Abstract
The antibiotic microcin C7 (McC) acts as a bacteriocide by inhibiting aspartyl-tRNA synthetase and stalling the protein translation machinery. McC is synthesized as a heptapeptide-nucleotide conjugate, which is processed by cellular peptidases within target strains to yield the biologically active compound. As unwanted processing of intact McC can result in self-toxicity, producing strains utilize multiple mechanisms for autoimmunity against processed McC. We have shown previously that the mccE gene within the biosynthetic cluster can inactivate processed McC by acetylating the antibiotic. Here, we present the characterization of this acetylation mechanism through biochemical and structural biological studies of the MccE acetyltransferase domain (MccEAcTase). We have also determined five crystal structures of the MccE-acetyl-CoA complex with bound substrates, inhibitor, and reaction product. The structural data reveal an unexpected mode of substrate recognition through π-stacking interactions similar to those found in cap-binding proteins and nucleotidyltransferases. These studies provide a rationale for the observation that MccEAcTase can detoxify a range of aminoacylnucleotides, including those that are structurally distinct from microcin C7.
Keywords: Aminoacyl-tRNA Synthetase, Antibiotics, Drug Resistance, Enzyme Structure, X-ray Crystallography
Introduction
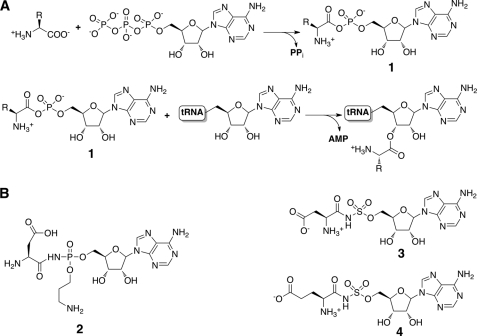
Microcin C7 (McC)3 is a member of the class of small molecule antimicrobials produced by Enterobacteriaceae that exert biological activity by specifically targeting aminoacyl-tRNA synthetases, thereby halting protein synthesis (1–4). Specifically, McC consists of a ribosomally synthesized heptapeptide precursor in which a carboxyl-terminal aspartate is linked to adenosine monophosphate (AMP) by a non-hydrolyzable phosphoramidate (5, 6). The peptidyl group mediates the active transport of McC into target bacterial cells (7). Subsequent cleavage of the peptide moiety within the target cell releases the aspartyl adenylate (8), which then inhibits translation by preventing the synthesis of aminoacylated tRNAAsp by aspartyl-tRNA synthetase (9) (Fig. 1A). Hence, McC utilizes a Trojan horse mechanism to target bacterial protein synthesis (10). The biosynthetic cluster for microcin C7 is harbored on a plasmid and consists of five genes (mccABCDE) (11). The 21-bp-long mccA gene encodes the heptapeptide precursor (12), which is post-translationally modified by MccB, MccD, and MccE, resulting in the attachment of the carboxyl-terminal adenosine monophosphate through a phosphoramidate linkage and esterification of the N-acyl phosphoramidate by a 3-aminopropyl group (13, 14). The resultant aspartyl-adenylated peptide (unprocessed McC) is secreted from the producer strain by the MccC efflux pump.
FIGURE 1.
tRNA synthetase mechanism and structures of inhibitors. A, the two-step reaction for the esterification of a specific amino acid onto its cognate tRNA to form a charged aminoacyl-tRNA proceeds through the production of a hydrolytically labile aminoacyl adenylate (compound 1). B, chemical structures of synthetic and naturally occurring aminoacyl adenylate mimics, including McC (compound 2), DSA (compound 3), and ESA (compound 4).
The biological activity of McC is contingent on the hydrolysis of the peptidyl moiety by intracellular aminopeptidases to release the toxic modified aspartyl adenylate (processed McC; Fig. 1B, compound 2). Consequently, the producer strains must harbor mechanisms to protect against the unwanted generation of processed McC within its own cytoplasm. Prior studies have established that disruption of the mccE gene from producer strains resulted in slow cell growth, suggesting a role for MccE in autoimmunity (15). The mccE locus encodes for a 400-residue bidomain protein with an amino-terminal domain showing significant homology to pyridoxal phosphate-dependent decarboxylases and a carboxyl-terminal region that bears sequence similarities to members of the GCN5 acetyltransferase superfamily. Although in vitro activity of the MccE amino-terminal domain remains to be determined, in vivo experiments suggest that MccE plays a role in the attachment of the propylamine adduct to McC. Further biochemical characterization of the MccE demonstrated that the immunity determinant is harbored within the carboxyl-terminal domain (16). Heterologously expressed and purified MccE carboxyl-terminal domain (MccECTD) showed acetyl-CoA-dependent acetyltransferase activity against processed McC and toward the structurally related aspartyl-sulfamoyl adenosine (DSA; Fig. 1B, compound 3) and glutamyl-sulfamoyl adenosine (ESA; Fig. 1B, compound 4) as well as other aminoacyl sulfamoyl adenylates except prolyl sulfamoyl adenylate. Lastly, overexpression of either full-length MccE or MccECTD conferred resistance against a structurally distinct antibiotic albomycin whose active part consists of a serine coupled to a thioxylofuranosyl pyrimidine and inhibits seryl-tRNA synthetase (16, 17).
To decipher the molecular basis for the broad substrate specificity range of the MccE acetyltransferase, here we have extended the biochemical analysis of the MccE acetyltransferase domain (encoded in plasmid pBM43 from Escherichia coli strain TG1) acting on its substrates. We discovered that adenosine monophosphate is a competitive inhibitor of the MccE-catalyzed acetylation of aminoacyl adenylates, suggesting that substrate recognition is likely mediated through the binding of the nucleotide moiety. To affirm these biochemical studies, we have determined five different crystal structures of the MccE acetyltransferase domain bound to either CoA or acetyl-CoA in the presence of various ligands (substrates, inhibitors, and product) at 1.6-Å resolution or higher. These co-crystal structures validate inferences from our biochemical analyses and provide a molecular rationale for understanding the ability of MccE to detoxify McC and other aminoacyl-tRNA synthetase inhibitors.
EXPERIMENTAL PROCEDURES
Cloning, Protein Expression, and Purification
The gene encoding the carboxyl-terminal acetyltransferase domain of MccE encoded by amino acids 405–589 was amplified using the polymerase chain reaction with the forward primer 5′-AAGCAGCCGCATATGCGGAAGTATGATGTATCTCTTACGCC bearing an engineered NdeI restriction site (underlined) and the reverse primer 5′-CTAGCTCGAGTTAACCAATTACTTTCGAATAAATATTTTGATC with an engineered XhoI restriction site (underlined). The PCR product was purified and following a restriction digest was ligated into pET28a(+) plasmid (Novagen) to yield an amino-terminal hexahistidine tag and a thrombin digestion site preceding the inserted gene. The plasmid was sequenced to confirm the integrity of the coding sequence.
Expression strain E. coli Rosetta2 (DE3) was transformed with the MccE acetyltransferase domain (MccEAcTase) expression plasmid. A single colony of transformed E. coli was used to inoculate 5 ml of LB medium supplemented with kanamycin (50 μg/ml) and chloramphenicol (34 μg/ml), and 14 h after induction, the small scale culture was added to 2 liters of LB medium similarly supplemented with antibiotics for growth at 37 °C with vigorous shaking. When the A600 nm of the culture reached 0.6, isopropyl β-d-thiogalactopyranoside was added to a final concentration of 0.5 mm, and the culture was shifted to 18 °C for a further 18 h of growth with vigorous shaking. Bacterial cells were pelleted by centrifugation (4000 × g for 20 min); resuspended in 20 mm Tris (pH 8.0), 500 mm NaCl, 10% glycerol buffer; and lysed by multiple passes through an Avestin C5 cell homogenizer. Insoluble aggregates and cell debris were removed by centrifugation (20,000 × g for 30 min). Clarified supernatant was applied to a 5-ml Nickel HiTrap column (GE Healthcare) equilibrated with the lysis buffer, and target protein was eluted by a continuous gradient of 30–200 mm imidazole over 20 column volumes after extensive washing with lysis buffer supplemented with 30 mm imidazole. The hexahistidine tag was removed using thrombin (1 unit/mg of protein) and further purified by size exclusion chromatography in a final buffer of 20 mm HEPES (pH 7.5), 100 mm KCl. The protein was estimated to be greater than 95% pure as judged by SDS-PAGE.
Preparation of Processed McC
McC-sensitive E. coli B cells were grown at 37 °C to A600 nm of 0.6 in 1 liter of M63 medium supplemented with 0.2% glucose and 0.1% yeast extract. Cells were collected by centrifugation; washed with 20 mm Tris-HCl (pH 8.0), 2 mm MgCl2; resuspended in 3 ml of the same buffer; and disrupted by sonication using a Branson Ultrasonics sonifier with a microtip (5 × 10-s pulses at maximal power setting). The lysate was centrifuged at 30,000 × g for 30 min. The supernatant (total protein concentration of 10 mg/ml) was divided into aliquots and stored at −70 °C until further use. For processing McC, 1 mg of McC was dissolved in 100 ml of 25 mm Tris-HCl (pH 8.0), 2 mm MgCl2 and combined with 400 ml of cell extract prepared as described above. The reaction was incubated for 2 h at 37 °C with continuous shaking. The reaction was terminated by the addition of an equal volume of 100% acetonitrile, 0.1% trifluoroacetic acid. After a 30-min incubation at 4 °C, proteins were removed by centrifugation, and the pellet was extracted three times with 200 ml of water. The extracts were pooled, lyophilized, dissolved in water, and fractionated on a Jupiter 300A C18 column (5 mm, 250 × 10 mm). The column was first developed isocratically (3 ml/min) with 10 mm triethylammonium bicarbonate (pH 7.0) followed by a linear gradient of acetonitrile (0–60%) in the same buffer. Fractions showing UV absorption were collected, lyophilized, dissolved in water, and tested by mass spectrometry. Processed McC (molecular mass of 519 Da) eluted at 27.3 min of the gradient. Processed McC prepared in this way was chromatographically and mass spectrometrically pure. The material was lyophilized and dissolved in water. The concentration was determined using UV absorption at 260 nm (using a calculated extinction coefficient of 16,500 g/mol for processed McC).
Isothermal Titration Calorimetry
Measurements were made at 25 °C using a VP-ITC calorimeter (MicroCal, Inc., Northampton, MA). Protein after size exclusion chromatography was extensively dialyzed against 50 mm potassium phosphate buffer (pH 7.0) and concentrated to a final concentration of 0.2 mm followed by incubation with a final concentration of 0.5 mm acetyl coenzyme A for 1 h on ice. A stock of adenosine monophosphate at a concentration of 50 mm was prepared in the spent dialysis buffer. The protein sample in a 1.4495-ml reaction cell was injected with 25 successive 10-μl aliquots of adenosine monophosphate at a spacing of 300 s with spent dialysis buffer in the reference cell. Based on the crystallographic results, the number of binding sites for adenosine monophosphate was presumed to be 1. Data were fitted by nonlinear regression using a single site binding model (MicroCal Origin), and thermodynamic parameters were calculated using the Gibbs free energy equation (ΔG = ΔH − TΔS) and the relation −RTlnKa = ΔG.
Measurement of Enzyme Activity
Reaction rates were measured spectrophotometrically by determining the rate of increase in absorbance at 412 nm due to the reaction between the free sulfhydryl group of coenzyme A, generated by the enzymatic transfer of the acetyl group from acetyl coenzyme A to the substrate molecule, and 5,5′-dithiobis(2-nitrobenzoic acid). The assays were performed at room temperature in a final volume of 100 μl and monitored continuously using a Cary UV-visible spectrophotometer. Each reaction consisted of a 2.5 mm final concentration of 5,5′-dithiobis(2-nitrobenzoic acid) and 1.5 mm acetyl coenzyme A. Five different substrate concentrations were used, the assays were initiated by the addition of the enzyme to a final concentration of 100 nm, and the increase in absorbance was monitored for 5 min. Rates for each substrate concentration were measured in triplicate.
Crystallization and X-ray Data Collection
Initial crystallization conditions were obtained by the sparse matrix sampling method using commercial screens. Crystals of MccEAcTase were grown using the hanging vapor drop diffusion method. For crystallization of the apoprotein, 1 μl of protein at 13.5 mg/ml concentration incubated with 2 mm acetyl coenzyme A or coenzyme A for 2 h on ice was mixed with 1 μl of precipitant solution (50 mm magnesium chloride, 50 mm HEPES (pH 7.0), 25% PEG550 monomethyl ether) and equilibrated over a well containing the precipitant solution at 9 °C. Co-crystallization for substrate complexes involved further incubation of the protein with 5 mm substrates for 1 h on ice. Crystals grew within 3 days and were briefly soaked in precipitant solution with PEG concentration increased to 30% prior to flash cooling in liquid nitrogen. Ternary complex co-crystal structures with AMP, McC, DSA, and ESA were grown under similar conditions and manipulated in the same manner.
Flash cooled crystals of MccEAcTase in complex with acetyl coenzyme A diffracted x-rays to 1.6-Å resolution at an insertion device synchrotron beam line (Life Sciences Collaborative Access Team, Sector 21 ID-D, Advanced Photon Source, Argonne, IL). These crystals occupy space group P21212 with unit cell parameters a = 78.9 Å, b = 96.7 Å, and c = 53.4 Å with two molecules in the crystallographic asymmetric unit. A 9-fold redundant data set was collected to 1.6-Å resolution with an overall Rmerge of 6.1% and I/σ(I) of 5 in the highest resolution shell. Crystals of the four ternary complexes diffracted to slightly higher resolutions, and all occupied the same space group albeit with changes in the unit cell parameters.
Phasing and Structure Determination
All data were indexed and scaled using the HKL2000 package (18). The structure of MccEAcTase-acetyl-CoA complex was determined by molecular replacement (MR) (19, 20) using the coordinates of a putative acetyltransferase (Protein Data Bank code 1NSL; 35% sequence identity) whose structure was determined by the Midwest Consortium for Structural Genomics (21). Use of the full-length structure as a search probe failed to yield a successful MR solution. Noting that the conservation in primary sequence was limited over the amino-terminal 70 residues, further efforts at MR utilized a model from which the amino-terminal 70 residues were removed. One such model in which all non-conserved residues were changed to serine resulted in two MR solutions without any packing clashes that were consistent with the self-rotation function data with minimal clashes. Following rigid body refinement of the initial MR solution, phases from the resultant model were further improved by 2-fold density averaging and the use of Prime-and-Switch maps (22, 23), allowing for a significant portion of the model to be manually rebuilt using XtalView (24). Further manual fitting was interspersed with automated rebuilding using ARP/wARP (25) and rounds of refinement using REFMAC5 (26, 27). Cross-validation using 5% of the data for the calculation of the free R factor (28) was utilized throughout the model building process to monitor building bias. Although clear density for acetyl-CoA could be observed prior to crystallographic refinement, the ligand was manually built into the model only after the free R factor dropped below 30%. The stereochemistry of the models was routinely monitored throughout the course of refinement using PROCHECK (29).
The crystal structures of the MccEAcTase-acetyl-CoA ternary complex with AMP, McC, DSA, and LSA were all determined by the molecular replacement method using the final refined coordinates of the MccEAcTase-acetyl-CoA complex as a search probe. Multiple rounds of manual model building were interspersed with refinement using REFMAC5 to complete structure refinement. Cross-validation used 5% of the data in the calculation of the free R factor. Crystal parameters, data collection parameters, and refinement statistics for each of the structures are summarized in Table 1. The refined coordinates have been deposited in the Protein Data Bank.
TABLE 1.
Data collection, phasing, and refinement statistics
| Acetyl-CoA | DSA | ESA | AMP | McC | |
|---|---|---|---|---|---|
| Data collection | |||||
| Space group | P21212 | P21212 | P21212 | P21212 | P21212 |
| Cell dimensions a, b, c (Å) | 78.9, 94.7, 53.9 | 78.9, 94.8, 53.3 | 78.9, 94.8, 53.4 | 78.5, 94.9, 53.3 | 77.9, 95.1, 53.2 |
| Resolution (Å)a | 50-1.6 (1.66-1.6) | 50-1.25 (1.29-1.25) | 50-1.2 (1.24-1.2) | 50-1.3 (1.35-1.3) | 50-1.35 (1.4-1.35) |
| Rsym (%) | 6.1 (36.1) | 6.3 (42.7) | 6.5 (44.7) | 6.5 (23.5) | 6.6 (49.7) |
| I/σ(I) | 28.7 (4.8) | 46.9 (2.9) | 51.7 (3.3) | 40.5 (11.2) | 45.7 (2.6) |
| Completeness (%) | 98.8 (96.9) | 99.5 (96.6) | 99.3 (96.0) | 99.8 (100.0) | 99.2 (95.2) |
| Redundancy | 7.2 (7.1) | 9.1 (5.4) | 8.6 (6.9) | 12.1 (11.8) | 6.5 (4.4) |
| Refinement | |||||
| Resolution (Å) | 25.0-1.6 | 25.0-1.25 | 25.0-1.2 | 25.0-1.3 | 25.0-1.35 |
| No. of reflections | 50,128 | 105,359 | 118,379 | 93,420 | 82,500 |
| Rwork/Rfreeb | 20.3/22.8 | 18.2/20.2 | 18.5/19.9 | 17.6/19.7 | 18.7/21.6 |
| No. of atoms | |||||
| Protein | 2,800 | 2,864 | 2,836 | 2,787 | 2,787 |
| (Ac)CoA | 102 | 96 | 96 | 102 | 96 |
| Substrate | 62 | 64 | 46 | 76 | |
| Water | 419 | 610 | 665 | 696 | 517 |
| B factors | |||||
| Protein | 17.8 | 12.7 | 13.3 | 11.5 | 15.7 |
| (Ac)CoA | 15.8 | 12.8 | 13.2 | 9.2 | 14.5 |
| Substrate | 17.1 | 14.4 | 10.5 | 14.9 | |
| Water | 30.6 | 27.7 | 28.6 | 26.5 | 28.9 |
| r.m.s.c deviations | |||||
| Bond lengths (Å) | 0.006 | 0.007 | 0.007 | 0.007 | 0.007 |
| Bond angles (°) | 1.14 | 1.43 | 1.52 | 1.35 | 1.49 |
a Highest resolution shell is shown in parentheses.
b R factor = Σ(|Fobs| − k|Fcalc|)/Σ|Fobs|, and Rfree is the R value for a test set of reflections consisting of a random 5% of the diffraction data not used in refinement.
c Root mean square.
RESULTS
Protein Expression and Purification
We have previously described a plasmid for overproduction of MccECTD (encompassing residues Lys-407 through Gly-590) that was used for in vitro analysis of acetyltransferase activity (16). However, as the resultant protein failed to yield diffraction quality crystals, a number of additional expression plasmids were generated based on results from multiple sequence alignments of the MccE acetyltransferase domain with other acetyltransferases. A construct encompassing residues Asp-409 through Ile-589 produced a protein (MccEAcTase) that demonstrated superior stability and yielded crystal amenable for structural studies. A thrombin-cleavable amino-terminal hexahistidine tag was used to facilitate purification, and all biochemical and structural studies were carried out using samples from which the affinity tag was removed.
Kinetic and Thermodynamic Analysis
We have shown previously that MccECTD covalently modified and inactivated processed McC as well as various aminoacyl sulfamoyl adenylates. To further characterize this activity, we carried out a kinetic analysis of MccEAcTase by reaction of 5,5′-dithiobis(2-nitrobenzoic acid) with the free sulfhydryl of the product coenzyme A that is formed upon acetylation of the substrate. Using ESA as a substrate, the spectrophotometric studies yielded a Km value of 55.71 ± 6.96 μm and kcat value of 23.68 ± 2.19 min−1. As neither the side chain of the sulfamoyl adenylate or the propylamine of McC makes any specific contacts with MccEAcTase (see below), the kinetic constants for DSA and processed McC are likely very similar to those experimentally determined for ESA.
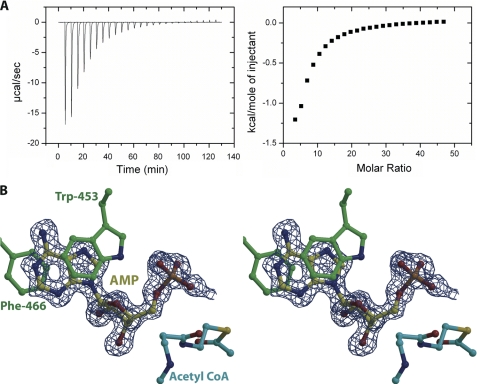
Because MccE acetyltransferase acetylates various aminoacyl adenylates irrespective of the nature of the aminoacyl moiety, we hypothesized that the enzyme may have a binding determinant for the shared AMP moiety of these substrates. Although spectrophotometric analysis indeed revealed that AMP competitively inhibited the DSA acetylation reaction, we were unable to accurately measure the inhibition constant. Therefore, to determine the binding characteristics of AMP as a competitive inhibitor of MccEAcTase, we used isothermal titration calorimetry. The binding isotherm for the interaction was fitted by nonlinear regression using a simple bimolecular interaction model. The data are consistent with a 1:1 interaction between the ligand and protein with a dissociation constant of about 1.03 ± 0.01 mm (see Fig. 4A).
FIGURE 4.
AMP acts as competitive inhibitor. A, raw data and corresponding binding isotherm for the interaction of MccEAcTase with AMP. B, stereoview of the active site of MccEAcTase in complex with CoA and AMP. Substrate and protein residues are colored and the superimposed difference Fourier electron density map (contoured at 3σ over background and shown in blue) was calculated as in Fig. 3.
Overall Structure
Although unliganded MccEAcTase failed to yield crystals, co-crystallization with either CoA or acetyl-CoA readily produced crystals that diffracted to 1.6-Å resolution at an insertion device synchrotron source (Life Sciences Collaborative Access Team, Station 21 ID-D at Argonne National Laboratory, Argonne, IL). The three-dimensional structure of MccEAcTase was solved by the molecular replacement method using the coordinates of the putative ribosomal N-acetyltransferase YdaF from Bacillus subtilis (Protein Data Bank code 1NSL; 35% sequence identity) (21). A successful molecular replacement solution could only be identified using a truncated search model from which the amino-terminal 70 residues were removed. Extensive manual and automated rebuilding led to a complete model of the protein chain encompassing residues Asp-409 through Ile-589. The addition of solvent molecules and acetyl-CoA at positions with suitably well defined electron density features resulted in the final model with a free R factor of 22.7% and good Ramachandran statistics. All data collection and refinement data are listed in Table 1.
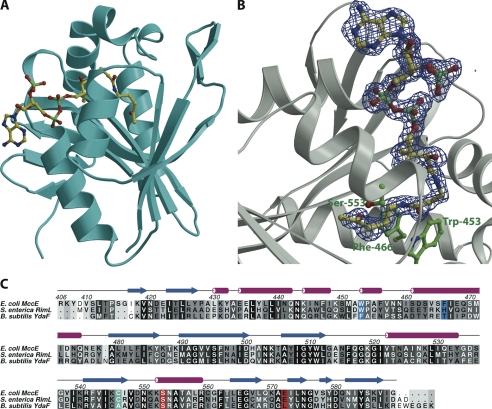
The overall structure of MccEAcTase consists of a GCN5-related N-acetyltransferase (GNAT) superfamily fold consisting of seven antiparallel β strands flanked by four α helices (30, 31) (Fig. 2A). The structure is similar to those of the uncharacterized acetyltransferase YdaF from B. subtilis (Protein Data Bank code 1NSL; root mean square deviation of 1.6 Å over 176 aligned Cα atoms) and Salmonella typhimurium RimL (Protein Data Bank code 1S7L; root mean square deviation of 1.2 Å over 174 aligned Cα atoms) (21, 32). The structure of the polypeptide can be roughly divided into two domains: an amino-terminal domain composed of two large α helices separated by a short 310 helix and a carboxyl-terminal α/β fold that corresponds to the acetyl-CoA-binding site. The most significant deviations from other GNAT family members are along the amino-terminal 70 residues that define the ligand-binding site of MccEAcTase. Unlike Salmonella RimL that has been shown to be a dimer both in solution and in the crystal (32) and B. subtilis YdaF that shares the same dimerization interface in the crystal (21), MccEAcTase is a monomer in the crystal as well as in solution as determined by the elution profile upon size exclusion chromatography with calibrated standards (Fig. S2).
FIGURE 2.
Overall structure and multiple sequence alignment. A, ribbon diagram showing the overall structure of MccEAcTase (in cyan) in complex with acetyl-CoA (yellow ball and stick). B, close-up view of the acetyl-CoA-binding site. Superimposed is a difference Fourier electron density map (in blue) calculated with coefficients |Fobs| − |Fcalc| and phases from the final refined model with the coordinates of acetyl-CoA deleted prior to one round of refinement. The map is contoured at 3σ over background. C, a structure-based sequence alignment of MccEAcTase along with the GNAT acetyltransferases RimL and YdeF. The active site general acid/base are colored in red, the cysteine residue that is disulfide-bonded to CoA is shown in cyan, and Trp-453 and Phe-466, which are involved in π-stacking interactions with the substrate, are shown in blue. S. enterica, Salmonella enterica.
Within the MccEAcTase co-crystal structure, clear electron density corresponding to acetyl-CoA can be observed at a position similar to that observed in other structures of GNAT family members (Fig. 2B). The adenosine group is located at the surface of the molecule and engages in crystal packing contacts with the adenine ring of a symmetry-related molecule of MccEAcTase. The pantetheine and pyrophosphate moieties engage in hydrogen bonding and van der Walls interactions with the polypeptide in a manner similar to that observed in the co-crystal structure of RimL (32). Electron density is evident for the entire ligand, and the acetyl group is situated at the base of the active site pocket where the carbonyl group hydrogen bonds with the backbone amide of Tyr-510 (O-N distance of 3.1 Å) and is flanked by residues Ser-553 and Glu-572. These residues are implicated to act as general acid/base catalysts for the acetyltransferase reaction, and substitution of either of these residues for alanine was shown to compromise the catalytic activity and biological function of MccE (16) (Fig. 2C).
Interactions with Sulfamoyl Adenylate Substrates
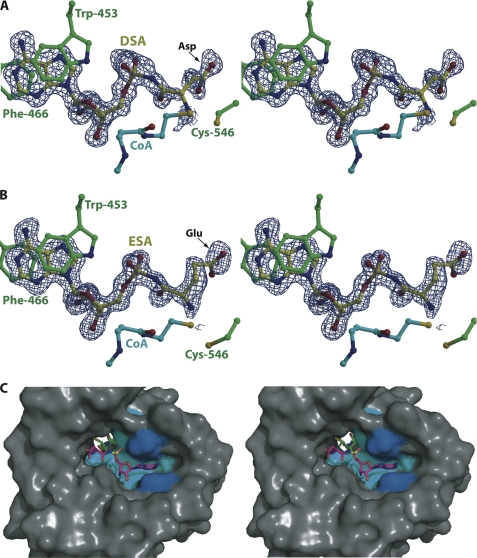
Co-crystals of MccEAcTase with the synthetic sulfamoyl adenylate substrates could only be produced in the presence of acetyl-CoA. In the resultant co-crystal structures with ESA (1.2-Å resolution) and DSA (1.25-Å resolution), clear electron density corresponding to each substrate can be located at the base of a hydrophobic cleft on the face of the molecule opposite from the acetyl-CoA-binding site (Fig. 3, A and B). The binding pocket for the substrate is composed of a number of hydrophobic residues. The substrate molecule is oriented perpendicular to the central β strands of the GNAT fold, resulting in the deposition of the tail of the CoA donor near the α-amino group of the peptide portion of the acceptor substrate. The CoA donor and the substrate are located on opposite sides of the polypeptide, and acetyl transfer occurs through a cavity in the center of MccEAcTase (Fig. 3C).
FIGURE 3.
Active site of MccEAcTase in complex with CoA and DSA/ESA. A, the DSA substrate is colored in yellow, the CoA molecule is colored in cyan, and polypeptide residues colored in green include Trp-453 and Phe-466 that form a π-stacked sandwich with the adenine ring of the substrate and Cys-546 that is disulfide-bonded to the CoA thiol. Superimposed is a difference Fourier electron density map (contoured at 3σ over background and shown in blue) calculated with coefficients |Fobs| − |Fcalc| and phases from the final refined model with the coordinates of DSA deleted prior to one round of refinement. B, the ESA substrate and protein residues are colored as above, and the superimposed difference Fourier electron density map (contoured at 3σ over background and shown in blue) is calculated as above. C, stereoview of a surface rendering of the substrate-binding hydrophobic pocket of MccEAcTase with the superimposed coordinates of DSA (in magenta) and CoA (in green). The hydrophobic cluster of residues (Ile-440, Met-451, Val-493, and Trp-511) that are in van der Waals contact with the substrate is colored in cyan, and Trp-453 and Phe-466 that form a π-stacking interaction with the adenine ring of the substrate are colored in blue.
In both the ESA and DSA co-crystal structures, the polypeptide makes minimal contacts with the amino acid side chain of the substrate. Instead, the most significant determinant for substrate specificity is the adenine ring of the substrate. The ring is sandwiched between two aromatic residues, Trp-453 and Phe-466, which are located within the two large helices in the amino terminus of MccEAcTase, resulting in a strong π-stacking interaction with the aromatic adenine ring of the substrate (Fig. 3, A and B). The two aromatic residues are part of a hydrophobic cluster that also includes Ile-440, Met-451, Val-493, and Trp-511, all of which engage in van der Waals contact with the adenine ring of the substrate.
Aside from this sandwich π-stacking interaction, there are minimal contacts between the polypeptide and the substrate molecule. With respect to the aminoacyl portion of the substrate, the carbonyl oxygen is within hydrogen bonding distance (2.6 Å) from Nδ2 of Asn-497, whereas for the side chain, the closest interacting residue is Lys-545 in which Nζ is 3.7 Å away from Oδ2 of DSA and 3.9 Å away from Oϵ2 of ESA. The ribose oxygens are engaged in hydrogen bonding interactions with Ser-495 (O2′-Oγ distance of 3.3 Å) and the backbone carbonyl of Tyr-510 (O3′-O distance of 3.3 Å). One of the sulfamate oxygens is within hydrogen bonding distance (2.9 Å) from the indole nitrogen of Trp-453. Other than the π-stacking interaction with Trp-453 and Phe-466, there are minimal contacts with any of the polar atoms of the adenine ring. Both N6 and N7 are solvent-exposed, whereas N3 is 3.1 Å away from the indole nitrogen of Trp-511.
In each of the two structures, no electron density can be observed for the acetyl group of the donor, suggesting that hydrolysis of the thioester has occurred in situ. In addition, the side chain of Cys-546 is rotated toward the active site pocket where it forms a covalent disulfide linkage with CoA. Although these structures suggest that MccEAcTase proceeds through an enzyme-acetyl intermediate, this is unlikely as mutational analysis of the equivalent residue in RimL suggests that this cysteine does not play a role in catalysis. Additionally, the equivalent residue in the B. subtilis YdaF acetyltransferase is an alanine (Fig. 2C). A similar cysteyl-acetyl linkage is observed in the RimL co-crystal structure (32).
Interactions with the Competitive Inhibitor AMP
The 1.3-Å resolution co-crystal structure of MccEAcTase-acetyl-CoA in complex with the competitive inhibitor AMP shows clear electron density for both the inhibitor and the intact acetyl group of acetyl-CoA (Fig. 4). The location of the AMP is nearly identical to that occupied by the adenosine sulfamate moieties in the co-crystal structures with DSA and ESA. The remainder of the substrate binding cleft is occupied by a number of solvent molecules. The polypeptide engages the adenosine monophosphate with a set of interactions similar to those observed in the co-crystal structure with DSA and ESA. Additionally, the side chain of Asn-497 is now oriented toward the AMP where Oδ1 is within hydrogen bonding distance (3.2 Å) from the O2′ of the ribose.
Interactions with the Product Acetylated McC
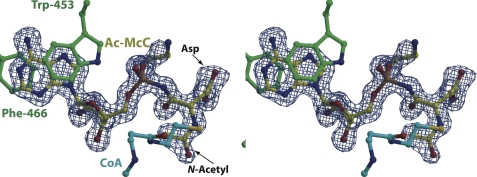
Processed McC was prepared by proteolytic digestion of intact McC in vitro (see “Experimental Procedures”) (Fig. S2) and used to obtain a 1.35-Å resolution co-crystal structure with MccEAcTase-acetyl-CoA. In the structure, clear electron density can be observed for the entire substrate molecule, including the aspartyl side chain and the propylamine decoration on the phosphoramidate oxygen. Unexpectedly, the structure also shows continuous electron density corresponding to the attachment of an acetyl group on the α-amino nitrogen of processed McC (Fig. 5). Density for the substrate molecule is equally clear for both molecules in the crystallographic asymmetric unit, confirming that the structure presents a view of the acetylated processed McC product complex.
FIGURE 5.
Co-crystal structure with acetylated McC product. Stereoview of the active site of MccEAcTase in complex with CoA and acetylated McC (colored as in Fig. 3) with a superimposed difference Fourier electron density map (contoured at 3σ over background and shown in blue) calculated with coefficients |Fobs| − |Fcalc| and phases from the final refined model with the coordinates of substrate deleted prior to one round of refinement.
MccEAcTase engages acetylated processed McC through contacts similar to those observed in co-crystal structures with AMP, DSA, and ESA. Although the 3-aminopropyl group of McC is directed outward to bulk solvent and does not make contacts with the polypeptide, electron density corresponding to this modification can clearly be observed in the co-crystal structure. The acetyl group is situated in a hydrophobic cleft defined by Ile-508, Tyr-510, and Ile-544 where the acetyl oxygen is located within hydrogen bonding distance (3.0 Å) from the backbone carbonyl of Ile-508.
DISCUSSION
The structural and biochemical studies presented here provide a molecular basis for understanding how MccEAcTase can detoxify processed microcin C7 and related aminoacyl adenylate mimics. Although the overall structure of MccEAcTase is similar to those of other GNAT family member, co-crystal structures with AMP, DSA, ESA, and processed McC reveal the presence of a novel, well defined hydrophobic pocket where these substrates are accommodated. The substrate-binding pocket is located on the opposite face of the molecule from the canonical acetyl-CoA-binding site, and the two ligands interact across a cavity near the center of the polypeptide where acetyl transfer likely occurs. The lack of specific contacts with substrate also explains the ability of MccEAcTase to detoxify the structurally distinct seryl-tRNA synthetase inhibitor albomycin (16). Processed albomycin is composed of an N4-carbamyl-5-methyl-4-imino cytidine conjugated to a thioxylofuranosyl pyrimidine (33), and the nucleoside moiety may be similarly accommodated into the MccEAcTase hydrophobic substrate-binding pocket. The rate and specificity for the modification of processed albomycin by MccEAcTase are not yet clear as these studies were conducted as end point assays using whole cell extracts (16).
Detoxification of microcin C7 occurs by the acetylation of the α-amino group of the amino acid adenylate. The co-crystal structure of E. coli AspRS in complex with an aspartate adenylate (Protein Data Bank code 3NEM) provides a rationale for understanding the basis for this detoxification. Addition of an acetyl moiety at the α-amino position would both disrupt favorable hydrogen bond interactions and result in steric clashes with the backbone of AspRS.
At the substrate-binding site, the major determinant for recognition is through π-stacking interactions between the aromatic adenine ring of the substrate and two aromatic residues, Trp-453 and Phe-466, which are located within the two large helices in the amino terminus of MccEAcTase. This mode of purine base engagement is reminiscent of recognition mechanisms used by eukaryotic and viral proteins that recognize the 7-methylguanosine cap at the 5′-end of host messenger RNA (34) and by bacterial and viral nucleotidyltransferases such as DNA/RNA ligases and RNA capping enzymes (35). With respect to mRNA cap-binding proteins, despite the fact that each contains structurally distinct folds, they all share a hydrophobic binding pocket where the methylated guanosine is sandwiched between two aromatic residues, one of which is usually a tryptophan or tyrosine (36–39). Preference for the methylated over non-methylated guanosine is dictated by a stronger interaction between the delocalized positive charge on the 7-methylguanosine and the π-electrons of the aromatic residues. In the nucleotidyltransferase superfamily, the purine nucleotide substrate is similarly sandwiched between a conserved aromatic residue and a conserved aliphatic residue with additional contacts with either a conserved glutamate (phage/bacterial DNA ligase), threonine (viral DNA ligase), or lysine (RNA capping enzymes) residue.
Notably, in the co-crystal structure of T4 phage RNA ligase Rnl2, specificity for the adenine base is conferred solely through contacts only with the protein backbone (40). Similarly, MccEAcTase recognizes its nucleotide substrate without making any direct side chain contacts with the adenine. Calorimetric studies demonstrate a lack of any appreciable binding between MccEAcTase and guanosine monophosphate,4 suggesting some level of discrimination against other purine nucleotides. Modeling studies suggest that steric clashes between the guanine 2-NH2 and the hydrophobic residues that align the binding pocket likely precluded binding. In addition, a thermodynamic argument can also be made for preferential binding as adenine is a more hydrophobic nucleobase than guanine. Studies of Rnl2 have led to the speculation that nucleotidyltransferases evolved from an undifferentiated ATP-dependent ancestral gene, and subsequently, substrate specificity resulted from minimal side chain alterations that resulted in specific contacts (40). Our biochemical and structural biological studies of MccEAcTase provide another example of a structurally unrelated polypeptide that utilizes a similar aromatic sandwich to achieve specificity for its adenylated substrate, suggesting a common ancestral origin for these enzymes.
Supplementary Material
Acknowledgments
We thank Gaston Vondenhoff and Arthur van Aerschot (Rega Institute for Medical Research, Leuven, Belgium) for generous gifts of DSA and ESA. We thank Dr. Heather Cooke for assistance with spectrophotometric assays and Dr. Susan Martinis for valuable scientific discussions. We also thank Keith Brister and Joseph Brunzelle at the Life Sciences Collaborative Access Team (23-ID at Argonne National Laboratory, Advanced Photon Source) for facilitating and technical assistance during crystallographic data collection.
This work was supported by a Technology Commercialization Fund grant from Rutgers University (to K. S.) and a Molecular and Cell Biology Program grant from the Russian Academy of Sciences. A. M. was supported by Grant 06-04-48865 from the Russian Foundation for Basic Research.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
The atomic coordinates and structure factors (codes 3R95, 3R96, 3R9E, 3R9F, and 3R9G) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
V. Agarwal and S. K. Nair, unpublished results.
- McC
- microcin C7
- MccECTD
- MccE carboxyl-terminal domain
- DSA
- aspartyl-sulfamoyl adenosine
- ESA
- glutamyl-sulfamoyl adenosine
- MccEAcTase
- MccE acetyltransferase domain
- MR
- molecular replacement
- GNAT
- GCN5-related N-acetyltransferase.
REFERENCES
- 1. Destoumieux-Garzón D., Peduzzi J., Rebuffat S. (2002) Biochimie 84, 511–519 [DOI] [PubMed] [Google Scholar]
- 2. Hurdle J. G., O'Neill A. J., Chopra I. (2005) Antimicrob. Agents Chemother. 49, 4821–4833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pohlmann J., Brötz-Oesterhelt H. (2004) Curr. Drug Targets Infect. Disord. 4, 261–272 [DOI] [PubMed] [Google Scholar]
- 4. Severinov K., Semenova E., Kazakov A., Kazakov T., Gelfand M. S. (2007) Mol. Microbiol. 65, 1380–1394 [DOI] [PubMed] [Google Scholar]
- 5. Guijarro J. I., González-Pastor J. E., Baleux F., San Millán J. L., Castilla M. A., Rico M., Moreno F., Delepierre M. (1995) J. Biol. Chem. 270, 23520–23532 [DOI] [PubMed] [Google Scholar]
- 6. Metlitskaya A. Z., Katrukha G. S., Shashkov A. S., Zaitsev D. A., Egorov T. A., Khmel I. A. (1995) FEBS Lett. 357, 235–238 [DOI] [PubMed] [Google Scholar]
- 7. Novikova M., Metlitskaya A., Datsenko K., Kazakov T., Kazakov A., Wanner B., Severinov K. (2007) J. Bacteriol. 189, 8361–8365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kazakov T., Vondenhoff G. H., Datsenko K. A., Novikova M., Metlitskaya A., Wanner B. L., Severinov K. (2008) J. Bacteriol. 190, 2607–2610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Metlitskaya A., Kazakov T., Kommer A., Pavlova O., Praetorius-Ibba M., Ibba M., Krasheninnikov I., Kolb V., Khmel I., Severinov K. (2006) J. Biol. Chem. 281, 18033–18042 [DOI] [PubMed] [Google Scholar]
- 10. Reader J. S., Ordoukhanian P. T., Kim J. G., de Crécy-Lagard V., Hwang I., Farrand S., Schimmel P. (2005) Science 309, 1533 [DOI] [PubMed] [Google Scholar]
- 11. González-Pastor J. E., San Millán J. L., Castilla M. A., Moreno F. (1995) J. Bacteriol. 177, 7131–7140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. González-Pastor J. E., San Millán J. L., Moreno F. (1994) Nature 369, 281 [DOI] [PubMed] [Google Scholar]
- 13. Metlitskaya A., Kazakov T., Vondenhoff G. H., Novikova M., Shashkov A., Zatsepin T., Semenova E., Zaitseva N., Ramensky V., Van Aerschot A., Severinov K. (2009) J. Bacteriol. 191, 2380–2387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Roush R. F., Nolan E. M., Löhr F., Walsh C. T. (2008) J. Am. Chem. Soc. 130, 3603–3609 [DOI] [PubMed] [Google Scholar]
- 15. Fomenko D. E., Metlitskaya A. Z., Péduzzi J., Goulard C., Katrukha G. S., Gening L. V., Rebuffat S., Khmel I. A. (2003) Antimicrob. Agents Chemother. 47, 2868–2874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Novikova M., Kazakov T., Vondenhoff G. H., Semenova E., Rozenski J., Metlytskaya A., Zukher I., Tikhonov A., Van Aerschot A., Severinov K. (2010) J. Biol. Chem. 285, 12662–12669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zeng Y., Roy H., Patil P. B., Ibba M., Chen S. (2009) Antimicrob. Agents Chemother. 53, 4619–4627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Otwinowski Z., Borek D., Majewski W., Minor W. (2003) Acta. Crystallogr. A 59, 228–234 [DOI] [PubMed] [Google Scholar]
- 19. McCoy A. J. (2007) Acta. Crystallogr. D. Biol. Crystallogr. 63, 32–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Brunzelle J. S., Wu R., Korolev S. V., Collart F. R., Joachimiak A., Anderson W. F. (2004) Proteins 57, 850–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) Acta. Crystallogr. D Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 23. Terwilliger T. C., Grosse-Kunstleve R. W., Afonine P. V., Moriarty N. W., Zwart P. H., Hung L. W., Read R. J., Adams P. D. (2008) Acta. Crystallogr. D Biol. Crystallogr. 64, 61–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. McRee D. E. (1999) J. Struct. Biol. 125, 156–165 [DOI] [PubMed] [Google Scholar]
- 25. Perrakis A., Sixma T. K., Wilson K. S., Lamzin V. S. (1997) Acta. Crystallogr. D Biol. Crystallogr. 53, 448–455 [DOI] [PubMed] [Google Scholar]
- 26. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta. Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 27. Murshudov G. N., Vagin A. A., Lebedev A., Wilson K. S., Dodson E. J. (1999) Acta. Crystallogr. D Biol. Crystallogr. 55, 247–255 [DOI] [PubMed] [Google Scholar]
- 28. Brünger A. T. (1992) Nature 355, 472–475 [DOI] [PubMed] [Google Scholar]
- 29. Laskowski R. A., Rullmannn J. A., MacArthur M. W., Kaptein R., Thornton J. M. (1996) J. Biomol. NMR 8, 477–486 [DOI] [PubMed] [Google Scholar]
- 30. Dyda F., Klein D. C., Hickman A. B. (2000) Annu. Rev. Biophys. Biomol. Struct. 29, 81–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vetting M. W., de Carvalho L. P., Yu M., Hegde S. S., Magnet S., Roderick S. L., Blanchard J. S. (2005) Arch. Biochem. Biophys. 433, 212–226 [DOI] [PubMed] [Google Scholar]
- 32. Vetting M. W., de Carvalho L. P., Roderick S. L., Blanchard J. S. (2005) J. Biol. Chem. 280, 22108–22114 [DOI] [PubMed] [Google Scholar]
- 33. Stefanska A. L., Fulston M., Houge-Frydrych C. S., Jones J. J., Warr S. R. (2000) J. Antibiot. 53, 1346–1353 [DOI] [PubMed] [Google Scholar]
- 34. Fechter P., Brownlee G. G. (2005) J. Gen. Virol. 86, 1239–1249 [DOI] [PubMed] [Google Scholar]
- 35. Shuman S., Lima C. D. (2004) Curr. Opin. Struct. Biol. 14, 757–764 [DOI] [PubMed] [Google Scholar]
- 36. Guilligay D., Tarendeau F., Resa-Infante P., Coloma R., Crepin T., Sehr P., Lewis J., Ruigrok R. W., Ortin J., Hart D. J., Cusack S. (2008) Nat. Struct. Mol. Biol. 15, 500–506 [DOI] [PubMed] [Google Scholar]
- 37. Hodel A. E., Gershon P. D., Shi X., Quiocho F. A. (1996) Cell 85, 247–256 [DOI] [PubMed] [Google Scholar]
- 38. Marcotrigiano J., Gingras A. C., Sonenberg N., Burley S. K. (1997) Cell 89, 951–961 [DOI] [PubMed] [Google Scholar]
- 39. Mazza C., Segref A., Mattaj I. W., Cusack S. (2002) EMBO. J. 21, 5548–5557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ho C. K., Wang L. K., Lima C. D., Shuman S. (2004) Structure 12, 327–339 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.