Abstract
Interleukin-6 (IL-6) is a pleiotropic cytokine that plays a central role in host defense. IL-6 expression can be regulated at both a transcriptional and a post-transcriptional level. We used a combination of bioinformatics and experimental techniques to demonstrate that the miR-365 is a direct negative regulator of IL-6. Overexpression of miR-365 mimics decreased activity of a luciferase reporter containing the IL-6 3′-UTR and led to repression of IL-6 protein. In contrast, ectopic expression of a miR-365 inhibitor elevated IL-6 expression. The negative regulation of miR-365 was strictly dependent on a microRNA binding element in the 3′-UTR of IL-6 mRNA. Deletion mutant analysis of the miR-365 promoter showed that two transcription factors, Sp1 and NF-κB, are essential for the transcriptional regulation of miR-365. We also demonstrate that the MAPK/ERK pathway contributes to the regulation of miR-365. Furthermore, miR-365 exhibited a greater negative regulatory effect on IL-6 than hsa-let-7a, a previously identified microRNA negatively regulating IL-6. Taken together, our results show that miR-365 is a novel negative regulator of IL-6.
Keywords: Interleukin, MicroRNA, NF-κB, Sp1, Transcription Factors
Introduction
MicroRNAs (miRNAs, miRs)3 are small RNA molecules that regulate gene expression at the post-transcriptional level (1). In mammals, miRNAs are initially transcribed by RNA polymerase II, and the primary miRNA transcripts are sequentially cut by RNase III enzymes Drosha and Dicer (2, 3). The resulting ∼23-nucleotide double-stranded mature miRNA molecules load into RNA-induced silencing complexes (4, 5), where they act to repress mRNA translation or reduce mRNA stability by interacting with miRNA recognition elements (MRE) within 3′-untranslated region (UTR) of target genes (6). The specificity of miRNAs is thought to be primarily mediated by residues 2–8 at the 5′ end of miRNA, also known as the seed region (7).
Growing evidence exists indicating that miRNAs play an important role in the regulation of inflammation. For example, miR-146a has been characterized as a negative regulator of the inflammatory response by targeting IL-1 receptor-associated kinase 1 (IRAK1) and TNF receptor associated factor 6 (8). Xiao et al. (9) reported that miR-155 is involved in negative regulation of Heliobacter pylori-induced inflammation, and miR-155 was further identified as targeting myeloid differentiation primary response gene 88 (MyD88) (10). miRNAs regulate inflammation not only by targeting adapter proteins involved in the signaling pathway but also inflammatory mediators. Many interleukins have been found to be targets of miRNAs. Interleukin-10 (IL-10) has been identified as the target of hsa-miR-106a (11). Kaposi sarcoma-associated herpesvirus-encoded miR-K12-3 and miR-K12-7 up-regulate IL-10 and IL-6 levels in murine macrophages and human myelomonocytic cells (12). miR-466l elevates IL-10 expression by preventing IL-10 mRNA degradation (13). IL-13, a cytokine essential for expression for allergic lung disease, was recently found to be regulated by mmu-let-7a (14). miR-346 indirectly regulates IL-18 release by indirectly inhibiting LPS-induced Bruton's tyrosine kinase expression in LPS-activated rheumatoid fibroblast-like synoviocytes (15).
IL-6 is a pleiotropic cytokine involved in the regulation of the immune response, hematopoiesis, and inflammation (16). It was previously considered to be a regulator of acute-phase responses and a lymphocyte-stimulatory factor (17–19). However, recent advances have documented a series of IL-6 activities that are critical for resolving innate immunity and promoting acquired immunity (20). IL-6 expression can be regulated at the transcriptional and post-transcriptional level (21, 22), but whether and how IL-6 is regulated at the post-transcriptional level by miRNA has so far been incompletely understood.
In this study we establish that miR-365 is a novel miRNA regulator of human IL-6. miR-365 regulates the expression of IL-6 by classic interaction with the MRE in the 3′-UTR of IL-6 and further represses mRNA translation, although mRNA degradation is not involved. We also investigated the transcriptional regulation mechanism of miR-365.
EXPERIMENTAL PROCEDURES
Prediction of Candidate miRNA Binding to IL-6 3′-UTR
The software programs Microcosm (23–25), miRanda (26, 27), and TargetScan (28–31) were used to predict potential binding sites for miRNA in the IL-6 3′-UTR. Only those miRNA target pairs detected by at least two of the three programs were used for further study.
Cell Culture and Reagents
HEK293 and HeLa cells were cultured in DMEM (Invitrogen) supplemented with 10% fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin at 37 °C and 5% CO2. miR-365 mimics and inhibitors were obtained from GenePharma (Shanghai, China). The sequence of mimics, inhibitors, or scrambled oligonucleotides are as follows: miR-365 mimics, 5′-UAA UGC CCC UAA AAA UCC UUA U-3′ (forward) and 5′-AAG GAU UUU UAG GGG CAU UAU U-3′ (reverse); mimics negative control, 5′-UUC UCC GAA CGU GUC ACG UTT-3′ (forward) and 5′-ACG UGA CAC GUU CGG AGA ATT-3′ (reverse); miR-365 inhibitor, 5′-AUA AGG AUU UUU AGG GGC AUU A-3′; inhibitor negative control, 5′-CAG UAC UUU UGU GUA GUA CAA-3′. Pharmacological inhibitors of JNK (SP600125), p38 MAP kinase (SB203580 and SB202190), NF-κB (BAY11-7082), and ERK (U0126) were obtained from Calbiochem-Merck. Phorbol 12-myristate 13-acetate, poly(I:C), and poly(dAT:dAT) were obtained from Sigma.
Plasmid Constructs
To construct pMIR-IL-6 3′-UTR, the 3′-UTR of IL-6 was amplified from cDNA derived from HeLa cells. The PCR product was digested with SacI and HindIII and cloned into the pMIR-REPORT luciferase reporter vector (Ambion). The luciferase reporter plasmids TSS-1348, TSS-1097, TSS-705, and TSS-336, which contains the1348-, 1097-, 705-, and 336-bp proximal promoter sequences of miR-365, respectively, were constructed by PCR amplification using genomic DNA of HeLa cells as templates and subsequent cloning into KpnI and BglII site of pGL3-basic (Promega, Madison, WI). To construct the miRNA expression vector, each miRNA precursor sequence plus 3′- and 5′-flanking region (about 150 bp) was amplified from genomic DNA of HeLa cells and cloned into pSilencer 4.1-CMV neo. The Sp1 expression construct was created by amplifying the whole human Sp1 coding sequence from HEK293 cells and then cloned into pEGFP-N1 at the XhoI and KpnI sites. The IL-6 3′-UTR reporter constructs lacking the predicted miR-365 binding site or having a 3-bp substitution in the MRE of IL-6 gene and miR-365 promoter reporter constructs containing site-specific mutations for transcription factor binding site NF-κB or Sp1 were constructed by introducing a point mutation or deletion with overlap-extension PCR (32). Primers used in this study are listed in Table 1. All DNA constructs were verified by sequencing.
TABLE 1.
Primers used in this study
F, forward; R, reverse.
| Primer | Sequence 5′ → 3′ |
|---|---|
| IL6 3′-UTR intact F | ATAGAGCTCCATGGGCACCTCAGATTG |
| IL6 3′-UTR deletion F | CGCGAGCTCGCATGGGCACCTCAGCTTCTTCTGGTCAGAAACCTGT |
| IL6 3′-UTR Point Mut F | CGCGAGCTCGCATGGGCACCTCAGATTGTTGTTGTTAATGACCAGTCCT |
| IL6 3′-UTR R | GCGAAGCTTGCTGAATTTTTTAAAATGCC |
| TSS-336 F | GGGGTACCCCGCTTGATAAAGCTTAATTGCATC |
| TSS-705 F | GGGGTACCCCAGGTCTAATTTTTATTATGCAAG |
| TSS-1097 F | GGGGTACCCCTTAAAATCACAGTGGAAACTGG |
| TSS-1348 F | GGGGTACCCCCTGCAGTCAGCGCAAGACCAACTG |
| TSS-Universal R | GAAGATCTTCAAAGAAAGAATGAATGTTAGCC |
| NF-κB mut1 F | TTTAAATTCCTAATTTAACCATTTCCCTTTGAGTCATTAGGAA |
| NF-κB mut1 R | TTCCTAATGACTCAAAGGGAAATGGTTAAATTAGGAATTTAAA |
| NF-κB mut2 F | GAGTCATTAACCATATCTCATAGGTCTAATTTTTATTATG |
| NF-κB mut2 R | CATAATAAAAATTAGACCTATGAGATATGGTTAATGACTC |
| NF-κB mut3 F | CAAACAATTACCAAATACCTTTCCAAGGAAAAATCAGGCACC |
| NF-κB mut3 R | GGTGCCTGATTTTTCCTTGGAAAGGTATTTGGTAATTGTTTG |
| Sp1 mut1 F | GCAAGAAAAATATTTTTCAGAGTCAACAGGAGTATTCCCC |
| Sp1 mut1 R | GGGGAATACTCCTGTTGACTCTGAAAAATATTTTTCTTGC |
| Sp1 mut2 F | CGAGAGAACAACTAGTCCACCATTCGATCATGCCACGTAAG |
| Sp1 mut2 R | CTTACGTGGCATGATCGAATGGTGGACTAGTTGTTCTCTCG |
| Sp1 mut3 F | CATCCCTAACGTGACAGAACATGACTTAGCTCTCCAAGAAG |
| Sp1 mut3 R | CTTCTTGGAGAGCTAAGTCATGTTCTGTCACGTTAGGGATG |
| TSS-1097 mut F | GGGGTACCCCTTAAAATCACAGTGGAAACTGGTTTGAATG |
| TSS-1097 mut R | GAAGATCTTCAAAGAAAGAATGAATGTTAGCCTAATTACAG |
| hsa-miR-338–3p F | ATAGGATCCGAGACAGACCCTGCTTCG |
| hsa-miR-338–3p R | GCGAAGCTTAAAAACCCCACATAAAACCCAT |
| hsa-miR-365 F | ATAGGATCCTGAGGTCCCTTTCGTG |
| hsa-miR-365 R | GCGAAGCTTAAAAACAGCGGAAGAGTTTGG |
| hsa-miR-548c F | ATAGGATCCGTCCTAACTTATTTTG |
| hsa-miR-548c R | CGCAAGCTTAAAAAGTAACTCTTCACATCT |
| hsa-miR-548d F | ATAGGATCCTTCATAGGCTCGAAAA |
| hsa-miR-548d R | GCGAAGCTTAAAAATACTCTGCCCCTGATG |
| hsa-miR-548o F | ATAGGATCCACCGACTACCACTTCT |
| hsa-miR-548o R | CGCAAGCTTAAAAACTGGGATTTGCTCTTG |
| Human Sp1 CDS R | GCCGGTACCGTGAAGCCATTGCCACTGATATTAATGG |
| Human Sp1 CDS F | GCCCTCGAGCCACCATGAGCGACCAAGATC |
Transient Transfections and Luciferase Assays
Transient transfections were performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. For luciferase assay, HEK293 cells were cultured in 24-well plates and co-transfected with 0.1 μg of an indicated reporter plasmid per well and 0.05 μg pRL-TK (Promega) per well along with miRNA expression vector (0.4 μg/well) or miRNA mimics (20, 40, and 60 nm) or inhibitors (60, 80, and 100 nm). In some selected experiments, 1 μg/well poly(dAT:dAT), 1 μg/well poly(I:C), 1 μg/well phorbol 12-myristate 13-acetate, or 10 hemagglutination units/well Sendai virus were transfected or treated at 24 h after the initial co-transfection. Luciferase activity was analyzed 24 h later using the dual luciferase reporter assay (Promega). Renilla luciferase activity in the lysates was used to normalize the firefly luciferase activity.
Small RNA Interference (siRNA)
siRNA duplexes consisting of 21 bp of oligonucleotides were purchased from GenePharma (Shanghai, China). The sequences of siRNA duplexes for Sp1 and negative control (NC) were as follows: Sp1 siRNA, 5′-AUC ACU CCA UGG AUG AAA UGA TT-3′(sense)/5′-UCA UUU CAU CCA UGG AGU GAU TT-3′(antisense); NC, 5′-UUC UCC GAA CGU GUC ACG UTT-3′ (sense)/5′-ACG UGA CAC GUU CGG AGA ATT-3′ (antisense). Transfection of siRNA duplexes was performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions.
Real-time PCR
HeLa cells were transfected with either miR-365 mimics or inhibitors using Lipofectamine 2000. Total RNA was extracted from the transfected cells with TRIzol (Invitrogen), and 0.4 μg of RNA was used to synthesize cDNA using a first-strand cDNA synthesis kit (TOYOBO). Quantitative real-time PCR analysis was performed using the Lightcycler 480 (Roche Applied Science). The primers used for quantitative real-time PCR are as follows: IL-6, 5′-AGG AGA CTT GCC TGG TGA AA-3′ (forward)/5′-CAG GGG TGG TTA TTG CAT CT-3′ (reverse); GAPDH, 5′-GCA CCG TCA AGG CTG AGA AC-3′ (forward)/5′-TGG TGA AGA CGC CAG TGG A-3′ (reverse). Data were normalized according to the level of GAPDH expression in each sample. For miRNA real-time PCR, a commercial Hirpin-itTM miRNAs qPCR Quantification kit (GenePharma) was used. Briefly, 2 μg of RNA was used as the template and then reverse-transcribed by using a miR-365-specific RT-primer. The resulting cDNA was further amplified in the quantitative real-time PCR with a universal reverse primer and a specific forward primer. The PCR procedure included predenaturation at 95 °C for 2 min and then 40 cycles of 94 °C for 10 s, 58 °C for 15 s, and 72 °C for 20 s followed by melting curve analysis.
IL-6 ELISA
Human IL-6 expression was measured in the supernatants of stimulated cells using a commercial human IL-6 ELISA kit (Invitrogen) according to manufacturer's instructions.
Statistical Analysis
Results obtained from three independent experiments were expressed as the mean ± S.D. For all statistical analyses, two-tailed Student's t test was used. A p value less than 0.05 was considered significant, and a p value less than 0.01 was considered highly significant.
RESULTS
Identification of miR-365 as a Negative Regulator of IL-6
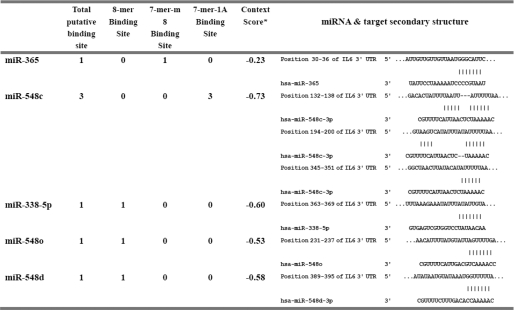
Potential miRNA binding sites to the IL-6 3′-UTR were predicted by Microcosm, miRanda, and TargetScan using the 3′-UTR of human IL-6 (NM_000600.3) as a query sequence. Five miRNAs, including miR-365, miR-548o, miR-548d, miR-548c, and miR-338–5p, were returned as candidates. The predicted binding sites for the individual miRNA in the 3′-UTR of IL-6 are listed in Table 2.
TABLE 2.
A list of predicted miRNAs possessing potential binding sites in the 3′-UTR of IL-6
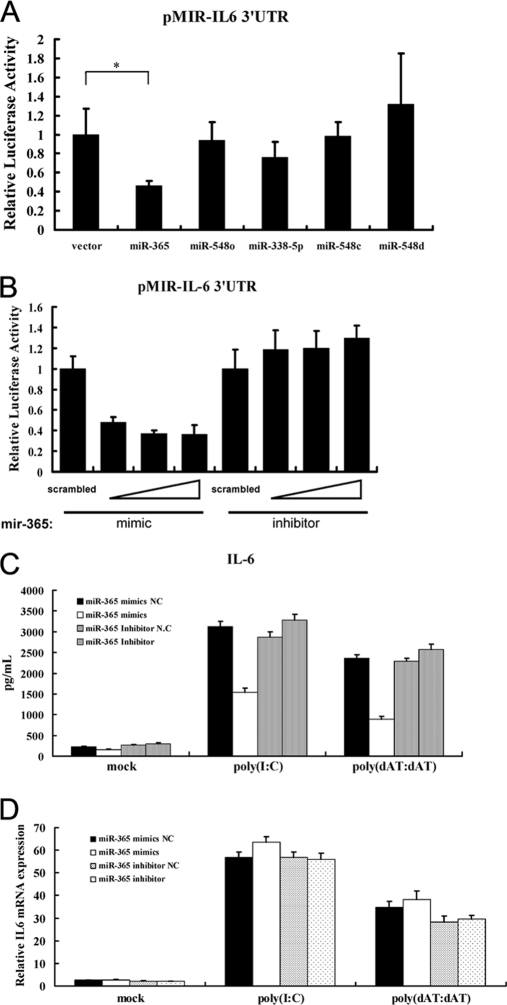
To investigate the roles of these predicted miRNAs on IL-6 expression, DNA constructs expressing the miRNA precursors were co-transfected with pMIR-IL-6 3′-UTR. As shown in Fig. 1A, only transfection with miR-365 expression vector significantly down-regulated luciferase activity compared with transfection with an empty vector, indicating that miR-365 is a potential regulator of IL-6.
FIGURE 1.
miR-365 is a negative regulator of IL-6. A, HEK293 cells were co-transfected with the indicated miRNA expression construct (0.4 μg), pMIR-IL6 3′-UTR reporter plasmid (0.2 μg), and pRL-TK (0.05 μg). At 24 h post-transfection, cells were collected, and luciferase activity was measured. Luciferase activity of the empty vector was regarded as 1. *, p < 0.01, as compared with the empty vector group. B, mimics (20, 40, and 60 nm) or inhibitors (60, 80, and 100 nm) of miR-365 were co-transfected with pMIR-IL-6 3′-UTR reporter plasmid into HEK293 cells. A scrambled mimic (60 nm) or inhibitor (100 nm) was used as control. Luciferase activity was measured at 24 h post-transfection. The luciferase activity of the scrambled mimic or inhibitor was regarded as 1. C and D, HeLa cells were transfected with the indicated miRNA oligonucleotides. Poly(I:C) or poly(dAT:dAT) were transfected at 24 h after the initial transfection. Culture supernatants were collected 24 h later, and IL-6 was measured using a human IL-6 ELISA kit (C). Total RNA was extracted from cells, and real-time PCR assay was performed with specific primers listed in “Experimental Procedures” (D).
To further evaluate the effect of mature miR-365 on IL-6 expression, chemosynthetic miR-365 mimic and inhibitor were employed. As shown in Fig. 1B, the miR-365 mimic also inhibited luciferase activity of pMIR-IL-6 3′-UTR in HEK293 cells. Conversely, the miR-365 inhibitor induced luciferase activity. In addition, the dose-dependent effects of the mimic and the inhibitor on inhibition and induction of luciferase activity, respectively, could be observed (Fig. 1B). To further validate the miR-365 inhibitory effect on IL-6 protein expression, HeLa cells were transfected with the miR-365 mimic (40 nm) or inhibitor (100 nm), and IL-6 levels in the culture supernatant were measured by ELISA. As shown in Fig. 1C, treatment with the miR-365 mimic remarkably repressed IL-6 expression, whereas the miR-365 inhibitor induced IL-6 expression. As a critical proinflammatory cytokine, IL-6 expression is highly inducible by some immunostimulatory molecules such as poly(I:C) (dsRNA analog) and poly(dAT:dAT) (dsDNA analog). Thus, we further evaluated the inhibitory effect of miR-365 on poly(I:C)- or poly(dAT:dAT)-induced IL-6 expression. As shown in Fig. 1C, transfection with poly(I:C) or poly(dAT:dAT) significantly induced IL-6 expression, but overexpression of miR-365 remarkably blocked poly(I:C)- or poly(dAT:dAT)-induced IL-6 expression.
We further investigated whether miR-365 acts to affect the stability of IL-6 mRNA. To this end, real-time PCR was performed by using total RNA prepared from cells transfected with the mimic or inhibitor oligonucleotides. As shown in Fig. 1D, neither miR-365 mimic nor inhibitor affected the expression of IL-6 mRNA. Thus, it can be concluded that miR-365 decreases IL-6 expression by repressing its mRNA translation but does not affect its mRNA stability.
Confirmation of the Response Element for miR-365 in the IL-6 3′-UTR
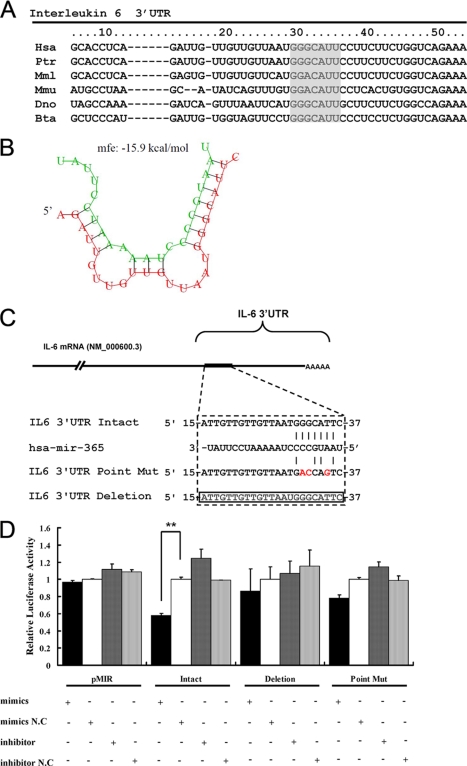
Post-transcriptional regulation of target genes by miRNA is mediated by binding of miRNA to the 3′-UTR of target gene, whereas the specificity of the individual miRNA is determined by the seed region, which located in nucleotides 2–8 of 5′ portion of the miRNA (7). Computational analysis suggested that there is a putative binding site for miR-365 in the 3′-UTR of the human IL-6 gene and the binding site for the seed region of miR-365 is located at 30–36 bp of IL-6 3′-UTR (Fig. 2A). The minimum free energy of hybridization between the target RNA and miR-365, as predicted by RNAhybrid (33), also supported the possibility that miR-365 can bind at that site (Fig. 2B). Furthermore, alignment of multiple IL-6 3′-UTRs showed that the binding site for the seed region of miR-365 is highly conserved among different species, including human, mouse, rhesus, chimpanzee, armadillo, and cow (Fig. 2A).
FIGURE 2.
Identification of the binding site for miR-365 in the IL-6 3′-UTR. A, alignment of the IL-6 3′-UTR showed that the MRE (gray) was conserved among different species. B, shown is a schematic representation of the hybridization between miR-365 and IL-6 3′-UTR. Green and red letters indicate miR-365 and IL-6 mRNA, respectively. C, shown is a schematic representation of mutant reporters of IL-6 3′-UTR. The frame and red letters indicate the deletion and the point mutation, respectively. D, intact, point mutant, or deletion mutants pMIR-IL-6 3′-UTR and pMIR-REPORT were co-transfected with the indicated oligonucleotides into HEK293 cells. 24 h post-transfection, luciferase activities were measured. Results are presented as -fold induction over control groups. Bar graph data are presented as the means ± S.D. (n = 3); **, p < 0.01, as compared with the transfection with the scrambled oligonucleotides.
To validate these bioinformatics predictions with experimental evidence, 3′-UTR reporter constructs lacking the predicted miR-365 binding site or having a 3-bp substitution in the MRE of IL-6 gene were generated and termed as pMIR-IL-6 3′-UTR deletion and point mutation, respectively (Fig. 2C). When the miR-365 mimic or inhibitor were co-transfected with these constructs, the miR-365 mimic was observed to inhibit by 45% the luciferase activity of the reporter with an intact 3′-UTR, as compared with luciferase activity after treatment with scrambled mimics. However, overexpression of miR-365 only inhibited 10 and 20% luciferase activity of the reporter pMIR-IL-6 3′-UTR deletion and point mutation, respectively (Fig. 2D). These results provided experimental support to the prediction that the miR-365 response element is located in the 3′-UTR of IL-6.
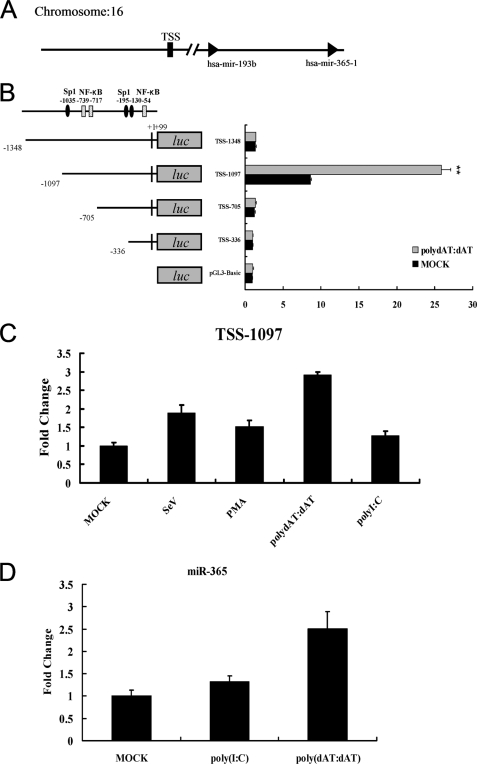
Analysis of miR-365 Promoter Activity
To investigate the transcriptional regulation of miR-365, the genomic information of miR-365, including the putative transcription start site and genomic location, were obtained from miRBase (23–25). A schematic representation of the miR-365 promoter is shown in Fig. 3A. For the deletion mutation assay, luciferase reporters with different lengths of the miR-365 promoter were transfected into HEK293 cells to determine the basal promoter activity. As shown in Fig. 3B, the highest luciferase activity was produced by reporter TSS-1097, with a 1.2-kb putative promoter region, which led to a 10-fold induction in luciferase activity with respect to pGL3-Basic, suggesting that TSS-1097 possesses fully intact promoter activity. Transfection with TSS-1348, TSS-705, or TSS-336 reporter plasmids resulted in a less basal promoter activity. These results indicated that the region from −1097 to −705 is required for intact promoter activity and the region from −1348 to −1097 may contain negative regulatory element(s). We also investigated the responses of the miR-365 promoter to different inflammatory stimuli. As shown in Fig. 3C, luciferase activity of reporter TSS-1097 could be induced by poly(dAT:dAT), poly(I:C), phorbol 12-myristate 13-acetate, and Sendai virus. The highest induction was up to 3-fold after stimulation with poly(dAT:dAT), whereas induction by poly(dAT:dAT) was not observed in transfections with TSS-1348, TSS-705, or TSS-336 (Fig. 3B). In addition, we examined whether endogenous miR-365 is inducible by stimulation with poly(dAT:dAT) or poly(I:C). As shown in Fig. 3D, the expression of miR-365 was significantly high in cells stimulated with poly(dAT:dAT) than with poly(I:C). The results were in agreement with that of luciferase activity of reporter TSS-1097 after stimulation with poly(dAT:dAT) and poly(I:C). Together, these results demonstrate that the region from −1097 to −705 is required for basal miR-365 promoter activity and that miR-365 promoter activity can be significantly induced by poly(dAT:dAT).
FIGURE 3.
Transcriptional regulation of miR-365. A, shown is a schematic representation of the region encoding hsa-miR-365. TSS is represented as a black box. B, HEK293 cells were cultured in 24-well plates and transfected with miR-365 promoter reporter mutants containing various lengths of the miR-365 promoter region. At 24 h post-transfection, cells were transfected or mock-transfected with poly(dAT:dAT) (1 μg/well). 24 h later luciferase activity was measured, and luciferase activity of the mock-transfected empty vector pGL3 Basic group was regarded as 1. **, p < 0.01. C, HEK293 cells were transfected with TSS-1097-Luc reporter. At 24 h post-transfection, cells were infected with Sendai virus (10 hemagglutinin activity units/well) or incubated with phorbol 12-myristate 13-acetate (PMA; 1 μg/well) or transfected with poly(I:C) (1 μg/well) or poly(dAT:dAT) (1 μg/well), respectively. Luciferase activities were measured 24 h later. D, HEK293 cells were transfected with poly(I:C) or poly(dAT:dAT). Total RNA were extracted at 24 h post-transfection. The expression of endogenous miR-365 was detected by miRNA real-time PCR. Results are presented as -fold induction over control groups.
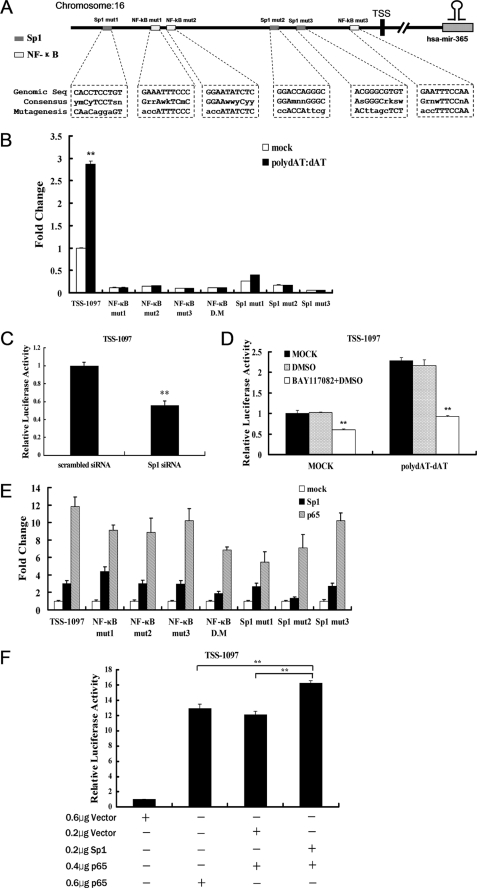
NF-κB and Sp1 Response Elements Are Essential for the Transcriptional Activity of the miR-365 Promoter
To investigate the transcription factors involved in the transcription of miR-365, a 1.1-kb region upstream of the putative transcription start site (TSS) of miR-365 was analyzed with prediction software Alibaba2.1. Sp1 and NF-κB binding sites were predicted to be separately located in the promoter region. We used site-directed mutagenesis to abolish each site (Fig. 4A). Mutations of either the NF-κB or Sp1 site significantly inhibited the transcription activity of TSS-1097 induced by poly(dAT:dAT) (Fig. 4B). We further investigated the involvement of Sp1 in the transcriptional regulation of miR-365 by using gain- and loss-of-function assays. Promoter activity of miR-365 was inhibited by 45% in the presence of 80 nm Sp1 siRNA (Fig. 4C), whereas forced expression of Sp1 induced promoter activity by 3-fold (Fig. 4E). Mutation of any NF-κB or Sp1 binding sites in the miR-365 promoter resulted in similar responses to the induction caused by Sp1. In contrast, the double mutation (mutations of NF-κB sites number 1 and 2 located in the region from −1097 to −705 bp) and Sp1-mut2 inhibited the transcriptional increase induced by ectopic expression of Sp1 by 50 and 60%, respectively (Fig. 4E).
FIGURE 4.
Both Sp1 and NF-κB are required for the transcription of miR-365. A, shown is a schematic representation of the binding sites for Sp1 and NF-κB. The mutated sequences are indicated in lowercase. B, HEK293 cells were transfected with the mutated miR-365 promoter reporters. At 24 h post-transfection, cells were stimulated with poly(dAT:dAT) as described in Fig. 3B. Luciferase activity was measured at 24 h after stimulation. Results are represented as -fold induction over non-stimulated control and are expressed as the mean ± S.D. for triplicate determinations where each experiment is representative of three separate experiments. **, p < 0.01, as compared with the control group. C, TSS-1097-Luc reporter was co-transfected into HEK293 cells with Sp1 specific siRNA or scrambled siRNA. Luciferase activities were measured 24 h post-transfection. D, TSS-1097-Luc reporter was transfected into HEK293 cells. At 24 h post-transfection, cells were treated with BAY11-7082 (15 μm) for 1 h, and then poly(dAT:dAT) (2 μg/ml) was transfected into HEK293 cells in some wells. E, the Sp1 or p65 expression constructs were co-transfected with TSS-1097-Luc reporter or individual mutant into HEK293 cells, respectively. Luciferase activities were measured at 24 h post-transfection. F, TSS-1097-Luc reporter (0.1 μg) was co-transfected with a total of 0.6 μg of a DNA mixture composed of pEGFP-N1-Sp1 and/or pEGFP-C1-p65 and/or empty vector at the indicated dose. Luciferase activities were measured 24 h post-co-transfection. In all cases results were represented as -fold induction over control. Data were represented as the mean ± S.D. (n = 3). **, p < 0.01.
The NF-κB signaling pathway has previously been implicated in the regulation of many miRNAs (8). We used gain- and loss-of-function assays to show that forced expression of NF-κB subunit p65 induced a 12-fold up-regulation of the miR-365 promoter activity (Fig. 4E). However, interfering with NF-κB activity by pretreatment with 10 μm NF-κB specific inhibitor BAY11-7082 inhibited both basal promoter activity and transcriptional increase by poly(dAT:dAT) stimulation (Fig. 4D). These results indicated a role for NF-κB in the transcription regulation of miR-365. In addition, the site-directed mutation assays showed that mutations of any of the NF-κB or Sp1 binding sites slightly inhibited the transcriptional increase induced by p65 overexpression, but the double mutation and Sp1-mut2 mutants inhibited by at least 45 and 50%, respectively, the transcriptional increase induced by p65 overexpression (Fig. 4E). These results indicate an important role for the NF-κB sites number 1 and 2 and Sp1 site number 2 in the transcription of miR-365.
Previous studies have revealed an interaction between Sp1 and NF-κB in gene regulation processes through synergistic or antagonistic effects (34). To investigate the relationship between Sp1 and NF-κB in the transcriptional regulation of miR-365, Sp1 and p65 expression constructs were co-transfected, and the luciferase activity of TSS-1097 was analyzed. As shown in Fig. 4F, forced expression of both Sp1 and NF-κB p65 up-regulated transcription of miR-365 to a greater degree (16-fold) than transfection with either one alone (12- or 13-fold, respectively). These results suggest a synergistic effect between Sp1 and NF-κB on the transcription of miR-365.
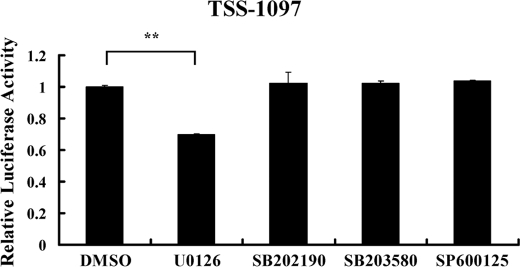
The Involvement of MAPK/ERK in miR-365 Transcription
The MAPK pathway is an important signaling pathway regulating numerous biological processes including miRNA-mediated effects (35). To address the question as to whether MAPK signaling is involved in the transcription of miR-365, we employed specific inhibitors of various intermediates of the pathway. As shown in Fig. 5, pretreatment with the ERK inhibitor U0126 inhibited miR-365 promoter activity by 30% with respect to DMSO pretreatment, whereas pretreatment with other inhibitors, including the p38 inhibitors SB202190 and SB203580 and the JNK inhibitor SP600125, had no appreciable inhibitory effect on the transcription of miR-365. These results indicated that MAPK/ERK may play a role in the transcription regulation of miR-365.
FIGURE 5.
The involvement of MAPK in the transcription of miR-365. HEK293 cells were transfected with TSS-1097-Luc reporter. At 24 h post-transfection cells were treated with indicated inhibitor (10 μm) for 1 h and then replaced with fresh medium. Luciferase activities were measured 24 h post-treatment. The results are represented as the percentage of the control group treated with DMSO, which was considered as 1. The values are represented as the means ± S.D. (n = 3). **, p < 0.01.
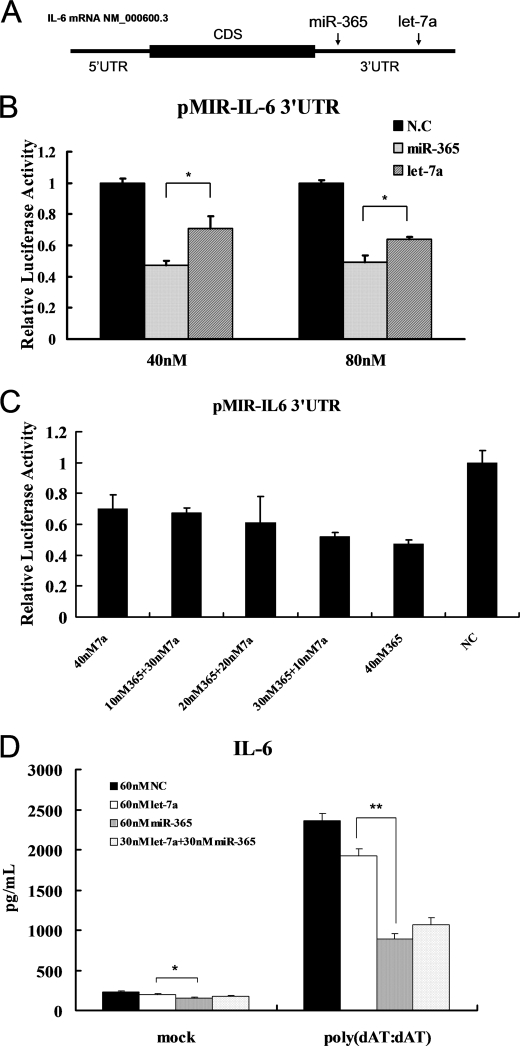
miR-365 Has a Stronger Regulatory Effect on IL-6 in Vitro Than Does Let-7a
Let-7a has recently been described as a regulator of IL-6 by direct interaction with IL-6 3′-UTR (36). Let-7a and miR-365 bind to different regions of the IL-6 3′-UTR (Fig. 6A). To compare the inhibitory effect of the two miRNAs on IL-6 expression, equal amounts of miR-365 or let-7a mimics along with pMIR-IL-6 3′-UTR were transfected into HEK293 cells. As shown in Fig. 6B, when 40 nm mimics were transfected, miR-365 inhibited by 50% the reporter activity, whereas let-7a only inhibited by 30%, as compared with transfection with the scrambled oligonucleotide. When the amount of miRNA mimics was raised to 80 nm, the reporter activities were inhibited by 50 and 40% in transfections with miR-365 and let-7a mimics, respectively. Next, we investigated the inhibitory effect on IL-6 expression by combining the two miRNAs. miR-365 and let-7a were mixed in various ratios, and the total amount of the mimics was limited to 40 nm. As shown in Fig. 6C, when the concentration of let-7a in the mixture was greater, the inhibitory effect dropped. ELISA was also performed to measure the protein level of IL-6 in the culture supernatants of HeLa cells pretreated with equal amounts of miR-365 or let-7a. The presence of miR-365 had a stronger inhibitory effect on the IL-6 protein level in vitro than did the presence of let-7a (Fig. 6D). Even if IL-6 was induced by stimulation with poly(dAT:dAT), miR-365 also exhibited stronger inhibitory effect on IL-6 expression than did let-7a (Fig. 6D). Together, these results demonstrated that miR-365 exhibited a greater negative regulatory effect on IL-6 than let-7a.
FIGURE 6.
Comparison of the effect of miR-365 and let-7a on IL6 expression in vitro. A, shown is a schematic representation of the MREs for let-7a and miR-365. The MREs for let-7a and miR-365 were obtained from the report by Iliopoulos et al. (36) and our study, respectively. CDS, coding sequence. B, pMIR-IL-6 3′-UTR was co-transfected with miR-365 or let-7a into HEK293 cells. Luciferase activities were measured at 24 h post-transfection. NC, negative control. *, p < 0.05. C, pMIR-IL-6 3′-UTR was co-transfected with a total of 40 nm miRNA composed of individual or mixed miR-365 and/or let-7a at the indicated ratios. Luciferase activities were measured 24 h post-co-transfection. D, HeLa cells were transfected with a total of 60 nm miR-365 and/or let-7a mimics or scrambled oligonucleotides as indicated. Cells were transfected with poly(dAT:dAT) at 24 h after the initial transfection. Culture supernatants were collected 24 h later, and IL-6 was measured using a human IL-6 ELISA kit. Bar graph data are presented as the means ± S.D. (n = 3). *, p < 0.05; **, p < 0.01.
DISCUSSION
In this study we established that miR-365 is a direct negative regulator of IL-6. We found that IL-6 protein expression is modulated by miR-365, and this regulatory effect is driven by miR-365 through conserved seed matches within the 3′-UTR of IL-6. The stability of IL-6 mRNA is not affected by miR-365, and it appears regulation of the target gene may be achieved by repressing mRNA translation. We further showed that the promoter region of miR-365 possesses several consensus Sp1 and NF-κB binding sites, that overexpression of NF-κB p65 or Sp1 leads to a significant up-regulation of miR-365 transcription, and co-expression of Sp1 and NF-κB p65 is required for the maximal induction of miR-365.
We provide multiple pieces of evidence to identify miR-365 as a regulator of IL-6. 1) A miR-365 response element was predicted in the IL-6 3′-UTR with a seed sequence match. (2) miR-365 and its seed region are strictly conserved across species. The putative binding site of miR-365 to the IL-6 3′-UTR is broadly (although not strictly) conserved among vertebrates. 3) The activity of a luciferase reporter in which the IL-6 3′-UTR was fused to the downstream of the reporter gene was repressed by miR-365 overexpression in a dose-dependent manner, whereas a luciferase reporter with MRE deletion or mutation in the IL-6 3′-UTR was no longer repressed by miR-365. And 4) the negative regulation of IL-6 at the protein level was further confirmed by ELISA.
Previous studies have suggested the possibility that IL-6 could be regulated by miRNAs. For example, miR-146a modulates IL-6 expression in response to rising IL-6 levels to restrain excessive senescence associated secretory phenotype activity (37), and miR-155 was shown to negatively regulate the induction of IL-6 expression in monocytes exposed to tumor culture supernatants (38). However, no evidence exists to support the direct regulation of IL-6 by miR-146a or miR-155. Instead, it appears that miR-146a and miR-155 represses the expression of IL-6 indirectly by targeting upstream molecules IRAK1 and CEBP-β, respectively (37, 38). In this study, we demonstrate that miR-365 directly regulates IL-6 level by classic interaction with its 3′-UTR. We further identified the MRE for miR-365 by using mutant reporters of IL-6 3′-UTR. More recently, Iliopoulos et al. (36) reported hsa-let-7a as a miRNA regulator of IL-6 that functions by direct interaction with 3′-UTR of IL-6. However, the MRE for let-7a is located in 316–322 bp of IL-6 3′-UTR, which is different from the region that occupied by miR-365 (Fig. 6A). Our data showed that, compared with let-7a, miR-365 exhibits a stronger inhibitory effect on IL-6 expression, as demonstrated by IL-6 3′-UTR luciferase reporter assay and IL-6 ELISA. Furthermore, the observation that the concentration of let-7a in a mixture of miR-365 and let-7a negatively correlated with the inhibitory effect on IL-6 expression further supported this conclusion (Fig. 6C). On the other hand, the result also showed that certain miRNA seems to exert function independently, and there is no synergistic or antagonistic action between the two miRNAs even they have the same target.
As the importance of miRNA activity in gene regulation becomes increasingly evident, interest has focused on the mechanisms by which miRNAs are governed (8, 11, 35, 39). In this study we investigated the transcriptional regulation of miR-365 and demonstrated that NF-κB and Sp1 are involved in miR-365 transcription. Overexpression of the NF-κB subunit p65 significantly induced activity of the miR-365 promoter, whereas pretreatment with an NF-κB-specific inhibitor inhibited the transcription of miR-365. Furthermore, the NF-κB transcription factor binding sites in the promoter of miR-365 were required for the transcription of miR-365, collectively suggesting that NF-κB plays a critical role in the biological processes of miR-365. Indeed, in addition to miR-365, miR-146a (8), miR-155 (40), let-7i (41), miR-17–92, miR-125b-1, miR-21, miR-23b-27b-24–1, miR-30b, and miR-130a (42) have all also been found to be under the control of NF-κB. Sp1 is a ubiquitously expressed transcription factor and is indispensable for the development and survival of animals (43–45). Interestingly, Sp1 elements are often found in the enhancers or promoters of NF-κB-regulated genes (46–49). Furthermore, the Sp1-NF-κB complex has been shown to be involved in the regulation of miR-29b and mouse TLR-2 (50, 51). Considering that our data show that both Sp1 and NF-κB contribute to the transcription of miR-365 and two NF-κB binding sites and one Sp1 binding site were found to be indispensable for miR-365 transcription, we speculated that cooperation between Sp1 and NF-κB in the transcriptional regulation of miR-365 might exist. The synergistic increase in the miR-365 promoter activity induced by co-expression of Sp1 and NF-κB p65 partly supported our hypothesis, but further studies are needed to test these possibilities.
In addition to the NF-κB pathway, previous studies have revealed that the MAPK signaling pathway also plays a regulatory role in the generation of some miRNAs (35, 52, 53). In our study, pretreatment with ERK inhibitor significantly inhibited miR-365 promoter activity, indicating a regulatory role for ERK in the transcription of miR-365. Although p38 and JNK have been demonstrated to regulate the expression of other miRNAs, for example JNK regulating miR-155 and p38 regulating miR-34c (54, 55), we did not observe any appreciable role in the regulation of miR-365 by p38 and JNK.
A single miRNA can regulate several or even hundreds of genes (1). With the exception of IL-6, bioinformatics analyses have indicated other molecules, such as ubiquitin-specific peptidase 48 (USP48), heat shock 70-kDa protein 8 (HSPA8), zinc finger protein 148 (ZNF-148), and serum/glucocorticoid-regulated kinase 1 (SGK-1), are potential targets of miR-365 (Table 3). Interestingly, a very close correlation between these targets and NF-κB activation has been reported (56–59). These molecules regulate NF-κB by several mechanisms, including specific promoter binding activity as well as phosphorylation and ubiquitination. For example, SGK-1 facilitates NF-κB activation by phosphorylation of IKKα, which in turn leads to the degradation of IκBα (59). NF-κB has been shown to play a major role in LPS-induced expression of IL-6 or other inflammatory cytokines (30). It is possible that, in addition to negative regulation of IL-6 expression by direct binding to the 3′-UTR of IL-6, miR-365 may also affect the activation of NF-κB and thereby indirectly inhibit IL-6 expression. Interestingly, we observed that overexpression of miR-365 significantly attenuated NF-κB activation in HEK293 cells (data not shown). Certainly, additional experiments should be performed to demonstrate the indirect regulation of IL-6 by miR-365.
TABLE 3.
List of putative target genes of miR-365
| Target gene | Gene name | Conserved sites |
Poorly conserved sites |
Total context score | Aggregate PCT | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | 8-Mer | 7-Mer-m8 | 7-Mer-1A | Total | 8-Mer | 7-Mer-m8 | 7-Mer-1A | ||||
| LPAR5 | Lysophosphatidic acid receptor 5 | 1 | 1 | 0 | 0 | 2 | 1 | 1 | 0 | −0.93 | 0.11 |
| NFIB | Nuclear factor I/B | 2 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | −0.63 | 0.56 |
| ZNF148 | Zinc finger protein 148 | 2 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | −0.53 | 0.36 |
| TMOD3 | Tropomodulin 3 (ubiquitous) | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | −0.53 | 0.24 |
| SGK1 | Serum/glucocorticoid regulated kinase 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | −0.52 | 0.42 |
| USP33 | Ubiquitin-specific peptidase 33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | −0.48 | 0.53 |
| ZNF644 | Zinc finger protein 644 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | −0.47 | 0.47 |
| USP48 | Ubiquitin-specific peptidase 48 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | −0.47 | 0.51 |
| ARRDC3 | Arrestin domain containing 3 | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | −0.47 | 0.43 |
| LOC283514 | Similar to seven in absentia 2 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | −0.47 | 0.55 |
| PIK3R3 | Phosphoinositide 3-kinase, regulatory subunit 3 (γ) | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | −0.43 | 0.24 |
| SGK3 | Serum/glucocorticoid-regulated kinase family, member 3 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | −0.29 | 0.24 |
| MAPK1IP1L | Mitogen-activated protein kinase 1 interacting protein 1-like | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | −0.29 | 0.24 |
| PRKAR2A | Protein kinase, cAMP-dependent, regulatory, type II, α | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | −0.27 | 0.24 |
| HSPA8 | Heat shock 70-kDa protein 8 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | −0.26 | 0.24 |
In conclusion, our studies identified miR-365 as a novel negative regulator of IL-6. In addition to direct negative regulation, miR-365 may regulate IL-6 expression indirectly. In vitro evidence indicated that miR-365 functions as a more powerful regulator of IL-6 than let-7a. NF-κB, Sp1, and ERK are involved in the expression of miR-365, suggesting that miR-365 is controlled by a complex network to exert its function in an appropriate manner.
This work was supported by the New Century Excellent Talent Project (NCET-07–0347), the National Natural Sciences Foundation of China (30972189, 30871871), and the Program for Changjiang Scholars and Innovative Research Team in University (IRT0726).
- miRNAs
- microRNA
- MRE
- miRNA recognition elements
- TSS
- transcription start site.
REFERENCES
- 1. Ambros V. (2004) Nature 431, 350–355 [DOI] [PubMed] [Google Scholar]
- 2. Lee Y., Jeon K., Lee J. T., Kim S., Kim V. N. (2002) EMBO J. 21, 4663–4670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yi R., Qin Y., Macara I. G., Cullen B. R. (2003) Genes Dev. 17, 3011–3016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lee Y., Ahn C., Han J., Choi H., Kim J., Yim J., Lee J., Provost P., Rådmark O., Kim S., Kim V. N. (2003) Nature 425, 415–419 [DOI] [PubMed] [Google Scholar]
- 5. Bartel D. P. (2004) Cell 116, 281–297 [DOI] [PubMed] [Google Scholar]
- 6. Kiriakidou M., Nelson P. T., Kouranov A., Fitziev P., Bouyioukos C., Mourelatos Z., Hatzigeorgiou A. (2004) Genes Dev. 18, 1165–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lewis B. P., Shih I. H., Jones-Rhoades M. W., Bartel D. P., Burge C. B. (2003) Cell 115, 787–798 [DOI] [PubMed] [Google Scholar]
- 8. Taganov K. D., Boldin M. P., Chang K. J., Baltimore D. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 12481–12486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Xiao B., Liu Z., Li B. S., Tang B., Li W., Guo G., Shi Y., Wang F., Wu Y., Tong W. D., Guo H., Mao X. H., Zou Q. M. (2009) J. Infect. Dis. 200, 916–925 [DOI] [PubMed] [Google Scholar]
- 10. Tang B., Xiao B., Liu Z., Li N., Zhu E. D., Li B. S., Xie Q. H., Zhuang Y., Zou Q. M., Mao X. H. (2010) FEBS Lett. 584, 1481–1486 [DOI] [PubMed] [Google Scholar]
- 11. Sharma A., Kumar M., Aich J., Hariharan M., Brahmachari S. K., Agrawal A., Ghosh B. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 5761–5766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Qin Z., Kearney P., Plaisance K., Parsons C. H. (2010) J. Leukoc. Biol. 87, 25–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ma F., Liu X., Li D., Wang P., Li N., Lu L., Cao X. (2010) J. Immunol. 184, 6053–6059 [DOI] [PubMed] [Google Scholar]
- 14. Polikepahad S., Knight J. M., Naghavi A. O., Oplt T., Creighton C. J., Shaw C., Benham A. L., Kim J., Soibam B., Harris R. A., Coarfa C., Zariff A., Milosavljevic A., Batts L. M., Kheradmand F., Gunaratne P. H., Corry D. B. (2010) J. Biol. Chem. 285, 30139–30149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Alsaleh G., Suffert G., Semaan N., Juncker T., Frenzel L., Gottenberg J. E., Sibilia J., Pfeffer S., Wachsmann D. (2009) J. Immunol. 182, 5088–5097 [DOI] [PubMed] [Google Scholar]
- 16. Akira S., Hirano T., Taga T., Kishimoto T. (1990) FASEB J. 4, 2860–2867 [PubMed] [Google Scholar]
- 17. Hirano T., Akira S., Taga T., Kishimoto T. (1990) Immunol. Today 11, 443–449 [DOI] [PubMed] [Google Scholar]
- 18. Kishimoto T. (1989) Blood 74, 1–10 [PubMed] [Google Scholar]
- 19. Kishimoto T. (2005) Annu. Rev. Immunol. 23, 1–21 [DOI] [PubMed] [Google Scholar]
- 20. Jones S. A. (2005) J. Immunol. 175, 3463–3468 [DOI] [PubMed] [Google Scholar]
- 21. Rambaldi A., Bettoni S., Rossi V., Tini M. L., Giudici G., Rizzo V., Bassan R., Mantovani A., Barbui T., Biondi A. (1993) Br. J. Haematol. 83, 204–211 [DOI] [PubMed] [Google Scholar]
- 22. Clark A. (2000) Arthritis Res. 2, 172–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Griffiths-Jones S., Grocock R. J., van Dongen S., Bateman A., Enright A. J. (2006) Nucleic Acids Res. 34, D140–D144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Griffiths-Jones S. (2004) Nucleic Acids Res. 32, D109–D111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Griffiths-Jones S., Saini H. K., van Dongen S., Enright A. J. (2008) Nucleic Acids Res. 36, D154–D158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. John B., Enright A. J., Aravin A., Tuschl T., Sander C., Marks D. S. (2004) PLoS Biol. 2, e363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Betel D., Wilson M., Gabow A., Marks D. S., Sander C. (2008) Nucleic Acids Res. 36, D149–D153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Grimson A., Farh K. K., Johnston W. K., Garrett-Engele P., Lim L. P., Bartel D. P. (2007) Mol. Cell 27, 91–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Friedman R. C., Farh K. K., Burge C. B., Bartel D. P. (2009) Genome Res. 19, 92–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kim B. H., Lee K. H., Chung E. Y., Chang Y. S., Lee H., Lee C. K., Min K. R., Kim Y. (2006) Eur. J. Pharmacol. 543, 158–165 [DOI] [PubMed] [Google Scholar]
- 31. Lewis B. P., Burge C. B., Bartel D. P. (2005) Cell 120, 15–20 [DOI] [PubMed] [Google Scholar]
- 32. Higuchi R., Krummel B., Saiki R. K. (1988) Nucleic Acids Res. 16, 7351–7367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rehmsmeier M., Steffen P., Hochsmann M., Giegerich R. (2004) RNA 10, 1507–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hirano F., Tanaka H., Hirano Y., Hiramoto M., Handa H., Makino I., Scheidereit C. (1998) Mol. Cell. Biol. 18, 1266–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Paroo Z., Ye X., Chen S., Liu Q. (2009) Cell 139, 112–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Iliopoulos D., Hirsch H. A., Struhl K. (2009) Cell 139, 693–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bhaumik D., Scott G. K., Schokrpur S., Patil C. K., Orjalo A. V., Rodier F., Lithgow G. J., Campisi J. (2009) Aging 1, 402–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. He M., Xu Z., Ding T., Kuang D. M., Zheng L. (2009) Cell. Mol. Immunol. 6, 343–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Liu G., Friggeri A., Yang Y., Park Y. J., Tsuruta Y., Abraham E. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 15819–15824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gatto G., Rossi A., Rossi D., Kroening S., Bonatti S., Mallardo M. (2008) Nucleic Acids Res. 36, 6608–6619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. O'Hara S. P., Splinter P. L., Gajdos G. B., Trussoni C. E., Fernandez-Zapico M. E., Chen X. M., LaRusso N. F. (2010) J. Biol. Chem. 285, 216–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhou R., Hu G., Gong A. Y., Chen X. M. (2010) Nucleic Acids Res. 38, 3222–3232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dynan W. S., Tjian R. (1983) Cell 35, 79–87 [DOI] [PubMed] [Google Scholar]
- 44. Narayan V. A., Kriwacki R. W., Caradonna J. P. (1997) J. Biol. Chem. 272, 7801–7809 [DOI] [PubMed] [Google Scholar]
- 45. Saffer J. D., Jackson S. P., Annarella M. B. (1991) Mol. Cell. Biol. 11, 2189–2199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Stade B. G., Messer G., Riethmüller G., Johnson J. P. (1990) Immunobiology 182, 79–87 [DOI] [PubMed] [Google Scholar]
- 47. Ma W., Lim W., Gee K., Aucoin S., Nandan D., Kozlowski M., Diaz-Mitoma F., Kumar A. (2001) J. Biol. Chem. 276, 13664–13674 [DOI] [PubMed] [Google Scholar]
- 48. Jones K. A., Kadonaga J. T., Luciw P. A., Tjian R. (1986) Science 232, 755–759 [DOI] [PubMed] [Google Scholar]
- 49. Perkins N. D., Edwards N. L., Duckett C. S., Agranoff A. B., Schmid R. M., Nabel G. J. (1993) EMBO J. 12, 3551–3558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wang T., Lafuse W. P., Zwilling B. S. (2001) J. Immunol. 167, 6924–6932 [DOI] [PubMed] [Google Scholar]
- 51. Liu S., Wu L. C., Pang J., Santhanam R., Schwind S., Wu Y. Z., Hickey C. J., Yu J., Becker H., Maharry K., Radmacher M. D., Li C., Whitman S. P., Mishra A., Stauffer N., Eiring A. M., Briesewitz R., Baiocchi R. A., Chan K. K., Paschka P., Caligiuri M. A., Byrd J. C., Croce C. M., Bloomfield C. D., Perrotti D., Garzon R., Marcucci G. (2010) Cancer Cell 17, 333–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Cardinali B., Castellani L., Fasanaro P., Basso A., Alemà S., Martelli F., Falcone G. (2009) PLoS ONE 4, e7607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zeng Y., Sankala H., Zhang X., Graves P. R. (2008) Biochem. J. 413, 429–436 [DOI] [PubMed] [Google Scholar]
- 54. O'Connell R. M., Taganov K. D., Boldin M. P., Cheng G., Baltimore D. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 1604–1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Cannell I. G., Kong Y. W., Johnston S. J., Chen M. L., Collins H. M., Dobbyn H. C., Elia A., Kress T. R., Dickens M., Clemens M. J., Heery D. M., Gaestel M., Eilers M., Willis A. E., Bushell M. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 5375–5380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tzimas C., Michailidou G., Arsenakis M., Kieff E., Mosialos G., Hatzivassiliou E. G. (2006) Cell. Signal. 18, 83–92 [DOI] [PubMed] [Google Scholar]
- 57. Dokladny K., Lobb R., Wharton W., Ma T. Y., Moseley P. L. (2010) Cell Stress Chaperones 15, 153–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Borghaei R. C., Gorski G., Javadi M. (2009) Biochem. Biophys. Res. Commun. 382, 269–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Tai D. J., Su C. C., Ma Y. L., Lee E. H. (2009) J. Biol. Chem. 284, 4073–4089 [DOI] [PubMed] [Google Scholar]