. Struct. Biol. 17, 726–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ordureau A., Smith H., Windheim M., Peggie M., Carrick E., Morrice N., Cohen P. (2008) Biochem. J. 409, 43–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rodrigo-Brenni M. C., Morgan D. O. (2007) Cell 130, 127–139 [DOI] [PubMed] [Google Scholar]
- 15. Kim H. T., Kim K. P., Uchiki T., Gygi S. P., Goldberg A. L. (2009) EMBO J. 28, 1867–1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Markson G., Kiel C., Hyde R., Brown S., Charalabous P., Bremm A., Semple J., Woodsmith J., Duley S., Salehi-Ashtiani K., Vidal M., Komander D., Serrano L., Lehner P., Sanderson C. M. (2009) Genome Res. 19, 1905–1911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ballinger C. A., Connell P., Wu Y., Hu Z., Thompson L. J., Yin L. Y., Patterson C. (1999) Mol. Cell. Biol. 19, 4535–4545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dickey C. A., Koren J., Zhang Y. J., Xu Y. F., Jinwal U. K., Birnbaum M. J., Monks B., Sun M., Cheng J. Q., Patterson C., Bailey R. M., Dunmore J., Soresh S., Leon C., Morgan D., Petrucelli L. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 3622–3627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Williams A. J., Knutson T. M., Colomer Gould V. F., Paulson H. L. (2009) Neurobiol. Dis. 33, 342–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schmidt B. Z., Watts R. J., Aridor M., Frizzell R. A. (2009) J. Biol. Chem. 284, 4168–4178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tetzlaff J. E., Putcha P., Outeiro T. F., Ivanov A., Berezovska O., Hyman B. T., McLean P. J. (2008) J. Biol. Chem. 283, 17962–17968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang M., Windheim M., Roe S. M., Peggie M., Cohen P., Prodromou C., Pearl L. H. (2005) Mol. Cell 20, 525–538 [DOI] [PubMed] [Google Scholar]
- 23. Xu Z., Kohli E., Devlin K. I., Bold M., Nix J. C., Misra S. (2008) BMC Struct. Biol. 8, 26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chiang A. N., Valderramos J. C., Balachandran R., Chovatiya R. J., Mead B. P., Schneider C., Bell S. L., Klein M. G., Huryn D. M., Chen X. S., Day B. W., Fidock D. A., Wipf P., Brodsky J. L. (2009) Bioorg. Med. Chem. 17, 1527–1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 26. Goddard T. D., Kneller D. G. (2006) SPARKY 3, University of California, San Francisco [Google Scholar]
- 27. Dimitrova Y. N., Li J., Lee Y. T., Rios-Esteves J., Friedman D. B., Choi H. J., Weis W. I., Wang C. Y., Chazin W. J. (2010) J. Biol. Chem. 285, 13507–13516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Murata S., Minami Y., Minami M., Chiba T., Tanaka K. (2001) EMBO Rep. 2, 1133–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Xu Z., Devlin K. I., Ford M. G., Nix J. C., Qin J., Misra S. (2006) Biochemistry 45, 4749–4759 [DOI] [PubMed] [Google Scholar]
- 30. Jiang J., Ballinger C. A., Wu Y., Dai Q., Cyr D. M., Höhfeld J., Patterson C. (2001) J. Biol. Chem. 276, 42938–42944 [DOI] [PubMed] [Google Scholar]
- 31. Dominguez C., Bonvin A. M., Winkler G. S., van Schaik F. M., Timmers H. T., Boelens R. (2004) Structure 12, 633–644 [DOI] [PubMed] [Google Scholar]
- 32. Shloush J., Vlassov J. E., Engson I., Duan S., Saridakis V., Dhe-Paganon S., Raught B., Sheng Y., Arrowsmith C. H. (2011) J. Biol. Chem. 286, 4796–4808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Eddins M. J., Carlile C. M., Gomez K. M., Pickart C. M., Wolberger C. (2006) Nat. Struct. Mol. Biol. 13, 915–920 [DOI] [PubMed] [Google Scholar]
- 34. Wang D., Xu W., McGrath S. C., Patterson C., Neckers L., Cotter R. J. (2005) J. Proteome Res. 4, 1554–1560 [DOI] [PubMed] [Google Scholar]
- 35. Brzovic P. S., Lissounov A., Christensen D. E., Hoyt D. W., Klevit R. E. (2006) Mol. Cell 21, 873–880 [DOI] [PubMed] [Google Scholar]
- 36. Sakata E., Satoh T., Yamamoto S., Yamaguchi Y., Yagi-Utsumi M., Kurimoto E., Tanaka K., Wakatsuki S., Kato K. (2010) Structure 18, 138–147 [DOI] [PubMed] [Google Scholar]
- 37. Qian S. B., McDonough H., Boellmann F., Cyr D. M., Patterson C. (2006) Nature 440, 551–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Brzovic P. S., Rajagopal P., Hoyt D. W., King M. C., Klevit R. E. (2001) Nat. Struct. Biol. 8, 833–837 [DOI] [PubMed] [Google Scholar]
- 39. Huang L., Kinnucan E., Wang G., Beaudenon S., Howley P. M., Huibregtse J. M., Pavletich N. P. (1999) Science 286, 1321–1326 [DOI] [PubMed] [Google Scholar]
- 40. Zheng N., Wang P., Jeffrey P. D., Pavletich N. P. (2000) Cell 102, 533–539 [DOI] [PubMed] [Google Scholar]
- 41. Huang A., de Jong R. N., Wienk H., Winkler G. S., Timmers H. T., Boelens R. (2009) J. Mol. Biol. 385, 507–519 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.