Abstract
Phosphatidylcholine (PC) is synthesized by two different pathways, the Lands cycle and the Kennedy pathway. The recently identified key enzymes of the Lands cycle, lysophosphatidylcholine acyltransferase 1 and 2 (LPCAT1 and -2), were reported to localize to the endoplasmic reticulum and to function in lung surfactant production and in inflammation response. Here, we show in various mammalian cell lines that both enzymes additionally localize to lipid droplets (LDs), which consist of a core of neutral lipids surrounded by a monolayer of phospholipid, mainly PC. This dual localization is enabled by the monotopic topology of these enzymes demonstrated in this study. Furthermore, we show that LDs have the ability to locally synthesize PC and that this activity correlates with the LPCAT1 and -2 expression level. This suggests that LPCAT1 and -2 have, in addition to their known function in specialized cells, a ubiquitous role in LD-associated lipid metabolism.
Keywords: Lipid Droplet, Lipid Synthesis, Membrane Enzymes, Membrane Lipids, Phosphatidylcholine
Introduction
In animals, triacylglycerol (TAG)2 is the major compound for long term storage of energy. On the cellular level, the hydrophobic TAG is stored in organelles called lipid droplets (LDs), which consist of a core of neutral lipids, mainly TAG and sterol esters, that is surrounded by a monolayer of polar lipids (1). Glycerophospholipids, in particular PC, are the main component of this monolayer (2, 3). Additionally, proteins are attached to or inserted into the monolayer. Numerous LD-associated proteins were identified in recent proteomics studies in a variety of cell lines (4–13), among them are the members of the perilipin-adipophilin-tail interacting protein 47 (TIP47) (PAT) protein family (14) and several enzymes of lipid metabolism.
LDs are very dynamic organelles, which grow in size upon neutral lipid synthesis, lipogenesis, or shrink, when the stored lipids are mobilized during lipolysis. The necessary coordinated change of LD volume and surface is dynamically controlled by an unknown mechanism (15, 16). We have recently shown that TAG biosynthesis can take place directly at the surface of LDs (17). This activity is catalyzed by diacylglycerol acyltransferase 2 (DGAT2), which localizes to the LD surface (17, 18). However, evidence for a concomitant local synthesis of the main LD surface lipids, in particular PC, is lacking.
Two conserved major biochemical pathways contribute to the synthesis of PC as follows: the Kennedy pathway for de novo PC synthesis and the Lands cycle for remodeling of the fatty acid composition of PC species (19, 20). A third pathway, which operates by methylation of phosphatidylethanolamine to PC, is restricted to liver cells (21). In the Kennedy pathway, choline phosphate is activated with cytidine triphosphate (CTP) and transferred to diacylglyceride (DAG) to form PC. The enzymes to catalyze these reactions are the cytoplasmic CTP:phosphocholine cytidylyltransferase and the membrane-embedded cholinephosphotransferase (CPT) or choline/ethanolamine phosphotransferase (CEPT) (22, 23). The Lands cycle includes the removal of fatty acids at the sn-2 position of PC by phospholipase A2 (PLA2) to yield lysophosphatidylcholine (LPC) followed by re-acylation by lysophosphatidylcholine acyltransferases (LPCATs) (24). The Lands cycle and the Kennedy pathway are interconnected by LPCAT1, whose expression level influences the degradation rate of CPT1 (25).
Recently, four LPCATs were cloned and characterized (26–30). LPCAT1 and LPCAT2 belong to the lysophosphatidic acid acyltransferase (LPAAT) family, characterized by the presence of four conserved motifs (31), and possess a C-terminal KKXX ER retention motif. In contrast, LPCAT3 and LPCAT4 lack LPAAT motifs and are related to proteins of the membrane-bound O-acyltransferase family (32), which include the cholesterol acyltransferase ACAT1/SOAT1 and the DAG acyltransferase 1 (DGAT1).
LPCAT1 is highly expressed in lung tissue, especially in type II alveolar cells (26), and it was also detected in red blood cells (29). LPCAT2 expression is highest in resident macrophages and casein-induced neutrophils (28). LPCAT1 is important for the production of lung surfactant (26, 27, 33, 34). Recently, lyso-platelet-activating factor acyltransferase activity of LPCAT1 was demonstrated (33). The activity and expression of LPCAT1 is independent of any inflammatory stimulation, in contrast to LPCAT2. Upon acute inflammation LPCAT2 transfers acetyl-CoA to lyso-platelet-activating factor to form platelet-activating factor. Under resting conditions, it participates in membrane remodeling through insertion of arachidonyl-CoA into PC (28, 35). Beside acetyl-CoA and arachidonyl-CoA, LPCAT2 exhibits a similarly broad substrate preference as LPCAT1 (36).
In this study, we show that LPCAT1 and LPCAT2 localize to the surface of LDs. Additionally, we demonstrate that both proteins are monotopic membrane proteins and that their expression level correlates with LD-associated LPCAT activity.
EXPERIMENTAL PROCEDURES
Antibodies
Polyclonal rabbit antisera against the C-terminal peptide of human LPCAT1 CNSDAGRKPVRKKLD, conjugated to keyhole limpet hemocyanin, and against purified recombinant His6-hLPCAT2(310–545), His6-hACSL3(504–721), and His6-hNSDHL(1–211) were raised by Eurogentec (Seraing, Belgium) and were affinity-purified against the respective antigen. The specificity and sensitivity of the LPCAT1 and LPCAT2 antibodies were evaluated by Western blotting against overexpressed tagged proteins and against lysates of nonoverexpressing A431 cells. Antiserum against human tail interacting protein 47 (TIP47) was a kind gift of Stefan Hoening (Cologne University), and antiserum against ACAT1 (DM10 (37)) was generously provided by Dr. Ta-Yuan Chang (Dartmouth College, Hanover, NH). We used anti-hemagglutinin (HA) mAb clone F-7 (Santa Cruz Biotechnology), polyclonal rabbit anti-calnexin and anti-protein-disulfide isomerase (PDI) antibody (StressGen), Alexa 555-coupled goat polyclonal anti-rabbit IgG, Alexa 647-coupled goat polyclonal anti-mouse IgG, and Alexa 488-coupled goat polyclonal anti-mouse IgG (Invitrogen), and HRP-coupled polyclonal goat anti-rabbit IgG and anti-mouse IgG antibody (Dianova).
Chemicals
The lipids 16:0-lysophosphatidic acid (16:0-LPC) and CDP-choline were from Fluka, and oleoyl-CoA, palmitoyl-CoA, and dioleoyl-DAG were from Sigma. Synthetic lipid standards were from Avanti Polar Lipids, Inc. (Alabaster, AL), Larodan Fine Chemicals (Malmö, Sweden), and Sigma. [14C]CDP-choline was from Biotrend, and [3H]oleate was from Hartmann Analytic. [3H]Acyl-CoAs were synthesized as described previously (38).
DNA Constructs
Human LPCAT1 and LPCAT2 were amplified by PCR from full-length ESTs and cloned into pCDNA3.1 vectors (Clontech) with or without inserted N- or C-terminal HA3 tags or pEGFP vectors (Clontech) with C- or N-terminally enhanced GFP fusion protein. For immunization, a fragment of LPCAT2 was cloned behind a His6 tag into the bacterial expression vector pHAT2. The supplemental Table 1 contains detailed information, including primer sequences and restriction sites. All constructs were verified by sequencing.
siRNA sequences were from Ambion. Sequences and siRNA IDs are listed in supplemental Table 1. Lipofectamine 2000 was from Invitrogen, and INTERFERinTM was from Peqlab.
Cell Culture
A431 and COS7 cells were maintained in Dulbecco's modified Eagle's medium (DMEM, Invitrogen 31966) supplemented with 10% fetal calf serum (FCS), HuH7 cells in RPMI 1640 medium (Invitrogen 31870) supplemented with 10 mm HEPES, 0.1 mm nonessential amino acids, 2 mm l-glutamine, and 10% FCS. Cells were grown at 37 °C and 5% CO2. If indicated, normal FCS was replaced by delipidated FCS, prepared by solvent extraction as described previously (39).
siRNA Transfection
For transfection with siRNA, cells were seeded in 24-well dishes at 5000 cells per well and treated with siRNA (9 nm) using INTERFERinTM according to the manufacturer's instructions. After 48 h, oleic acid was added to the cells to a final concentration of 50 μm and grown for a further 24 h.
Purification of LDs
Cells were grown in 10-cm dishes in medium supplemented with 50–100 μm oleate for 16 h, washed, and scraped in ice-cold disruption buffer (20 mm HEPES/NaOH, pH 7.4, 0.25 m sucrose) followed by homogenization in a cooled EMBL cell cracker (HGM, Heidelberg, Germany) with 5 strokes using a maximum clearance of 18 μm. The lysate was centrifuged at 1000 × g for 10 min, and the post-nuclear supernatant was adjusted to 1.1 m sucrose. Four ml of the post-nuclear supernatants were loaded to the bottom of a 13-ml centrifuge tube and overlaid with ice-cold disruption buffer. The gradients were centrifuged in a swing-out rotor at 100,000 × g at 4 °C for 2.5 h. Fractions were taken from the top as follows: top 2 ml, LD fraction; next 3.5 ml, intermediate fraction; next 4 ml, including the phase boundary between 0.25 and 1.1 m sucrose, floating membranes; and last 3.5 ml, bottom fraction.
Acyltransferase Assays
Whole cell lysates (15 μl, adjusted to 200 μl with disruption buffer) or LD fractions from sucrose gradient centrifugation (200 μl) were mixed with 100 μl of assay buffer (200 mm Tris/HCl, pH 7.5, 10 mm MgCl2, 2 mg/ml fatty acid-free bovine serum albumin (BSA), 10 μm oleoyl-CoA, 10 μm palmitoyl-CoA, 1 μCi/ml each of [3H]oleoyl-CoA, [3H]palmitoyl-CoA, and [3H]myristoyl-CoA). For the LPAAT assay 100 μm sn-1-palmitoyl-sn-2-lyso-PA and for the LPCAT assay 100 μm sn-1-palmitoyl-sn-2-lyso-PC (final concentration) were added followed by incubation for 30 min at 30 °C. The assays for LPCAT overexpression or knockdown experiments were performed with 100 μm sn-1-palmitoyl-sn-2-lyso-PC and 6.5 μm oleoyl-CoA and 3 μCi/ml [3H]oleoyl-CoA (final concentration). These conditions ensure substrate saturation and linearity with respect to the amount of added enzyme, as determined in separate experiments (data not shown). Reactions were stopped by shaking with a mixture of 700 μl of chloroform/methanol, 1:3, and 400 μl of water. The phases were separated by centrifugation, and the chloroform phase was analyzed by TLC in ethanol/triethylamine/chloroform/water, 50:35:35:10 (17). Plates were sprayed with scintillant and exposed to x-ray film at −78 °C. Signals were quantified by scraping from TLC plate and scintillation counting or from the scanned x-ray films with Fiji software, corrected for total protein content, and normalized to the respective control.
Diacylglycerol Cholinephosphotransferase Assay
Whole A431 cell lysates (50 μl, adjusted to 200 μl with disruption buffer) or a mixture of bottom (50 μl) and floating membrane fractions (50 μl) (adjusted to 200 μl with disruption buffer) or LD fractions from sucrose gradient centrifugation (200 μl in disruption buffer) were mixed with 100 μl of assay buffer (50 mm Tris/HCl, pH 8.0, 10 mm MgCl2, 100 μm dioleoyl-DAG, 0.5 μCi of [14C]CDP-choline, 200 μm CDP-choline) and incubated for 30 min at 30 °C. The same procedure as described for acyltransferase assay was followed.
Glucose-6-phosphatase Assay
A standard assay adopted from Arden et al. (40) was used for quantification of glucose-6-phosphatase (Glc-6-Pase) activity. LD fraction or lysates of HuH7, A431, or COS7 cells (200 μl) were added to 100 μl of RB buffer (20 mm sodium tartrate, 10 mm EDTA, pH 6.5) either with or without 100 mm glucose 6-phosphate. As a standard for free phosphate, different amounts of sodium dihydrogen phosphate were used. The samples were incubated for 5 h at 30 °C. The reaction was stopped by the addition of 60 μl of 10% (v/v) trichloroacetic acid. Samples were centrifuged, and supernatants were mixed with 250 μl of color reagent (1 part 4.2% (w/v) ammonium molybdate in 5 m HCl and 2 parts 0.2% (w/v) malachite green in H2O). The absorbance at 650 nm was used to calculate the amounts of free phosphate formed using the standard curve.
Proteomic Analysis of LD Proteins
A431 cells (8 × 10-cm dishes) were grown as above and supplemented with 100 μm oleate for the final 16 h. Cells were lysed, and LDs were purified exactly as described above with the exception of the addition of Complete protease inhibitor tablets (Roche Applied Science) to all buffers. Proteins from pooled LD fractions were precipitated using chloroform/methanol (41), subjected to one-dimensional SDS-PAGE (10% gel), and visualized by Coomassie Brilliant Blue staining. The lane was cut into 34 bands that were separately digested with trypsin, and recovered peptides were analyzed by liquid chromatography-tandem mass spectrometry on LTQ linear ion trap mass spectrometer as described previously (42).
Fluorescence Microscopy
For transfection with expression vectors, cells were grown on glass coverslips to 80% confluency, transfected with DNA using Lipofectamine 2000 according to the manufacturer's instructions, and further cultivated. If indicated, media were supplemented with 100 μm oleate. After 24 h, cells were fixed with 3% (w/v) paraformaldehyde in PBS for 30 min, washed with PBS, blocked, and permeabilized for 30 min in PBS containing 0.5% BSA and 0.1% saponin (blocking buffer, BB). If indicated, saponin in the BB was replaced by 0.001% digitonin (Applichem A1905,0100). Cells were incubated with primary antibodies for 1 h in BB, washed three times with BB, incubated with secondary antibody in BB, washed two times in BB and two times in PBS, and counterstained with Bodipy 493/503 in PBS, followed by three washes in PBS. After rinsing in water, coverslips were mounted in Mowiol 4-88 containing 2.5% 1,4-diazabicyclo(2.2.2)octane. Images were acquired with a Zeiss LSM 510 confocal microscope equipped with a 100× NA 1.3 oil objective using laser excitation at 488, 543, and 633 nm.
Fluorescence Protease Protection Assay
A431 cells were grown in 3-cm glass bottom dishes and co-transfected with pDsRed2-ER (Clontech) and either pLPCAT1-EGFP, pEGFP-LPCAT1, pLPCAT2-EGFP, or pEGFP-LPCAT2. Cells were washed two times in KHM buffer (110 mm potassium acetate, 20 mm HEPES, pH 7, 2 mm MgCl2). The dish was placed on the microscope stage and incubated with 60 μm digitonin in KHM buffer for 1 min, and an image was recorded. Buffer was exchanged for KHM buffer containing 50 μg/ml proteinase K or 125 μg/ml trypsin (1:40 dilution of 10× trypsin, Sigma T4174). Immediately after, images were recorded every 10 s for 2 min. In Fig. 3C, images taken after 30 s are shown. Images were acquired at a Zeiss Axio Observer.Z1 with a 63× NA 1.2 objective.
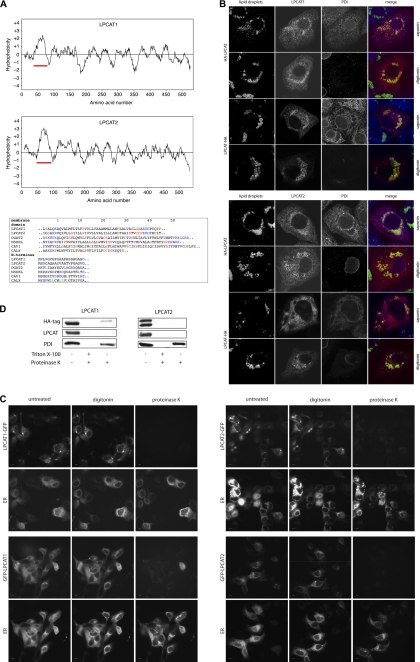
FIGURE 3.
LPCAT1 and LPCAT2 contain a long hydrophobic stretch and insert into LDs and membranes in a monotopic conformation. A, upper panels, hydrophobicity profiles of human LPCAT1 and LPCAT2 were calculated using the Kyte-Doolittle algorithm with a window of 17 amino acids. Red bar, N-terminal long hydrophobic stretch. Lower panel, sequence comparison between the hydrophobic sequences of LPCAT1 and LPCAT2 and the hairpin hydrophobic domains of the LD proteins DGAT2, NSDHL, and caveolin 1 (CAV1) and transmembrane domain of the ER protein calnexin (CALX). Position 1 indicates the first residue of the hydrophobic stretch or the N-terminal methionine. Color code in the membrane domains is as follows: charged residues, blue; helix-braking (Pro, Gly, Ser, and Cys), red. Color code for N termini: charged residues, blue; leucine, green. Sequences are taken from SwissProt with the following accession numbers: LPCAT1, Q8NF37; LPCAT2, Q7L5N7; DGAT2, Q96PD7; NSDHL, Q15738; CAV1, Q03135; CALX, P27842. B, A431 cells were transfected with vectors coding for LPCAT1 or LPCAT2 as indicated, both with either the N-terminal HA3 tag (HA-LPCAT) or C-terminal HA3 tag (LPCAT-HA) and cultivated in the presence of 100 μm oleate. Cells were fixed and processed for immunofluorescence microscopy using either saponin (permeabilizes all membranes) or digitonin (permeabilizes plasma membrane but not the ER membrane) for permeabilization. Cells were stained with Bodipy 493/503 to stain LDs (green), anti-HA to detect transfected LPCAT proteins (red), and anti-PDI (blue) as a lumenal ER marker (scale bar, 10 μm). C, living A431 cells overexpressing LPCAT1-GFP or GFP-LPCAT1 (upper panel) or LPCAT2-GFP or GFP-LPCAT2 (lower panel), all cultivated in the presence of oleate, were treated with digitonin. Cells were exposed to proteinase K, and images were recorded. Representative images before treatment (untreated), after 1 min digitonin (digitonin), and after 30 s of proteinase K exposure (proteinase K) are shown. The integrity of the ER at all conditions was monitored via the lumenal DsRed2-ER (ER). Scale bar, 10 μm. D, A431 cells expressing N-terminally HA3-tagged LPCAT1 or LPCAT2 were lysed, and microsomes were isolated. Microsomes were incubated in PBS with either no addition (negative digestion control), addition of 1% Triton X-100 and proteinase K (positive digestion control), or addition of proteinase K alone. The integrity of the microsomes was analyzed by Western blotting against the lumenal ER marker PDI. The N terminus of LPCAT1 and LPCAT2 was detected with a HA-specific antibody and the C terminus with LPCAT1 and LPCAT2 antibodies. No smaller fragments were detected on the blots (data not shown).
Proteinase Protection Assay in Isolated Microsomes
N-terminally HA3-tagged LPCAT1 or LPCAT2 was expressed in A431 cells for 16 h. Cells were lysed, and microsomes were collected by centrifugation. Microsomes were incubated in PBS and treated as indicated in Fig. 3D using 50 μg/ml proteinase K in PBS for 60 min. Digestion was stopped by the addition of 10 mm PMSF. Samples were boiled in reducing SDS-PAGE sample buffer and analyzed by SDS-PAGE/Western blotting.
RESULTS
LPCAT1 and LPCAT2 Localize to LDs
LD-associated proteins were isolated from A431 cells, separated by SDS-PAGE (supplemental Fig. 1A), and analyzed by protein mass spectrometry (42). Identified proteins (supplemental Fig. 1B) include the abundant PAT protein TIP47, the long chain acyl-CoA synthetase 3 (ACSL3), and the sterol dehydrogenase NAD(P)-dependent steroid dehydrogenase-like (NSDHL), which were observed in previous studies of LDs (4, 43, 44). In addition, we identified the two enzymes LPCAT1 (26, 27) and LPCAT2 (28) whose abundance (as judged by their abundance indices (45)) was comparable with that of bona fide LD proteins such as Rab18 (8, 46) and CGI-58 (47). No additional long chain acyltransferases were identified with the exception of much less abundant LPAAT-θ/AGPAT9, which is a distant relative of LPCAT1 and LPCAT2.
Overexpressed N-terminally tagged LPCAT1 and LPCAT2 were previously reported to localize to the ER in CHO K1 cells (26, 28). To study the subcellular localization of the endogenous proteins, we produced specific antibodies against human LPCAT1 and LPCAT2. Western blot analysis of different cell lines showed high expression levels of both LPCAT1 and LPCAT2 in A431 cells, whereas COS7 and HuH7 cells expressed much lower amounts of both LPCATs or only LPCAT1, respectively (Fig. 1). Confocal immunofluorescence microscopy of A431 cells revealed a distinct localization to LDs and ER structures for both endogenous and overexpressed LPCAT1 and LPCAT2 (Figs. 2A and 3B). Also in COS7 cells, both proteins localized to LDs (Fig. 2B). Unlike in A431 cells, where continuous rings around LDs were seen, the staining in COS7 cells showed discrete patches on LDs, likely a consequence of the lower expression levels of endogenous LPCAT1 and LPCAT2.
FIGURE 1.
Expression of LPCAT1 and LPCAT2 in different cell lines. Total lysates of HuH7 cells, A431 cells, and COS7 cells were subjected to SDS-PAGE/Western blotting for LPCAT1, LPCAT2, and GAPDH (loading control). Longer exposure times did not reveal any signal for LPCAT2 in HuH7 cells (data not shown).
FIGURE 2.
Endogenous LPCAT1 and LPCAT2 localize to LDs. A, A431 cells were cultivated in DMEM supplemented with 10% of either delipidated FCS (DL), normal FCS (FCS), or normal FCS + 100 μm oleate (Ole). Cells were fixed and processed for immunofluorescence microscopy using Bodipy 493/503 to stain neutral lipids (lipid droplets) and anti-LPCAT1 (LPCAT1, upper panels) or anti-LPCAT2 (LPCAT2, lower panels) to stain endogenous LPCAT proteins. In the merged pictures, Bodipy 493/503 staining is in green and anti-LPCAT in red. Scale bar, 10 μm. B, COS7 cells were cultivated in DMEM supplemented with 10% normal FCS + 100 μm oleate. Fixation, staining, and imaging were performed as described for A. C, lysates of A431 cells, grown in DMEM/FCS + 100 μm oleate, or HuH7 cells, grown in supplemented RMPI/FCS + 100 μm oleate, were subjected to floatation in a sucrose density gradient. The gradient was fractionated from the top into LDs, intermediate, floating membranes, and bottom fractions. Proteins were analyzed by SDS-PAGE/Western blotting for LPCAT1 and LPCAT2, the known integral LD proteins ACSL3 and NSDHL, the peripheral LD proteins TIP47 and ADRP, and the integral ER membrane proteins ACAT1 and calnexin.
Localization to the vicinity of LDs could represent direct localization to the LD surface but also to adjacent organelles, in particular to ER structures that surround LDs. To address this issue, we separated LDs from other membranes by sucrose gradient centrifugation and analyzed the fractions by Western blotting (Fig. 2C). In A431 cells (Fig. 2C, left panel), a significant fraction of both LPCAT1 and LPCAT2 was floating with the LD marker proteins NSDHL, ACSL3, and TIP47 to the top of the gradient, which in turn was devoid of immunoreactivity against the ER membrane proteins calnexin and ACAT1 (37). For all tested LD-associated proteins (LPCAT1, LPCAT2, ACSL3, NSDHL, and TIP47), 10–30% of the total protein floated with the LDs, with the remainder residing in both the bottom fraction and the floating membrane fraction. In HuH7 cells (Fig. 2C, right panel), 50% of LPCAT1 was found in the LD fraction, which was devoid of ER marker proteins but contained more than 96% of the LD marker ADRP. Note that HuH7 cells contain abundant ADRP, whereas A431 cells are devoid of ADRP (supplemental Fig. 1B).
LPCAT1 and LPCAT2 Are Monotopic Membrane Proteins
Both LPCAT1 and LPCAT2 possess a prominent hydrophobic region starting at amino acid 44 in LPCAT1 and amino acid 58 in LPCAT2 (Fig. 3A, upper panels). These hydrophobic regions span ∼35 amino acid residues, which is much longer than a typical transmembrane helix of 15–25 amino acids. They contain regularly interspaced helix-braking amino acids and lack N-terminal hydrophobic leucine-rich signal sequences (Fig. 3A, lower panel). A hydrophobic stretch with similar length is also found in NSDHL, DGAT2, and the caveolin proteins, all of which display complex multiple localization to LDs plus other cellular organelles (17, 44, 48–50). In NSDHL (51), DGAT2 (52), and caveolin 1 (53), the long hydrophobic domain does not fully span a membrane but forms a turn with both the N and C termini facing the cytoplasm. This monotopic mode of membrane insertion allows protein integration into the surface monolayer of LDs (54). To characterize the topology of LPCAT1 and LPCAT2, we expressed fusion constructs with either an N- or C-terminal HA3 tag. Cells were permeabilized using saponin, which permeabilizes all cellular membranes, or digitonin, which permeabilizes the plasma membrane but not the ER membrane (55), and stained with anti-HA antibody to detect expressed LPCAT proteins. An antibody against the lumenal ER protein, protein-disulfide isomerase (PDI), was used to monitor the selective permeabilization of the plasma membrane by digitonin. Although the anti-HA antibody detected the LPCAT constructs irrespective of the mode of permeabilization or the position of the tag, the anti-PDI only showed specific staining after saponin permeabilization (Fig. 3B). This observation was also independent of medium supplementation with additional oleate (data not shown). This result indicates that both termini of LPCAT1 and LPCAT2 are directed toward the cytoplasm. The same result was obtained by the use of a fluorescence protease protection assay (56), which was performed in living cells (Fig. 3C). In this assay, the N- or C-terminal GFP fusion constructs of LPCATs were expressed in A431 cells, which were treated with digitonin in the presence or absence of proteinase K. The GFP signal on both protein termini was susceptible to protease treatment, indicating localization in the cytoplasm, although a fluorescent lumenal ER protein (DsRed2-ER) was unaffected. Again, this observation was independent of medium supplementation with additional oleate (data not shown). These imaging-based results were corroborated by biochemical analysis (Fig. 3D). A431 cells were transfected with N-terminal HA3-tagged LPCAT1 or LPCAT2, and microsomes were isolated and treated with proteinase K. The integrity of the microsomes was analyzed with the antibody against PDI. The accessibility of the N terminus of LPCAT1 and LPCAT2 for proteinase K was detected by anti-HA antibody and the accessibility of the C terminus by our specific LPCAT1 and LPCAT2 antibodies. Although microsomes remained mostly intact after the isolation and digestion procedure, the signal of both LPCAT1 and LPCAT2 termini vanished or was strongly reduced after proteinase K digestion (Fig. 3D). The same results were obtained using microsomes from oleate-treated cells (data not shown). We conclude that LPCAT1 and LPCAT2 display a monotopic conformation with both termini facing the cytoplasm, irrespective of supplementation with oleic acid.
LDs Can Synthesize PC via the Lands Cycle but Not by the Kennedy Pathway
Having established the presence of LPCATs on LDs, we studied whether LDs have the ability to synthesize PC. We obtained lysates of A431, COS7, and HuH7 cells, which were cultured in the presence of oleate to enhance LD formation. Lysates were subjected to floatation in a low salt sucrose gradient to obtain pure LD fractions, which were assayed for LPCAT activity. To evaluate the specificity of the LD-localized LPCAT activity, in particular to exclude contamination from ER activities, we compared LPCAT activity to LPAAT activity and glucose-6-phosphatase (Glc-6-Pase) activity, which are abundant in the ER (40, 57). Although cell lysates showed comparable LPCAT and LPAAT activities (Fig. 4A, lanes 1 and 2, 5 and 6, and 9 and 10), purified LDs displayed a much weaker activity for LPAAT but a strong activity for LPCAT (Fig. 4A, lanes 3 and 4, 7 and 8, and 11 and 12). To obtain quantitative data of enzyme activity on LDs, radioactive spots were scraped from the TLC plates, and lipids were extracted and quantified by scintillation counting. In addition, Glc-6-Pase in LDs and lysates was quantified by a colorimetric assay. From the values obtained, we calculated ratios of LPCAT/LPAAT activity and LPCAT/Glc-6-Pase activity in LDs and lysates. By normalization of the ratios on LDs to the corresponding ratios in the lysate, values for enrichment of LPCAT activity on the LDs relative to LPAAT and Glc-6-Pase activities were obtained (Fig. 4B). The enrichment values of 9–15-fold (relative to LPAAT activity) and 25–45-fold (relative to Glc-6-Pase activity) show that LD-localized LPCAT activity is specific and not due to ER contaminations.
FIGURE 4.
LDs contain LPCAT activity. A, lysates of HuH7, COS7, and A431 cells as indicated, cultured in the presence of 100 μm oleic acid, were subjected to floatation in a sucrose density gradient. Lysates (lysate) or purified LDs (LD) were incubated in the presence of [3H]acyl-CoA and lysophosphatidylcholine (LPC) or lysophosphatidic acid (LPA). Lipids were extracted and analyzed by TLC followed by autoradiography. Radioactive spots were identified by co-migrating standards; PC, phosphatidylcholine; PA, phosphatidic acid. B, table shows the enrichment of LPCAT activity on LDs relative to LPAAT or glucose-6-phosphatase activity, defined as the ratio of LPCAT/LPAAT activities or LPCAT/Glc-6-Pase activities in the LDs, divided by the same ratio in the lysates.
This raised the question whether LPCAT expression levels correlate with LPCAT activity on LDs. Because previous findings showed that overexpression of LPCAT1 or LPCAT2 increased LPCAT activity in cell homogenates (26–28), we performed LPCAT assays with purified LDs from COS7, A431, and HuH7 cells overexpressing LPCAT1 or LPCAT2 (Fig. 5). LDs from cells overexpressing LPCAT1 or LPCAT2 displayed an increase in LPCAT activity in all experiments (Fig. 5). This increase reached statistical significance for LPCAT1 overexpression in all three cell lines but for LPCAT2 overexpression only in A431 cells.
FIGURE 5.
Overexpression of LPCAT1 or LPCAT2 increases LD-associated LPCAT activity. A, C, and E, normalized LPCAT activity on LDs isolated from oleate-treated COS7, A431, and HuH7 cells, transfected with either empty vector (Control), LPCAT1-overexpression vector (LPCAT1), or LPCAT2-overexpression vector (LPCAT2). Data are mean ± S.D. from triplicate determinations of four independent experiments. Significance was obtained by unpaired t test analysis. B, D, and F, Western blot against LPCAT1, LPCAT2, and GAPDH (loading control) in the lysates.
If overexpression of LPCATs leads to an increase of LPCAT activity on LDs, one would expect a corresponding decrease upon knockdown of LPCAT1 and LPCAT2. The advantage of this approach is the lack of overexpression artifacts such as mislocalization of excess protein or down-regulation of its activity. Therefore, we performed double knockdown of LPCAT1 and LPCAT2 in A431 (Fig. 6, A and B) or single knockdown of LPCAT1 in HuH7 cells (Fig. 6, C and D), each with two different siRNAs. We observed a strong reduction of LD-associated LPCAT activity, which was statistically highly significant (Fig. 6, B and D). Reduction of LPCAT1 activity in HuH7 cells was associated with a morphological alteration in the LD pool; the mean LD size increased (Fig. 6E), although the total stored TAG remained constant (data not shown).
FIGURE 6.
Knockdown of LPCAT1 and LPCAT2 decreases LD-associated LPCAT activity. A and C, Western blots against LPCAT1, LPCAT2, and GAPDH (loading control) in cell lysates from oleate-treated A431 and HuH7 cells, transfected with either nontargeting siRNA (Control), or siRNA targeting LPCAT1 (L1-A/B/C) and LPCAT2 (L2-A/B) (A and B) or siRNA targeting LPCAT1 only (C and D). B and D, normalized LPCAT activity on LDs isolated from oleate-treated A431 and HuH7 cells, transfected as described in A and C. Data are mean ± S.D. from triplicate determinations of four independent experiments. Significance was obtained by unpaired t test analysis. E, mean area of LDs from HuH7 cells, treated with siRNA as indicated (no oleate supplementation), was quantified from fluorescence microscopy images. Each bar contains data from at least 12 images with an average of 10 cells per image (***, p ≤ 0.001).
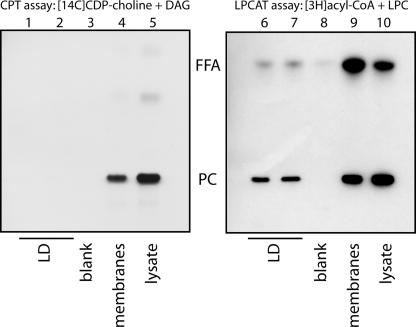
Apart from the LPCAT pathway, PC can also be synthesized from the CDP-choline and DAG, mediated by CPT/CEPT, in the Kennedy pathway. To reveal whether the Kennedy pathway is active on LDs, we subfractionated cells and assayed lysates, membranes, and isolated LDs for CPT/CEPT and LPCAT activity (Fig. 7). Although lysates and membrane fractions displayed both CPT/CEPT and LPCAT activity (Fig. 7, lanes 4 and 5 and 9 and 10), purified LDs showed only a strong LPCAT activity (Fig. 7, lanes 6 and 7) but little CPT/CEPT activity (Fig. 7, lanes 1 and 2). Quantification of two experiments revealed that 13.0 ± 1.2% of the LPCAT activity but only 0.07 ± 0.01% of the CPT/CEPT activity in the lysates was recovered in the LD fraction, resulting in an enrichment of LPCAT over CPT/CEPT activity of 179-fold.
FIGURE 7.
LDs can locally synthesize PC by the Lands cycle but not by the Kennedy pathway. Lysates of A431 cells, cultured in the presence of 100 μm oleic acid, were subjected to floatation in a sucrose density gradient. Samples of the LD fraction (LD), the floating membrane fraction + bottom fraction (membranes), and the total lysates (lysate) were incubated in the presence of either [14C]CDP-choline and DAG (left panel, CPT assay) or of [3H]acyl-CoA and LPC (right panel, LPCAT assay). Lipids were extracted and analyzed by TLC followed by autoradiography. Radioactive spots were identified by co-migrating standards. PC, phosphatidylcholine; FFA, free fatty acid.
DISCUSSION
The synthesis of PC and other membrane lipids generally takes place at the ER, where enzymes of both the Kennedy pathway and the Lands cycle reside (20, 35) In this study, we show that PC can also be synthesized at the surface of LDs via the Lands cycle. This activity is mediated by LPCAT1 and LPCAT2, which localize to both LDs and the ER. Identification of LPCAT1 and LPCAT2 on LDs is somewhat unexpected because both proteins have a C-terminal KKXX motif, which serves to retrieve protein that escaped the ER to the Golgi apparatus, and normally are found on ER-localized proteins. However, addition of such a KKXX motif to caveolin 1 led to the accumulation of the protein on LDs (48), suggesting that retrieval to the ER actually favors trafficking to LDs. Also, previous studies have shown localization of LPCAT1 and LPCAT2 to the ER (26, 33, 34, 58, 59). The apparent discrepancy to our results can be explained by the fact that these studies were performed under conditions that did not favor detection of LDs, which usually requires addition of fatty acids into the growth media.
Structurally, the dual localization to the ER and to LDs is supported by the monotopic (hairpin) topology, which enables insertion into both bilayer (ER) and monolayer (LD) membranes (Ref. 60 and references therein). In a recent study, Bridges et al. (34) concluded from a trypsin protection assay that LPCAT1 is a type II transmembrane protein with the C terminus inside the lumen of the ER. By three different experimental approaches (imaging-based accessibility assays in living and fixed cells and a protease protection assay in microsomes), we demonstrate a monotopic topology for both LPCAT1 and LPCAT2 in our cell system. In the protease protection assay, we experienced a reduced efficiency in protein degradation by trypsin compared with proteinase K (data not shown but were seen by the reviewers), which may explain the results seen by Bridges et al. (34).
In all three cell lines tested, purified LDs displayed LPCAT activity, which is in line with the presence of either only LPCAT1 (in HuH7 cells) or LPCAT1 and LPCAT2 (in A431 and COS7 cells) on the LDs. It appears that most of this LPCAT activity can be attributed to LPCAT1 and LPCAT2, because siRNA-mediated knockdown of these proteins leads to a strong reduction of the LPCAT activity on LDs. Our data do not exclude the possibility that other proteins with LPCAT activity may additionally contribute to the LD-associated LPCAT activity. However, the most obvious candidates, LPCAT3 and LPCAT4, are unlikely to reside on LDs in an active form. Both proteins are members of the membrane-bound O-acyltransferase family with multiple transmembrane domains (32), which precludes insertion into a lipid monolayer surrounding a hydrophobic lipid core.
Apart from synthesis by LPCATs, PC can also be produced via the Kennedy pathway. This pathway requires two enzymes as follows: CTP:phosphocholine cytidylyltransferase for the formation of CDP-choline and CPT/CEPT for the transfer of phosphocholine onto DAG. Although the presence of CTP:phosphocholine cytidylyltransferase on LDs of oleate-supplemented S2 cells was reported by Guo et al. (61), there is no publication that would report CPT/CEPT activity on LDs. In line with this, purified LDs in our cell system were devoid of CPT/CEPT activity, which would be required for PC synthesis on LDs by the Kennedy pathway.
LPCAT-mediated PC synthesis on LDs requires the presence of a long chain acyl-CoA and of LPC as substrates. The former is possibly produced locally by ACSL3, an abundant constituent of LDs (43, 62). The other substrate, LPC, is likely produced by a PLA2 activity either on the LD (9) or at other membrane compartments, in particular the ER, followed by transport to the LDs. Candidates for the PLA2 activity include iPLA2, the key activity in the Lands cycle of membrane lipid remodeling, or cPLA2α, an important player in LD biogenesis (63). Another possibility is the adipocyte-specific PLA2 (64), whose genetic ablation in mice results in a strong reduction of TAG storage (65). LPC is spontaneously released from membranes with a half-time of about 20 ms (66). Its desorption from membranes and transport in the cytosol are supported by binding to fatty acid-binding proteins (67). It should be noticed that the same principle, i.e. monomeric transport of a lysophospholipid followed by vectorial acylation at the target membrane, is also used for the synthesis of PA. Here, lysophosphatidic acid is produced in mitochondria, transported to the ER by fatty acid-binding protein (68), and converted to PA by an ER-resident LPAAT activity (57).
The existence of local LPCAT activity on LDs raises the question of its physiological role. PC is the major surface lipid of LDs (2, 3), and expansion of the LD surface during LD growth requires a source of PC. Local PC production would allow the growth of LDs independent of a physical connection to the ER or other routes of PC transport. This would be consistent with the observed increase of LD size upon LPCAT1 knockdown. Another possible function of LPCAT activity on LDs is to modify the existing PC at the sn-2 side chain. This would enable the cell to adjust the biophysical properties of the LD surface monolayer to different conditions, e.g. to prevent or promote LD fusion or support LD growth or degradation during lipolysis.
The results presented here contribute to the emerging picture of the complexity of cellular PC synthesis regarding the existence of isoenzymes and their compartmentalization. Future studies will have to address in more detail the influence of LPCAT1 and LPCAT2 on the cellular LD pool and on the balance of TAG and PC formation.
Supplementary Material
Acknowledgments
We thank the MPI-CBG light microscopy facility for support of image acquisition, Anna Shevchenko for protein mass spectrometry, and Xabier Contreras and Britta Bruegger (Heidelberg University) for analysis of TLC plates with the BioImager. We thank Ta-Yuan Chang (Dartmouth Medical School, Hanover, NH), for the ACAT1 antibody DM10 and Stefan Hoening (Institute of Biochemistry, Cologne University) for antiserum against TIP47.
This work was supported by Deutsche Forschungsgemeinschaft (Transregio TR83), the European Union Framework 7 Project “LipidomicNet,” and the German National Academic Foundation.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1 and Table 1.
- TAG
- triacylglyceride
- CEPT
- choline/ethanolamine phosphotransferase
- CPT
- cholinephosphotransferase
- CTP
- cytidine triphosphate
- DAG
- diacylglyceride
- Glc-6-Pase
- glucose-6-phosphatase
- LD
- lipid droplet
- LPC
- lysophosphatidylcholine
- LPAAT
- lysophosphatidic acid acyltransferase
- LPCAT
- lysophosphatidylcholine acyltransferase
- NSDHL
- NAD(P)dependent steroid dehydrogenase like
- PA
- phosphatidic acid
- PC
- phosphatidylcholine
- PLA2
- phospholipase A2
- PDI
- protein-disulfide isomerase
- ADRP
- adipose differentiation-related protein
- ER
- endoplasmic reticulum.
REFERENCES
- 1. Walther T. C., Farese R. V., Jr. (2009) Biochim. Biophys. Acta 1791, 459–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tauchi-Sato K., Ozeki S., Houjou T., Taguchi R., Fujimoto T. (2002) J. Biol. Chem. 277, 44507–44512 [DOI] [PubMed] [Google Scholar]
- 3. Bartz R., Li W. H., Venables B., Zehmer J. K., Roth M. R., Welti R., Anderson R. G., Liu P., Chapman K. D. (2007) J. Lipid Res. 48, 837–847 [DOI] [PubMed] [Google Scholar]
- 4. Brasaemle D. L., Dolios G., Shapiro L., Wang R. (2004) J. Biol. Chem. 279, 46835–46842 [DOI] [PubMed] [Google Scholar]
- 5. Fujimoto Y., Itabe H., Sakai J., Makita M., Noda J., Mori M., Higashi Y., Kojima S., Takano T. (2004) Biochim. Biophys. Acta 1644, 47–59 [DOI] [PubMed] [Google Scholar]
- 6. Liu P., Ying Y., Zhao Y., Mundy D. I., Zhu M., Anderson R. G. (2004) J. Biol. Chem. 279, 3787–3792 [DOI] [PubMed] [Google Scholar]
- 7. Umlauf E., Csaszar E., Moertelmaier M., Schuetz G. J., Parton R. G., Prohaska R. (2004) J. Biol. Chem. 279, 23699–23709 [DOI] [PubMed] [Google Scholar]
- 8. Ozeki S., Cheng J., Tauchi-Sato K., Hatano N., Taniguchi H., Fujimoto T. (2005) J. Cell Sci. 118, 2601–2611 [DOI] [PubMed] [Google Scholar]
- 9. Sato S., Fukasawa M., Yamakawa Y., Natsume T., Suzuki T., Shoji I., Aizaki H., Miyamura T., Nishijima M. (2006) J. Biochem. 139, 921–930 [DOI] [PubMed] [Google Scholar]
- 10. Turró S., Ingelmo-Torres M., Estanyol J. M., Tebar F., Fernández M. A., Albor C. V., Gaus K., Grewal T., Enrich C., Pol A. (2006) Traffic 7, 1254–1269 [DOI] [PubMed] [Google Scholar]
- 11. Bartz R., Zehmer J. K., Zhu M., Chen Y., Serrero G., Zhao Y., Liu P. (2007) J. Proteome Res. 6, 3256–3265 [DOI] [PubMed] [Google Scholar]
- 12. Cermelli S., Guo Y., Gross S. P., Welte M. A. (2006) Curr. Biol. 16, 1783–1795 [DOI] [PubMed] [Google Scholar]
- 13. Beller M., Riedel D., Jänsch L., Dieterich G., Wehland J., Jäckle H., Kühnlein R. P. (2006) Mol. Cell. Proteomics 5, 1082–1094 [DOI] [PubMed] [Google Scholar]
- 14. Brasaemle D. L. (2007) J. Lipid Res. 48, 2547–2559 [DOI] [PubMed] [Google Scholar]
- 15. Ohsaki Y., Cheng J., Suzuki M., Shinohara Y., Fujita A., Fujimoto T. (2009) Biochim. Biophys. Acta 1791, 399–407 [DOI] [PubMed] [Google Scholar]
- 16. Murphy S., Martin S., Parton R. G. (2009) Biochim. Biophys. Acta 1791, 441–447 [DOI] [PubMed] [Google Scholar]
- 17. Kuerschner L., Moessinger C., Thiele C. (2008) Traffic 9, 338–352 [DOI] [PubMed] [Google Scholar]
- 18. Stone S. J., Levin M. C., Zhou P., Han J., Walther T. C., Farese R. V., Jr. (2009) J. Biol. Chem. 284, 5352–5361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Carman G. M., Han G. S. (2009) J. Lipid Res. 50, S69–S73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Vance J. E., Vance D. E. (2004) Biochem. Cell Biol. 82, 113–128 [DOI] [PubMed] [Google Scholar]
- 21. Vance D. E., Ridgway N. D. (1988) Prog. Lipid Res. 27, 61–79 [DOI] [PubMed] [Google Scholar]
- 22. Kennedy E. P., Weiss S. B. (1956) J. Biol. Chem. 222, 193–214 [PubMed] [Google Scholar]
- 23. Henneberry A. L., McMaster C. R. (1999) Biochem. J. 339, 291–298 [PMC free article] [PubMed] [Google Scholar]
- 24. Lands W. E. (1958) J. Biol. Chem. 231, 883–888 [PubMed] [Google Scholar]
- 25. Butler P. L., Mallampalli R. K. (2010) J. Biol. Chem. 285, 6246–6258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nakanishi H., Shindou H., Hishikawa D., Harayama T., Ogasawara R., Suwabe A., Taguchi R., Shimizu T. (2006) J. Biol. Chem. 281, 20140–20147 [DOI] [PubMed] [Google Scholar]
- 27. Chen X., Hyatt B. A., Mucenski M. L., Mason R. J., Shannon J. M. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 11724–11729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shindou H., Hishikawa D., Nakanishi H., Harayama T., Ishii S., Taguchi R., Shimizu T. (2007) J. Biol. Chem. 282, 6532–6539 [DOI] [PubMed] [Google Scholar]
- 29. Soupene E., Fyrst H., Kuypers F. A. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 88–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhao Y., Chen Y. Q., Bonacci T. M., Bredt D. S., Li S., Bensch W. R., Moller D. E., Kowala M., Konrad R. J., Cao G. (2008) J. Biol. Chem. 283, 8258–8265 [DOI] [PubMed] [Google Scholar]
- 31. Lewin T. M., Wang P., Coleman R. A. (1999) Biochemistry 38, 5764–5771 [DOI] [PubMed] [Google Scholar]
- 32. Hofmann K. (2000) Trends Biochem. Sci. 25, 111–112 [DOI] [PubMed] [Google Scholar]
- 33. Harayama T., Shindou H., Ogasawara R., Suwabe A., Shimizu T. (2008) J. Biol. Chem. 283, 11097–11106 [DOI] [PubMed] [Google Scholar]
- 34. Bridges J. P., Ikegami M., Brilli L. L., Chen X., Mason R. J., Shannon J. M. (2010) J. Clin. Invest. 120, 1736–1748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shindou H., Shimizu T. (2009) J. Biol. Chem. 284, 1–5 [DOI] [PubMed] [Google Scholar]
- 36. Hishikawa D., Shindou H., Kobayashi S., Nakanishi H., Taguchi R., Shimizu T. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 2830–2835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chang C. C., Chen J., Thomas M. A., Cheng D., Del Priore V. A., Newton R. S., Pape M. E., Chang T. Y. (1995) J. Biol. Chem. 270, 29532–29540 [DOI] [PubMed] [Google Scholar]
- 38. Taylor D. C., Weber N., Hogge L. R., Underhill E. W. (1990) Anal. Biochem. 184, 311–316 [DOI] [PubMed] [Google Scholar]
- 39. Thiele C., Hannah M. J., Fahrenholz F., Huttner W. B. (2000) Nat. Cell Biol. 2, 42–49 [DOI] [PubMed] [Google Scholar]
- 40. Arden S. D., Zahn T., Steegers S., Webb S., Bergman B., O'Brien R. M., Hutton J. C. (1999) Diabetes 48, 531–542 [DOI] [PubMed] [Google Scholar]
- 41. Wessel D., Flügge U. I. (1984) Anal. Biochem. 138, 141–143 [DOI] [PubMed] [Google Scholar]
- 42. Shevchenko A., Tomas H., Havlis J., Olsen J. V., Mann M. (2006) Nat. Protoc. 1, 2856–2860 [DOI] [PubMed] [Google Scholar]
- 43. Fujimoto Y., Itabe H., Kinoshita T., Homma K. J., Onoduka J., Mori M., Yamaguchi S., Makita M., Higashi Y., Yamashita A., Takano T. (2007) J. Lipid Res. 48, 1280–1292 [DOI] [PubMed] [Google Scholar]
- 44. Ohashi M., Mizushima N., Kabeya Y., Yoshimori T. (2003) J. Biol. Chem. 278, 36819–36829 [DOI] [PubMed] [Google Scholar]
- 45. Shevchenko A., Roguev A., Schaft D., Buchanan L., Habermann B., Sakalar C., Thomas H., Krogan N. J., Shevchenko A., Stewart A. F. (2008) Genome Biol. 9, R167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Martin S., Driessen K., Nixon S. J., Zerial M., Parton R. G. (2005) J. Biol. Chem. 280, 42325–42335 [DOI] [PubMed] [Google Scholar]
- 47. Yamaguchi T., Omatsu N., Matsushita S., Osumi T. (2004) J. Biol. Chem. 279, 30490–30497 [DOI] [PubMed] [Google Scholar]
- 48. Ostermeyer A. G., Paci J. M., Zeng Y., Lublin D. M., Munro S., Brown D. A. (2001) J. Cell Biol. 152, 1071–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Pol A., Luetterforst R., Lindsay M., Heino S., Ikonen E., Parton R. G. (2001) J. Cell Biol. 152, 1057–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Fujimoto T., Kogo H., Ishiguro K., Tauchi K., Nomura R. (2001) J. Cell Biol. 152, 1079–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Caldas H., Herman G. E. (2003) Hum. Mol. Genet. 12, 2981–2991 [DOI] [PubMed] [Google Scholar]
- 52. Stone S. J., Levin M. C., Farese R. V., Jr. (2006) J. Biol. Chem. 281, 40273–40282 [DOI] [PubMed] [Google Scholar]
- 53. Dupree P., Parton R. G., Raposo G., Kurzchalia T. V., Simons K. (1993) EMBO J. 12, 1597–1605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ostermeyer A. G., Ramcharan L. T., Zeng Y., Lublin D. M., Brown D. A. (2004) J. Cell Biol. 164, 69–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Plutner H., Davidson H. W., Saraste J., Balch W. E. (1992) J. Cell Biol. 119, 1097–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lorenz H., Hailey D. W., Lippincott-Schwartz J. (2006) Nat. Methods 3, 205–210 [DOI] [PubMed] [Google Scholar]
- 57. Aguado B., Campbell R. D. (1998) J. Biol. Chem. 273, 4096–4105 [DOI] [PubMed] [Google Scholar]
- 58. Mansilla F., da Costa K. A., Wang S., Kruhøffer M., Lewin T. M., Orntoft T. F., Coleman R. A., Birkenkamp-Demtröder K. (2009) J. Mol. Med. 87, 85–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Morimoto R., Shindou H., Oda Y., Shimizu T. (2010) J. Biol. Chem. 285, 29857–29862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Thiele C., Spandl J. (2008) Curr. Opin. Cell Biol. 20, 378–385 [DOI] [PubMed] [Google Scholar]
- 61. Guo Y., Walther T. C., Rao M., Stuurman N., Goshima G., Terayama K., Wong J. S., Vale R. D., Walter P., Farese R. V. (2008) Nature 453, 657–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Yao H., Ye J. (2008) J. Biol. Chem. 283, 849–854 [DOI] [PubMed] [Google Scholar]
- 63. Gubern A., Casas J., Barceló-Torns M., Barneda D., de la Rosa X., Masgrau R., Picatoste F., Balsinde J., Balboa M. A., Claro E. (2008) J. Biol. Chem. 283, 27369–27382 [DOI] [PubMed] [Google Scholar]
- 64. Duncan R. E., Sarkadi-Nagy E., Jaworski K., Ahmadian M., Sul H. S. (2008) J. Biol. Chem. 283, 25428–25436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Jaworski K., Ahmadian M., Duncan R. E., Sarkadi-Nagy E., Varady K. A., Hellerstein M. K., Lee H. Y., Samuel V. T., Shulman G. I., Kim K. H., de Val S., Kang C., Sul H. S. (2009) Nat. Med. 15, 159–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Massey J. B., Bick D. H., Pownall H. J. (1997) Biophys. J. 72, 1732–1743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Thumser A. E., Voysey J. E., Wilton D. C. (1994) Biochem. J. 301, 801–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Vancura A., Haldar D. (1992) J. Biol. Chem. 267, 14353–14359 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.