Abstract
Metalloprotease-disintegrin ADAM12 is overexpressed and frequently mutated in breast cancer. We report here that ADAM12 expression in cultured mammalian cells is up-regulated by Notch signals. Expression of a constitutively active form of Notch1 in murine fibroblasts, myoblasts, or mammary epithelial cells or activation of the endogenous Notch signaling by co-culture with ligand-expressing cells increases ADAM12 protein and mRNA levels. Up-regulation of ADAM12 expression by Notch requires new transcription, is activated in a CSL-dependent manner, and is abolished upon inhibition of IκB kinase. Expression of a constitutively active Notch1 in NIH3T3 cells increases the stability of Adam12 mRNA. We further show that the microRNA-29 family, which has a predicted conserved site in the 3′-untranslated region of mouse Adam12, plays a critical role in mediating the stimulatory effect of Notch on ADAM12 expression. In human cells, Notch up-regulates the expression of the long form, but not the short form, of ADAM12 containing a divergent 3′-untranslated mRNA region. These studies uncover a novel paradigm in Notch signaling and establish Adam12 as a Notch-related gene.
Keywords: ADAM ADAMTS, MicroRNA, NF-kappa B, Notch Pathway, Protease, Post-transcriptional Regulation
Introduction
ADAM12, a member of the metalloprotease-disintegrin family, is emerging as a breast cancer-associated gene. ADAM12 expression is strongly up-regulated in pre-invasive and invasive carcinomas (1–3). Somatic mutations in the coding region of the ADAM12 gene are found at significant frequencies in human breast tumors (4, 5). Importantly, breast cancer-associated mutations inhibit the intracellular processing and function of the ADAM12 protein (6). In human mammary epithelial cells propagated in vitro, ADAM12 expression is the highest in a pool of stem/progenitor cells (7, 8). These cells maintain high levels of Notch signaling, which critically regulates their self-renewal and cell fate determination (8–10). Intriguingly, ADAM12 expression is also up-regulated in mesenchymal stem cells (11), neural stem cells (12), stem cells of the spleen (13), and stem cells of the liver (14). Each of these stem cell types is associated with high levels of Notch activity (13–18). Furthermore, ADAM12 is expressed in regenerating skeletal muscle (19, 20). ADAM12 expression is most prominent in muscle satellite cells undergoing expansion and self-renewal, where Notch activity is high, and it is down-regulated in differentiating myotubes, where Notch signaling is off (21, 22).
The canonical Notch signaling pathway is activated when a ligand, a transmembrane protein present at the surface of a signal-sending cell, binds to a Notch receptor in a signal-receiving cell. In mammals, there are five ligands (Delta-like 1, 3, and 4 and Jagged 1 and 2) and four Notch receptors (Notch 1–4) (23, 24). The ligand-receptor interaction is followed by a sequential cleavage of the receptor by an ADAM2 protease and by presenilin-dependent γ-secretase. Two different ADAMs have been reported to cleave Notch, ADAM10 and ADAM17 (25–28). Recent studies have established that although ADAM10 is absolutely required for ligand-induced signaling by wild-type Notch1, both proteases can cleave Notch1 mutants associated with leukemia (29). Whether any other ADAMs, including ADAM12, are also capable of Notch cleavage under certain pathological conditions has not been established. Cleavage by γ-secretase leads to release of the intracellular domain of Notch (NICD) and its translocation to the nucleus. Inside the nucleus, NICD forms a complex with the CSL (CBF1 in mammals/Su(H) in Drosophila/Lag-1 in Caenorhabditis elegans) transcription factor and a Mastermind-like co-activator and activates target gene expression (30, 31).
Because of the apparent correlation between the level of Notch activation and ADAM12 expression in several types of stem-like cells, we asked whether the expression of ADAM12 is directly regulated by Notch signals. We report here that Notch signaling increases the level of ADAM12 expression at both the protein and the mRNA levels. The regulation of ADAM12 expression by Notch involves NF-κB-dependent down-regulation of microRNA-29a, a member of the microRNA-29 family, and increased stability of Adam12 mRNA. These studies demonstrate for the first time the effect of Notch on the expression level of a member of the ADAM family of proteases.
EXPERIMENTAL PROCEDURES
Cell Culture
NIH3T3, C2C12, and HEK293 cells (American Tissue Culture Collection) and retroviral packaging cell lines Phoenix Eco and Phoenix Ampho (G. P. Nolan, Stanford University) were grown in DMEM supplemented with 10% FBS. SMAD2−/− MEFs (E. Bottinger, Mount Sinai School of Medicine), SMAD3−/− MEFs (K. Flanders, NCI, National Institutes of Health), and wild-type MEFs were grown in DMEM containing 10% FBS and 1% penicillin/streptomycin. NMuMG and MCF7 cells (ATCC) were grown in DMEM with 10% FBS and 10 μg/ml insulin. OT11 (CSL−/−) and OT13 (CSL+/+) mouse embryonic fibroblasts (RIKEN Cell Bank, obtained with permission of T. Honjo, Kyoto University) were grown in DMEM with 10% FBS and 100 units/ml interferon-γ. CHO cells stably transfected with Myc-tagged mouse Delta-like 1 (CHO-Dll1) or with empty vector (CHO-V) were grown in F12K nutrient mixture supplemented with 10% FBS and 800 μg/ml G418, as described (32). In co-culture experiments, CHO-Dll1 or CHO-V cells (5 × 105 cells/well in a 6-well plate) were added to ∼50% confluent NIH3T3 cells and incubated for an additional 24 h in DMEM with 10% FBS, without G418. Cell treatments were as follows: 5 μm N-[N-(3,5-di-fluorophenacetyl)-l-alanyl]-S-phenylglycine t-butyl ester (DAPT; EMD Biosciences) for 24 h, 5 μg/ml actinomycin D (Sigma-Aldrich) for 8 h, 10 μm MG132 (EMD Biosciences) for 16 h, and 5 or 10 μm BMS-345541 (EMD Biosciences) for 24 h. Control incubations with vehicle alone were included in all experiments.
Expression Constructs
Fragments of the Adam12 promoter located upstream of the translation initiation site (see Fig. 5A) were amplified from mouse genomic DNA using Pfu Turbo DNA polymerase and inserted into the multiple cloning site of pGL4.10luc2 vector (Promega). The NICD fragment of mouse Notch1 (amino acids 1747–2184) was amplified using a full-length Notch1 plasmid as a template and cloned into the pcDNA3.1 vector. caNotch1-AP retroviral vector (provided by R. Kageyama and C. Takahashi, Kyoto University) directed expression of the constitutively active mouse Notch1 spanning the transmembrane region, the RAM domain, the ankyrin repeats, and the nuclear localization signals (amino acids 1704–2192); AP vector lacking the RAM and ankyrin repeats sequences was used as a negative control (33).
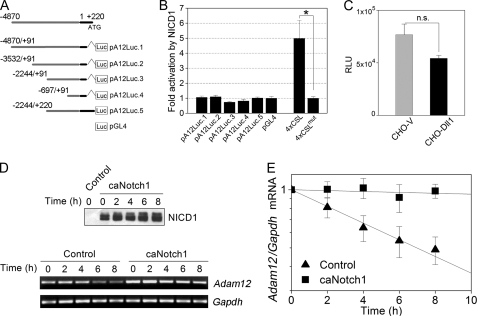
FIGURE 5.
caNotch1 increases Adam12 mRNA stability. A, diagram of gene reporter constructs. The indicated fragments of the promoter region of the mouse Adam12 gene were cloned into pGL4 luciferase reporter vector. The transcription start site is located at position +1, the translation initiation site is at position +220, the 5′-UTR region in the first exon is shown in black, and the genomic upstream sequence is shown in gray. B, NIH3T3 cells were co-transfected with the indicated luciferase reporters, pRL-TK, and either NICD1 or empty pcDNA3.1 vector. Firefly luciferase activity, normalized to Renilla luciferase, was measured 24 h after transfection, and -fold activation by NICD1 was calculated. The data represent the means ± S.E. from 2–4 independent experiments, *, p < 0.05. C, NIH3T3 cells stably transfected with pA12Luc.1 reporter were co-cultured with CHO-Dll1 or CHO-V cells. Firefly luciferase activity was measured 24 h later (relative luminescence units (RLU)). The data represent the means ± S.E. from 2 independent experiments. n.s., non-significant. D, NIH3T3 cells were infected with caNotch1 or control retroviruses. Actinomycin D was added 24 h after infection (time 0). The level of NICD1 at the indicated times after adding actinomycin D was evaluated by Western blotting using the Val1744 antibody. Adam12 and Gapdh mRNA levels were evaluated by semiquantitative RT-PCR. E, the band intensities in panel D were quantified by densitometry using the ImageJ software. The -fold change in Adam12 mRNA, normalized to Gapdh, is shown; the Adam12/Gapdh mRNA ratios at time 0 are set as 1. The data represent the means ± S.E. from 3 independent experiments.
Plasmid Transfection and Luciferase Reporter Assays
NIH3T3 cells grown in 6-well plates were co-transfected at 50% confluence with 0.5 μg of pA12.Luc reporters and NICD1 or empty pcDNA3.1 vector, 4×CSL reporters (Diane Hayward, Johns Hopkins University), and Igκ2-INF, a reporter containing two copies of the NF-κB binding site upstream of the interferon-β minimal promoter (Addgene plasmid 14886), together with 0.05 μg of Renilla luciferase vector (pRL-TK), using FuGENE 6 transfection reagent. After 24 h, firefly and Renilla luciferase activities were measured using the Dual-Luciferase reporter assay system (Promega). The assays were performed in duplicates. Stable pA12Luc.1 transfectants were selected for 7 days in the presence of 2 μg/ml puromycin; pooled populations of cells were used without isolation of individual clones.
Retroviral Infection
Virus packaging Phoenix Eco cells (for infection of murine cells) or Phoenix Ampho cells (for infection of human cells) were transfected with retroviral vectors (15 μg of plasmid DNA per 100-mm plate) using the calcium phosphate precipitation method. Viral supernatants were harvested 48 h later, supplemented with 5 μg/ml Polybrene, and used for cell infection without dilution. Cells were analyzed 48 h after infection, unless indicated otherwise.
Transfection of miRNA Mimic and miRNA Inhibitor
mmu-miR-29a miRIDIAN mimic, miRIDIAN microRNA mimic negative control 1, mmu-miR-29a miRIDIAN hairpin inhibitor, and miRIDIAN microRNA hairpin inhibitor negative control 1 were obtained from Dharmacon. Transfection of NIH3T3 cells grown in 6-well plates was performed using 50 nm miR-29a mimic or negative control, 100 nm inhibitor or negative control, and 4 μl of DharmaFECT 1 transfection reagent (Dharmacon), according to the manufacturer's instructions. Cells were analyzed 72 h after transfection, unless indicated otherwise.
Immunoblotting
Immunoblotting was performed as described (32). For ADAM12 detection, cell extracts were enriched for glycoproteins using concanavalin A-agarose prior to SDS-PAGE and Western blotting (34). The following primary antibodies were used: rabbit anti-ADAM12 cytoplasmic peptide antibody (1:3000, Ref. 34, used in Figs. 1, 3, A and D, and 4A) or rabbit anti-ADAM12 cysteine-rich domain (rb122, 1:5,000; a gift from M. Kveiborg and U. M. Wewer, University of Copenhagen, used in Figs. 3C, 4, D and E, 6, and 7), rabbit anti-cleaved Notch1 (Val1744, Cell Signaling, 1:500), mouse anti-β1 integrin (BD Transduction Laboratory, 1:3,000), mouse anti-tubulin (DM 1A, Sigma, 1:100,000), mouse anti-c-Myc (9E10, Upstate Biotech Millipore, 1:1,000). Secondary antibodies were horseradish peroxidase-conjugated anti-rabbit or anti-mouse IgG antibodies. Signal detection was performed using the West Pico chemiluminescence detection kit (Pierce).
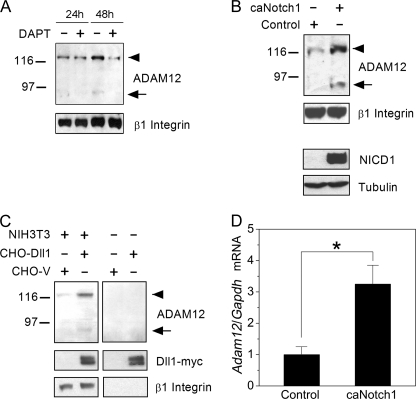
FIGURE 1.
Notch signaling up-regulates ADAM12 protein and mRNA levels. A, subconfluent NIH3T3 cells (24 h after plating) or confluent NIH3T3 cells (48 h after plating) were treated for 24 h with 5 μm DAPT, a γ-secretase inhibitor that blocks Notch activation. The level of ADAM12 protein was evaluated by Western blotting after glycoprotein enrichment on concanavalin A-agarose. The nascent form of ADAM12 is indicated by an arrowhead, the mature form of ADAM12 is shown by an arrow, and β1 Integrin indicates a gel loading control. B, NIH3T3 cells were infected with retroviruses expressing caNotch1 or control retroviruses. After 48 h, ADAM12 protein was measured as in panel A. The activated form of Notch1, NICD1, was detected in total cell lysates with Val1744 antibody, and Tubulin indicates a gel loading control. C, NIH3T3 cells were co-cultured for 24 h with CHO cells stably transfected with Dll1-Myc (CHO-Dll1) or with empty vector (CHO-V). Glycoprotein-enriched cell fractions were analyzed by Western blotting using anti-ADAM12 and anti-Myc antibodies, and β1 Integrin indicates a gel loading control. Anti-ADAM12 and anti-β1 integrin antibodies are specific for mouse antigens and do not react with ADAM12 or β1 integrin in CHO cells. In panels A–C, each experiment was repeated at least three times, with similar results. D, NIH3T3 cells were infected with caNotch1 or control retroviruses, and Adam12 mRNA level, normalized to Gapdh mRNA, was measured by quantitative RT-PCR. The data represent the means ± S.E. from 3 independent determinations; *, p < 0.05, indicates statistically significant difference.
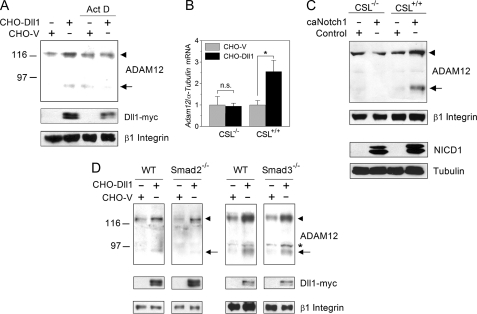
FIGURE 3.
Up-regulation of ADAM12 expression by Notch requires new transcription and is activated in a CSL-dependent, Smad2/3-independent manner. A, NIH3T3 cells were co-cultured with CHO-Dll1 or CHO-V cells for 8 h, in the absence or presence of actinomycin D (Act D), an inhibitor of transcription. The levels of ADAM12 protein and Dll1-Myc were evaluated by Western blotting after glycoprotein enrichment. The nascent form of ADAM12 is indicated by an arrowhead, the mature form of ADAM12 is shown by an arrow, and β1 Integrin indicates a gel loading control. B, CSL−/− or CSL+/+ mouse fibroblasts were co-cultured for 24 h with CHO-Dll1 or CHO-V cells. The level of Adam12 in mouse cells, normalized to α-tubulin, was evaluated by quantitative RT-PCR. The conditions of quantitative RT-PCR were optimized to ensure that the contribution of Adam12 or α-tubulin from CHO cells was negligible. The data represent the means ± S.E. from 3 independent measurements, *, p < 0.05, n.s., non-significant. C, CSL−/− or CSL+/+ fibroblasts were infected with caNotch1 retroviruses. NICD1 and the endogenous ADAM12 protein were detected by Western blotting. D, Smad2−/−, Smad3−/−, or the corresponding wild-type (WT) mouse embryonic fibroblasts were co-cultured for 24 h with CHO-Dll1 or CHO-V cells. Glycoprotein-enriched cell fractions were analyzed by Western blotting using anti-ADAM12 and anti-Myc antibodies, β1 Integrin indicates a gel loading control, and the asterisk indicates a nonspecific band. The experiments in panels A, C, and D were repeated three times with similar results.
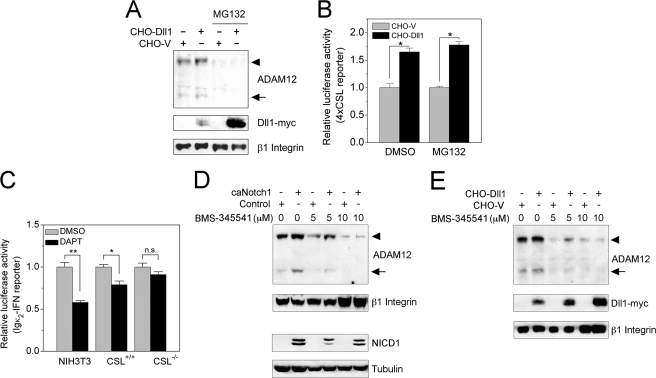
FIGURE 4.
Up-regulation of ADAM12 by Notch requires an active NF-κB pathway. A, NIH3T3 cells were incubated with CHO-Dll1 or CHO-V cells for 16 h in the absence or presence of MG132, a proteasomal inhibitor. Glycoprotein-enriched cell fractions were analyzed by Western blotting using anti-ADAM12 and anti-Myc antibodies, and β1 Integrin indicates a gel loading control. The nascent form of ADAM12 is indicated by an arrowhead, and the mature form of ADAM12 is shown by an arrow. B, NIH3T3 cells were co-transfected with the 4×CSL luciferase reporter and pRL-TK, an internal control. After 24 h, MG132 or vehicle (DMSO) was added, and the relative luciferase activity was determined 16 h later. The data represent the means ± S.E. from 2 independent measurements; *, p < 0.05. C, NIH3T3, CSL+/+, and CSL−/− cells were transfected with the Igκ2-IFN luciferase reporter, together with pRL-TK. After 18 h, 10 μm DAPT or DMSO was added to transfected cells, and the relative luciferase activity was determined 24 h later. The data represent the means ± S.E. from 4 independent measurements, *, p < 0.05, **, p < 0.01, n.s., non-significant. D, NIH3T3 cells were infected with caNotch1 or control viruses and incubated for 24 h with the indicated concentrations of BMS-345541, an inhibitor of IκB kinase. ADAM12 protein levels were examined by Western blotting after glycoprotein enrichment, and β1 Integrin indicates a gel loading control. NICD1 levels were measured in total cell lysates, and Tubulin indicates a gel loading control. E, NIH3T3 cells were co-cultured with CHO-Dll1 or CHO-V cells for 24 h with the indicated concentrations of BMS-345541. ADAM12 and Dll1-Myc levels were analyzed in glycoprotein-enriched fractions, and β1 Integrin indicates a loading control. The experiments in panels A, D, and E were repeated three times with similar results.
FIGURE 6.
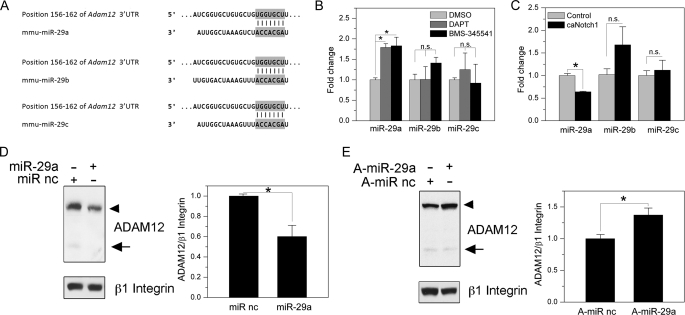
The role of miR-29a in the regulation of ADAM12 expression. A, predicted miR-29 target sites in Adam12 3′-UTR, based on TargetScan Release 5.1. Positions of the nucleotides in the 3′-UTRs that show exact matches to residues 2–8 of the mature miR-29a, miR-29b, or miR-29c are indicated. B, NIH3T3 cells were treated with DMSO, DAPT, or BMS-345541 for 6 h, and the levels of miR-29a, miR-29b, and miR-29c were determined by quantitative RT-PCR. C, NIH3T3 cells were infected with retroviruses expressing caNotch1 or control retroviruses. After 48 h, miR-29a, miR-29b, and miR-29c were determined by quantitative RT-PCR. D, left, NIH3T3 cells were transfected with miR-29a mimic or miRNA mimic negative control (miR nc). After 72 h, ADAM12 protein levels were examined by Western blotting after glycoprotein enrichment, and β1 Integrin indicates a gel loading control. The nascent form of ADAM12 is indicated by an arrowhead, and the mature form of ADAM12 is shown by an arrow. Right, ADAM12 level, normalized to β1 integrin, was determined by densitometry and the ImageJ software. The data represent the mean ± S.E. from 3 independent measurements. E, left, NIH3T3 cells were transfected with miR-29a hairpin inhibitor anti-miR-29a (A-miR-29a) or miRNA inhibitor negative control (A-miR nc). Right, ADAM12 level, normalized to β1 integrin, was determined as in panel D. The data represent the mean ± S.E. from 3 independent measurements, *, p < 0.05, n.s., non-significant.
FIGURE 7.
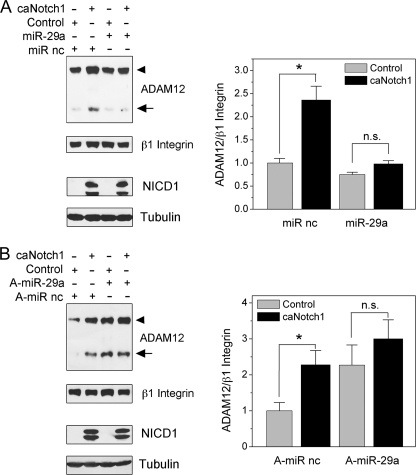
The role of miR-29a in the up-regulation of ADAM12 expression by Notch. A, NIH3T3 cells were transfected with miR-29a mimic or miR mimic negative control (miR nc). After 24 h, cells were infected with caNotch1 or control viruses and analyzed 48 h later. ADAM12 protein levels were examined by Western blotting after glycoprotein enrichment, and β1 Integrin indicates a gel loading control. The nascent form of ADAM12 is indicated by an arrowhead, and the mature form of ADAM12 is shown by an arrow. NICD1 levels were measured in total cell lysates, and Tubulin indicates a gel loading control. B, NIH3T3 cells were transfected with miR-29a hairpin inhibitor anti-miR-29a (A-miR-29a) or miR inhibitor negative control (A-miR nc). After 24 h, cells were infected with caNotch1 or control viruses and analyzed 48 h later, as in panel A. The bar graphs on the right show quantification of the signal intensities of ADAM12, normalized to β1 integrin; the data represent the means ± S.E. from 2 independent measurements, *, p < 0.05, n.s., not significant.
mRNA Analysis
Total RNA was extracted using the PureLink Micro-to-Midi total RNA purification system containing TRIzol (Invitrogen). RNA (1 μg) was treated with deoxyribonuclease I (Invitrogen) followed by reverse transcription using the SuperScript III first-strand synthesis system for RT-PCR (Invitrogen) and oligo(dT) primers. Semiquantitative PCR was performed in 50-μl reaction volumes using 1 μl of cDNA, 0.2 mm dNTPs, 2 units of BIO-X-ACT short DNA polymerase (Bioline), and 1 μm primers (supplemental Table S1). The PCR reaction conditions were: 94 °C, 30 s; 55 °C, 30 s; 72 °C, 45 s; 29–32 cycles for Adam12, 32–35 cycles for ADAM12L, 36–38 cycles for ADAM12S, 24–26 cycles for Gapdh, and 21–24 cycles for β-actin. PCR products were resolved in 2% agarose gels, visualized after ethidium bromide staining and UV illumination, and quantified by densitometry. Quantitative RT-PCR was performed in a total volume of 25 μl in a 96-well spectrofluorometric thermal cycler (iCycler, Bio-Rad). The final reaction mix contained 10.5 μl of 1:10 diluted cDNA, 12.5 μl of iQSYBR Green Supermix, and 0.4 μm primers. The PCR conditions were: 95 °C, 30 s; 55 °C, 30 s; 72 °C, 40 s for Adam12 and 95 °C, 30 s; 63 °C, 30 s; 72 °C, 40 s for α-tubulin. Under these conditions, in co-culture experiments in which mouse cells were mixed with CHO cells, only mouse Adam12 and α-tubulin were detected, and signals from CHO cells were negligible. The relative expression of Adam12 mRNA, normalized to α-tubulin, was calculated using the 2(−ΔΔCt) method.
Quantitative miRNA Analysis
Total RNA was extracted with miRNeasy kit (Qiagen) and quantified with a NanoDrop spectrophotometer (NanoDrop Technologies). Quantitative RT-PCR for analysis of mature miR-29a, -29b, or -29c was performed using the miRCURY LNATM universal RT microRNA PCR system (Exiqon), with specific LNATM PCR primer sets for miR-29a, -29b, or -29c, and the reference gene U6, according to the manufacturer's instructions. Quantitative RT-PCR was carried out in an iCycler. The relative expression of mature microRNA 29 (miR-29) levels, normalized to U6, was determined using the 2(−ΔΔCt) method.
Statistical Analysis
Each experiment was repeated at least two times; representative Western blots are shown. Paired t test was used to compare mean values of two groups (“treated” versus “untreated” controls from independent experiments; GraphPad Prism software).
RESULTS
Activation of ADAM12 Expression by Notch Signaling
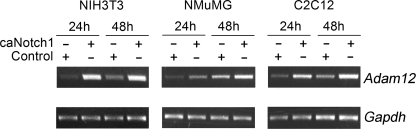
Two forms of ADAM12 were detected by immunoblotting in NIH3T3 cells: the ∼120-kDa nascent full-length form and the ∼90-kDa mature form lacking the N-terminal prodomain. Inhibition of the endogenous Notch signaling in subconfluent cells by DAPT, an inhibitor of γ-secretase, did not have a significant effect on the level of ADAM12 protein. In contrast, treatment of confluent cells with DAPT significantly decreased the amount of both forms of ADAM12 (Fig. 1A). This raised the possibility that ADAM12 expression was stimulated by cell-cell contacts and ligand-dependent activation of the endogenous Notch. Infection of NIH3T3 cells with retroviruses expressing a constitutively active form of mouse Notch1 (caNotch1) lacking the major portion of the extracellular domain resulted in higher levels of ADAM12 protein (Fig. 1B). Similarly, enhancement of Notch signaling by co-culture of subconfluent NIH3T3 cells with CHO cells stably transfected with Notch ligand Delta-like 1 (Dll1) increased the abundance of ADAM12 protein in NIH3T3 cells (Fig. 1C). Quantitative RT-PCR analysis showed that Adam12 mRNA level in cells expressing caNotch1 was about three times higher than in control cells (Fig. 1D). Up-regulation of Adam12 expression by caNotch1 was not limited to NIH3T3 fibroblasts and was also observed in murine mammary epithelial cell line NMuMG and in murine myoblasts C2C12 (Fig. 2).
FIGURE 2.
Up-regulation of Adam12 by Notch is not cell-specific. NIH3T3, NMuMG, or C2C12 cells were infected with caNotch1 or control retroviruses. After 24 or 48 h, the level of Adam12 mRNA was evaluated by semiquantitative RT-PCR; Gapdh is an internal control. The experiment was repeated twice, with similar results.
Up-regulation of ADAM12 Expression by Notch Requires New Transcription and Is CSL-dependent
Co-culture of NIH3T3 cells with ligand-expressing CHO-Dll1 cells in the presence of actinomycin D, a transcriptional inhibitor, effectively blocked the up-regulation of ADAM12 in NIH3T3 cells (Fig. 3A). In this experiment, we used a shorter co-culture time (8 h instead of 24 h) to ensure that cells were viable and healthy and that Dll1 was still expressed in CHO cells. This result indicates that new transcription is required for up-regulation of ADAM12 expression. Activation of Notch signaling in CSL−/− fibroblasts by either co-culture with CHO-Dll1 cells or expression of caNotch1 failed to up-regulate ADAM12 expression at the mRNA (Fig. 3B) or protein level (Fig. 3C). These results demonstrate that up-regulation of ADAM12 expression by Notch proceeds via the canonical CSL-dependent pathway.
We and others have recently demonstrated that ADAM12 expression is induced by transforming growth factor β (TGFβ) via Smad-dependent and Smad-independent signaling pathways (35–39). Consistently, ADAM12 induction by TGFβ is significantly attenuated in Smad2−/− or Smad3−/− MEFs (37). Several studies showed that Smad2/3 can form complexes with Notch1/4 and CSL (40, 41) and that these complexes may be more active toward promoters containing Smad-binding elements (42, 43). Therefore, we asked whether the up-regulation of ADAM12 expression by Notch could be Smad2/3-dependent. We observed, however, that the extent of ADAM12 up-regulation by Notch in Smad2−/− and Smad3−/− MEFs was similar to that in wild-type MEFs (Fig. 3D). These results argue against a direct role for Smad2/3 in the regulation of ADAM12 expression by Notch.
Up-regulation of ADAM12 by Notch Requires an Active NF-κB Pathway
We observed that the effect of Notch on ADAM12 expression was completely abolished in the presence of MG132, an inhibitor of proteasomal degradation (Fig. 4A). Importantly, MG132 did not impair the canonical Notch signaling, as indicated by a CSL luciferase reporter assay (Fig. 4B). This result suggests that an MG132-dependent step in the regulation of ADAM12 expression by Notch takes place downstream of CSL. Signaling by the nuclear factor-κB (NF-κB) family of transcription factors inherently depends on phosphorylation-induced proteasomal degradation of inhibitors of NF-κB (IκBs) (44, 45). Furthermore, many recent studies uncovered a cross-talk between Notch and NF-κB (Refs. 46–50 and reviewed in Refs. 51 and 52). Therefore, we asked whether up-regulation of ADAM12 expression could be mediated by NF-κB. First, we confirmed that there is a cross-talk between the Notch and NF-κB pathways in NIH3T3 cells. Inhibition of the endogenous Notch signaling in confluent NIH3T3 cultures by DAPT decreased the level of NF-κB activation, as evaluated by the Igκ2-INF luciferase reporter assay (53) (Fig. 4C). Similarly, treatment of CSL+/+ embryonic fibroblasts with DAPT led to a modest, but statistically significant, decrease in the NF-κB activity. DAPT had no effect on the NF-κB reporter in CSL−/− fibroblasts (Fig. 4C), suggesting that the Notch-NF-κB cross-talk is CSL-dependent. Second, BMS-345541, a highly selective inhibitor of IκB kinase, IKK, blocked Notch-induced up-regulation of ADAM12 in a dose-dependent manner (Fig. 4, D and E). Similar results were obtained using Bay 11-7082, another inhibitor of IκB phosphorylation (results not shown). Collectively, these results indicate that ADAM12 up-regulation by Notch requires an active NF-κB pathway.
Up-regulation of ADAM12 Expression by Notch Occurs via Stabilization of Adam12 mRNA
The mouse Adam12 promoter contains several potential NF-κB and CSL binding sites that are conserved between species. It has been reported recently that NF-κB interacts with certain regions of the human ADAM12 promoter (39). Thus, it seemed feasible that Notch might activate transcription of the Adam12 gene, either through stimulation of the NF-κB pathway or via direct binding of CSL to Adam12 promoter. To test this possibility, we performed gene reporter assays using a series of Adam12 promoter constructs comprising various fragments of the −4870/+220 genomic region upstream of the Adam12 translation initiation site (Fig. 5A). We have recently shown that the core Adam12 promoter is located in the −187/+91 region (37). None of the Adam12 promoter constructs were significantly activated by ectopic expression of NICD1 (Fig. 5B). In contrast, a control reporter containing four CSL binding sites in the promoter region was activated severalfold by NICD1, and this activation was abolished by mutating the CSL binding sites (Fig. 5B). To examine the possibility that activation of the Adam12 promoter by CSL occurs only when the promoter is incorporated into chromatin, NIH3T3 cells were stably transfected with pA12Luc.1, the reporter containing the ∼5-kb region upstream of the translation initiation site. Co-culture of stably transfected cells with CHO-Dll1 did not stimulate the activity of the luciferase reporter (Fig. 5C), indicating that Notch signaling does not seem to activate the ∼5-kb promoter region in the mouse Adam12 gene.
To determine whether Notch might regulate Adam12 at the post-transcriptional level, we examined the effect of Notch1 on the stability of Adam12 mRNA. NIH3T3 cells were infected with caNotch1 or control retroviruses; 24 h later, new transcription was blocked by actinomycin D, and Adam12 mRNA levels were measured at 0–8 h after adding the inhibitor. The abundance of NICD1 derived from caNotch1 was similar during the time of measurement of Adam12 mRNA (Fig. 5D). The stability of Adam12 mRNA was significantly higher in cells infected with caNotch1 than in cells infected with control viruses (Fig. 5, D and E), consistent with the post-transcriptional regulation of ADAM12 by Notch.
The Role of MicroRNA-29 in the Up-regulation of ADAM12 Expression by Notch
Post-transcriptional gene regulation is frequently mediated by microRNAs (54). The 3′-UTR of the mouse Adam12 gene contains a well conserved potential target site for the miR-29 family, which includes miR-29a, miR-29b, and miR-29c (Fig. 6A). According to a recent study, miR-29b regulates expression of ADAM12 in human trabecular meshwork cells (55), and at least two family members, miR-29a and miR-29b, are present in NIH3T3 cells (56). Furthermore, expression of the miR-29 appears to be negatively regulated by NF-κB in different cell types (57–59). We then wondered whether ADAM12 expression in NIH3T3 cells is modulated by miR-29 and whether miR-29 may be involved in the regulation of ADAM12 expression by Notch.
First, we examined the levels of miR-29a, miR-29b, and miR-29c in response to perturbations in Notch signaling in NIH3T3 cells. Inhibition of the Notch pathway resulted in the elevation of miR-29a by ∼80% and no significant change in miR-29b or miR-29c (Fig. 6B). Similar effects on miR-29a, miR-29b, and miR-29c were observed in response to inhibition of the NF-κB pathway (Fig. 6B). In contrast, activation of the Notch pathway led to ∼40% decrease in miR-29a, with no change in miR-29c (Fig. 6C). The level of miR-29b after activation of Notch was somewhat elevated, but this effect was not statistically significant. Based on these results, we concluded that miR-29a is the most likely miR-29 family member to regulate ADAM12 expression in response to Notch signaling.
Next, we used gain-of-function and loss-of-function approaches to change the level/activity of miR-29a followed by measuring ADAM12 expression levels. Transfection of miR-29a mimic, a double-stranded RNA oligonucleotide designed to imitate the endogenous miR-29a function, diminished the level of ADAM12 expression by ∼50% when compared with an miRNA mimic negative control (Fig. 6D). In contrast, transfection of miR-29a hairpin inhibitor, a synthetic inhibitor targeting the endogenous miR-29a, increased the level of ADAM12 expression by ∼30% when compared with an miRNA inhibitor negative control, and this effect was statistically significant (Fig. 6E). These results show that in NIH3T3 cells, ADAM12 expression is negatively regulated by miR-29a.
Second, we examined the effect of miR-29a mimic and inhibitor on Notch-mediated up-regulation of ADAM12 expression. When cells were first transfected with miR-29a mimic and then infected with caNotch1 viruses, the induction of ADAM12 was dramatically reduced when compared with the induction observed in the presence of miR mimic negative control (Fig. 7A). When cells were transfected with miR-29a inhibitor, the expression level of ADAM12 level was elevated, and the subsequent expression of caNotch1 did not further elevate ADAM12 expression (Fig. 7B). These results indicate that miR-29a represents an essential component of the pathway by which Notch modulates the expression of ADAM12 in mouse NIH3T3 cells.
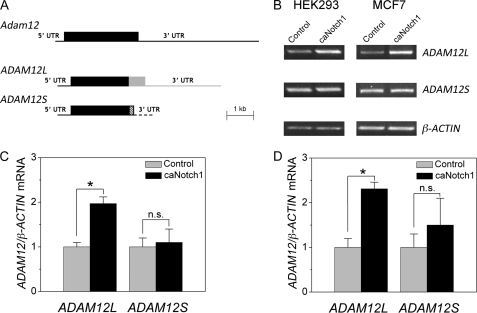
In humans, two isoforms of ADAM12 exist as a result of alternative mRNA splicing: the long transmembrane form (ADAM12L) and the short secreted form (ADAM12S), in which the transmembrane and cytoplasmic domains are replaced with a unique C-terminal sequence. In mice, there is only one form of ADAM12 that corresponds to the human ADAM12L isoform. Importantly, the 3′-UTRs of mouse Adam12 and human ADAM12L mRNAs are ∼90% similar, whereas the 3′-UTR region of human ADAM12S is distinct (Fig. 8A). Because mouse Adam12 is induced by Notch, we asked whether Notch exerts the same effect on human ADAM12 and whether this effect is isoform-specific. As shown in Fig. 8, B–D, expression of caNotch1 in two different human cell lines, HEK293 (human embryonic kidney) and MCF7 (breast cancer), resulted in increased levels of ADAM12L mRNA and did not have a significant effect on ADAM12S mRNA level. This result is consistent with the notion that Notch signaling selectively targets the 3′-UTRs of the mouse Adam12 and human ADAM12L transcripts.
FIGURE 8.
caNotch1 specifically increases the level of ADAM12L splice variant mRNA. A, the diagram of mouse Adam12 transcript and two alternatively spliced human ADAM12 mRNAs, ADAM12L and ADAM12S. The two human variants have the common 5′-UTRs and most of the coding region (black boxes) and divergent sequences in the C-terminal end of the coding region (gray and hatched boxes) and in the 3′-UTRs (gray and hatched lines). B, HEK293 and MCF7 cells were infected with caNotch1 or control retroviruses, and the levels of ADAM12L and ADAM12S were examined by semiquantitative RT-PCR 48 h after infection, using splice variant-specific primer sets; β-ACTIN indicates an internal control. C and D, the intensities of ADAM12L and ADAM12S bands shown in panel B were quantified by densitometry using the ImageJ software. The -fold changes in ADAM12 mRNA, normalized to β-actin, in HEK293 cells (C) and MCF7 cells (D) are shown. The data represent the means ± S.E. from 3 independent experiments. *, p < 0.05, n.s., not significant.
DISCUSSION
Mechanisms regulating ADAM12 expression are not well understood. As many recent studies have consistently found (8, 11–14, 21, 22) elevated levels of ADAM12 in cells with stem-like properties and as Notch activity is often required to maintain the stem-like phenotype, we investigated whether Notch signaling may regulate ADAM12 expression. Our studies demonstrate for the first time that ADAM12 expression is indeed positively regulated by Notch. According to our knowledge, this is the first report of any ADAM whose expression is responsive to Notch signaling.
We find that up-regulation of ADAM12 expression by Notch depends on the CSL transcription factor, a downstream effector of Notch signaling. We also find that stimulation of ADAM12 expression requires an active NF-κB pathway. Using an NF-κB gene reporter assay, we show that Notch up-regulates the basal level of the NF-κB activity in NIH3T3 cells and in CSL+/+, but not in CSL−/− embryonic fibroblasts. Ectopic expression of a constitutively active Notch increases the stability of Adam12 mRNA. Consistent with a recent study for human trabecular meshwork cells (55), we find that ADAM12 expression level is negatively regulated by miR-29 and that miR-29a plays an important role in up-regulation of ADAM12 by Notch. Based on the results presented in this study and reported by others, we propose a model of ADAM12 regulation by Notch shown in Fig. 9.
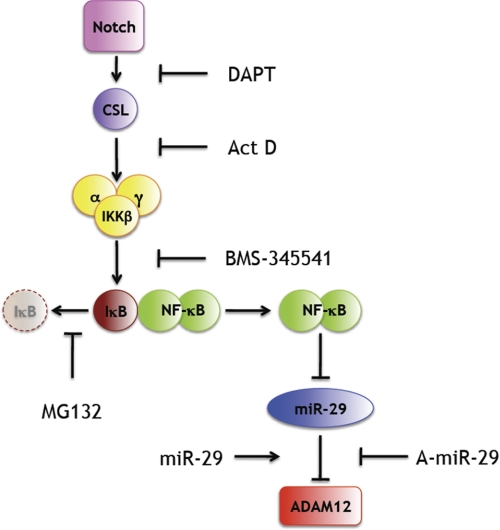
FIGURE 9.
Proposed mechanism of ADAM12 up-regulation by Notch. Notch activates CSL and stimulates IKK activity, leading to phosphorylation and proteasomal degradation of IκB and activation of NF-κB. NF-κB represses the transcription of miR-29, a negative regulator of ADAM12, and thus, increases ADAM12 expression. Points of action of inhibitors or activators used in this study are indicated. For details, see under “Discussion.” Act D, actinomycin D.
The model in Fig. 9 is further supported by several other studies on the signaling along the Notch/CSL/NF-κB/mir-29 axis. Espinosa et al. (50) have recently shown that Notch activation leads to a CSL-mediated expression of Hes1, a transcriptional repressor that blocks expression of the deubiquitinase CYLD. CYLD, in turn, is a negative regulator of IKK (60, 61). Thus, Notch signaling culminates in sustained NF-κB activation (50). In zebrafish embryos, inhibition of Notch signaling by DAPT leads to marked increase of miR-29b, indicating that Notch is a negative regulator of miR-29 (62). Our results further indicate that Notch also suppresses miR-29a in mouse NIH3T3 cells. Finally, activation of NF-κB in various cell types leads to repression of miR-29, as discussed below.
miRNAs are short, ∼22-nucleotide RNAs that bind to 3′-UTRs of mRNAs and promote mRNA degradation or inhibit translation. Targeting of an miRNA to an mRNA requires a 7-nucleotide “seed” sequence (positions 2–8) in the miRNA (54). The miR-29a belongs to the miR-29 family, which consists of miR-29a, -29b, and -29c. These three members are expressed from two bicistronic miRNA clusters; the first cluster contains miR-29a and miR-29b-1, and the second cluster contains miR-29b-2 and miR-29c. The miR-29b-1 and miR-29b-2 precursors are processed to generate the single mature miR-29b. Interestingly, both clusters seem to be repressed by NF-κB (57–59). This explains why blockage of the NF-κB pathway might increase the miR-29 level and inhibit ADAM12 expression, as depicted in Fig. 9. Intriguingly, we find that modulation of the Notch and/or NF-κB pathways in NIH3T3 cells has the most prevalent effect on miR-29a, with few changes in the miR-29b and miR-29c levels. Although this result is somewhat unexpected, differential regulation of individual miRNAs within miRNA clusters has been observed before and has been attributed to differences in the efficiency of post-transcriptional processing of miRNA precursors (63, 64).
All three members of the miR-29 family, i.e. miR-29a/b/c, contain the same seed sequence, and thus, all three may potentially repress ADAM12. Evaluation of the individual levels of miR-29a/b/c after Notch inhibition or activation suggests, however, that miR-29a may play the most important role in the regulation of ADAM12 expression, at least in NIH3T3 cells. Interestingly, studies showed that miR-29b is predominantly localized to the nucleus, where it might regulate gene expression by a different mechanism than RNA-induced silencing (65). In contrast, miR-29a and miR-29c appear to localize to the cytosol, and thus, they are more likely to regulate gene expression at the post-transcriptional level.
We have recently reported that inhibition of protein synthesis in NIH3T3 cells with cycloheximide results in a marked elevation of the Adam12 mRNA level (37). As protein turnover rates for IκB are much higher than the turnover rates for NFκB (44, 45), inhibition of the new protein synthesis leads to imbalance between IκB and NF-κB and preferential accumulation of NF-κB. Thus, an increase in Adam12 mRNA following cycloheximide treatment is fully consistent with the model depicted in Fig. 9.
Several studies demonstrated that ADAM12 expression is induced by TGFβ, via both Smad-dependent and Smad-independent pathways (35–39). Interestingly, ADAM12 induction by TGFβ, similar to the induction by Notch, is completely abolished by MG132 (37, 39). We showed that the effect of MG132 on Smad-dependent induction of ADAM12 is most likely due to the inability to degrade SnoN, a transcriptional repressor (37). On the other hand, the effect of MG132 on Smad-independent induction of ADAM12 may be explained by the inhibition of the NF-κB pathway, as proposed by Ray et al. (39). Interestingly, the latter study proposed that the regulation of ADAM12 expression by NF-κB occurs at the transcriptional level because TGFβ activated human ADAM12 promoter constructs in an NF-κB-dependent manner (39). In contrast, we were not able to detect a significant activation of mouse Adam12 promoter constructs by either TGFβ (37) or Notch (the current study). There are several possible explanations for this apparent discrepancy. The first possibility is that mouse Adam12 and human ADAM12 genes may be regulated by distinct mechanisms. However, Notch exerts different effects on the expression level of ADAM12L and ADAM12S variants, which are under control of the same promoter but utilize different 3′-UTRs. We believe, therefore, that regulation of ADAM12 expression in both mouse and human cells by Notch, and possibly by TGFβ, is mediated in large part by miR-29. Second, our luciferase reporter constructs contained the Adam12 promoter region and the entire 5′-UTR of the Adam12 gene, whereas the constructs used by Ray et al. (39) lacked a major portion of the 5′-UTR of the human ADAM12 gene. It has been reported recently that the 5′-UTR of the human ADAM12 gene contains a highly conserved Z-DNA-forming negative regulatory region that acts as a potent silencer of transcription (66). Whether or not the silencing effect of the 5′-UTR can be abolished by Notch or TGFβ signaling remains to be determined. Third, it is possible that transcriptional activation of the Adam12 gene is mediated by an enhancer that is located outside the genomic region included in our luciferase reporter constructs. Finally, it has to be stressed that the regulation of ADAM12 expression at the post-transcriptional level by miR-29 is not mutually exclusive with the transcriptional regulation of the Adam12 gene.
Barter et al. (38) have recently provided evidence that induction of ADAM12 in mouse C3H10T1/2 fibroblasts by TGFβ via Smad-independent pathways is completely blocked by histone deacetylase (HDAC) inhibitors. They also showed that depletion of HDAC3 by RNA interference significantly diminished TGFβ-induced ADAM12 expression. Interestingly, HDAC3 is also required for NF-κB-mediated repression of the miR-29a/miR-29b-1 cluster (59). Thus, Smad-independent induction of ADAM12 expression by TGFβ may be mediated, at least in part, by repression of miR-29a by NF-κB/HDAC3. Finally, the induction of ADAM12 by TGFβ is partially dependent on phosphatidylinositol 3-kinase/Akt pathway (36, 38). Down-regulation of miR-29 exerts a positive impact on the level of Akt activation (67), which further suggests that NF-κB-mediated repression of miR-29 may play a role in ADAM12 induction by TGFβ. Interestingly, the only other ADAM containing predicted conserved target sites for miR-29 is ADAM19, and this ADAM is also induced by TGFβ (68). It remains to be seen whether miR-29 plays a role in regulation of ADAM19 expression.
In addition to ADAM12, miR-29 represses many other important genes, and its dysregulation is implicated in several pathological conditions. For example, miRNA-29 targets many genes coding extracellular matrix proteins, and its down-regulation plays a key role in cardiac fibrosis (69). miR-29 is up-regulated in type II diabetes, and it represses insulin-stimulated glucose uptake in adipocytes (67). Other targets of miR-29 are DNA methyltransferases (DNMT)3A and -3B (70, 71) and progranulin, a secreted glycoprotein implicated in frontotemporal dementia (72). Finally, miR-29 is down-regulated in hepatocellular carcinoma (73), acute myeloid leukemia (74), and breast cancer (75). Although direct correlation between miR-29 and ADAM12 has not been established for individual breast tumors, miR-29 down-regulation is consistent with ADAM12 up-regulation, which is commonly seen in breast cancer.
Interestingly, post-transcriptional regulation by miR-29 should provide means for differential regulation of ADAM12L and ADAM12S, the transmembrane and secreted forms of ADAM12, respectively, in human tissues. ADAM12S stimulates signaling through the insulin-like growth factor receptor (IGFR) by cleaving IGF-binding protein (IGFBP)-3 and IGFBP-5 and thereby releasing IGF for receptor binding (1). This variant does not have miR-29 target sites in its 3′-UTR, and thus, should not be susceptible to the post-transcriptional regulation by Notch. ADAM12L is the variant that is up-regulated in stem-like cells (8). This variant contains an miR-29 target site in the its 3′-UTR, and thus, its up-regulation in stem-like cells seems consistent with high levels of Notch signaling in these cells. The biological role of ADAM12L in stem-like cells is an urgent and intriguing question that needs to be solved.
Our studies demonstrate for the first time that expression of ADAM12 is responsive to Notch signals. We have shown previously that ADAM12 is capable of shedding Dll1, and thus, modulating Notch signaling via ligand depletion (32). This creates a possibility of a feedback loop that regulates both the status of Notch signaling and the expression of ADAM12 in systems where Dll1 represents the major ligand for Notch. High activity of Notch in one cell could up-regulate ADAM12 expression, and high levels of ADAM12 protein may in turn increase Dll1 shedding and down-regulate Notch in a neighboring cell. Thus, ADAM12 may help establish a heterogeneity in Notch signaling within cell populations. Given the critical role of Notch signaling in the maintenance of mammary stem cells, mammary epithelial differentiation, and mammary oncogenesis and given the fact that ADAM12 is overexpressed and frequently mutated in breast tumors, the link between ADAM12 and Notch should remain in the focus of further studies on the role of ADAM12 in breast cancer.
Supplementary Material
Acknowledgments
We thank Dr. Diane Hayward for the CSL reporter, Drs. Ryoichiro Kageyama and Chiaki Takahashi for the caNotch1 retroviral vector, Dr. Garry P. Nolan for Phoenix Eco and Phoenix Ampho cells, Dr. Tasuku Honjo for OT11 and OT13 cells, Dr. Erwin Bottinger for SMAD2−/− MEFs, Dr. Kathy Flanders for SMAD3−/− MEFs, and Drs. Ulla M. Wewer and Marie Kveiborg for the rb122 antibody.
This work was supported, in whole or in part, by National Institutes of Health Grant CA151065 (to A. Z.). This work was also supported by an Innovative Research Award from Terry C. Johnson Center for Basic Cancer Research at Kansas State University.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1.
- ADAM
- a disintegrin and metalloprotease
- caNotch1
- constitutively active Notch1
- NICD1
- Notch1 intracellular domain
- miRNA
- microRNA
- miR-29
- microRNA 29
- DAPT
- N-[N-(3,5-di-fluorophenacetyl)-l-alanyl]-S-phenylglycine t-butyl ester
- Dll1
- Delta-like 1
- MEF
- mouse embryonic fibroblast
- DMSO
- dimethyl sulfoxide
- IKK
- IκB kinase.
REFERENCES
- 1. Kveiborg M., Albrechtsen R., Couchman J. R., Wewer U. M. (2008) Int. J. Biochem. Cell Biol. 40, 1685–1702 [DOI] [PubMed] [Google Scholar]
- 2. Kveiborg M., Fröhlich C., Albrechtsen R., Tischler V., Dietrich N., Holck P., Kronqvist P., Rank F., Mercurio A. M., Wewer U. M. (2005) Cancer Res. 65, 4754–4761 [DOI] [PubMed] [Google Scholar]
- 3. Lendeckel U., Kohl J., Arndt M., Carl-McGrath S., Donat H., Röcken C. (2005) J. Cancer Res. Clin. Oncol. 131, 41–48 [DOI] [PubMed] [Google Scholar]
- 4. Sjöblom T., Jones S., Wood L. D., Parsons D. W., Lin J., Barber T. D., Mandelker D., Leary R. J., Ptak J., Silliman N., Szabo S., Buckhaults P., Farrell C., Meeh P., Markowitz S. D., Willis J., Dawson D., Willson J. K., Gazdar A. F., Hartigan J., Wu L., Liu C., Parmigiani G., Park B. H., Bachman K. E., Papadopoulos N., Vogelstein B., Kinzler K. W., Velculescu V. E. (2006) Science 314, 268–274 [DOI] [PubMed] [Google Scholar]
- 5. Wood L. D., Parsons D. W., Jones S., Lin J., Sjöblom T., Leary R. J., Shen D., Boca S. M., Barber T., Ptak J., Silliman N., Szabo S., Dezso Z., Ustyanksky V., Nikolskaya T., Nikolsky Y., Karchin R., Wilson P. A., Kaminker J. S., Zhang Z., Croshaw R., Willis J., Dawson D., Shipitsin M., Willson J. K., Sukumar S., Polyak K., Park B. H., Pethiyagoda C. L., Pant P. V., Ballinger D. G., Sparks A. B., Hartigan J., Smith D. R., Suh E., Papadopoulos N., Buckhaults P., Markowitz S. D., Parmigiani G., Kinzler K. W., Velculescu V. E., Vogelstein B. (2007) Science 318, 1108–1113 [DOI] [PubMed] [Google Scholar]
- 6. Dyczynska E., Syta E., Sun D., Zolkiewska A. (2008) Int. J. Cancer 122, 2634–2640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Stingl J., Eirew P., Ricketson I., Shackleton M., Vaillant F., Choi D., Li H. I., Eaves C. J. (2006) Nature 439, 993–997 [DOI] [PubMed] [Google Scholar]
- 8. Dontu G., Abdallah W. M., Foley J. M., Jackson K. W., Clarke M. F., Kawamura M. J., Wicha M. S. (2003) Genes Dev. 17, 1253–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bouras T., Pal B., Vaillant F., Harburg G., Asselin-Labat M. L., Oakes S. R., Lindeman G. J., Visvader J. E. (2008) Cell Stem Cell 3, 429–441 [DOI] [PubMed] [Google Scholar]
- 10. Raouf A., Zhao Y., To K., Stingl J., Delaney A., Barbara M., Iscove N., Jones S., McKinney S., Emerman J., Aparicio S., Marra M., Eaves C. (2008) Cell Stem Cell 3, 109–118 [DOI] [PubMed] [Google Scholar]
- 11. Barberi T., Willis L. M., Socci N. D., Studer L. (2005) PLoS Med. 2, e161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pollard S. M., Wallbank R., Tomlinson S., Grotewold L., Smith A. (2008) Mol. Cell Neurosci. 38, 393–403 [DOI] [PubMed] [Google Scholar]
- 13. Dieguez-Acuña F., Kodama S., Okubo Y., Paz A. C., Gygi S. P., Faustman D. L. (2010) Int. J. Biochem. Cell Biol. 42, 1651–1660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Conigliaro A., Colletti M., Cicchini C., Guerra M. T., Manfredini R., Zini R., Bordoni V., Siepi F., Leopizzi M., Tripodi M., Amicone L. (2008) Cell Death Differ. 15, 123–133 [DOI] [PubMed] [Google Scholar]
- 15. Mareddy S., Dhaliwal N., Crawford R., Xiao Y. (2010) Tissue Eng. Part A 16, 749–758 [DOI] [PubMed] [Google Scholar]
- 16. Kageyama R., Ohtsuka T., Shimojo H., Imayoshi I. (2009) Curr. Opin. Cell Biol. 21, 733–740 [DOI] [PubMed] [Google Scholar]
- 17. Pierfelice T. J., Schreck K. C., Eberhart C. G., Gaiano N. (2008) Cold Spring Harb. Symp. Quant. Biol. 73, 367–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Aguirre A., Rubio M. E., Gallo V. (2010) Nature 467, 323–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Borneman A., Kuschel R., Fujisawa-Sehara A. (2000) J. Muscle Res. Cell Motil. 21, 475–480 [DOI] [PubMed] [Google Scholar]
- 20. Zhao P., Hoffman E. P. (2004) Dev. Dyn. 229, 380–392 [DOI] [PubMed] [Google Scholar]
- 21. Cao Y., Zhao Z., Gruszczynska-Biegala J., Zolkiewska A. (2003) Mol. Cell Biol. 23, 6725–6738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sun D., Li H., Zolkiewska A. (2008) J. Cell Sci. 121, 3815–3823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kopan R., Ilagan M. X. (2009) Cell 137, 216–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gordon W. R., Arnett K. L., Blacklow S. C. (2008) J. Cell Sci. 121, 3109–3119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Brou C., Logeat F., Gupta N., Bessia C., LeBail O., Doedens J. R., Cumano A., Roux P., Black R. A., Israël A. (2000) Mol. Cell 5, 207–216 [DOI] [PubMed] [Google Scholar]
- 26. Mumm J. S., Schroeter E. H., Saxena M. T., Griesemer A., Tian X., Pan D. J., Ray W. J., Kopan R. (2000) Mol. Cell 5, 197–206 [DOI] [PubMed] [Google Scholar]
- 27. Hartmann D., de Strooper B., Serneels L., Craessaerts K., Herreman A., Annaert W., Umans L., Lübke T., Lena Illert A., von Figura K., Saftig P. (2002) Hum. Mol. Genet. 11, 2615–2624 [DOI] [PubMed] [Google Scholar]
- 28. van Tetering G., van Diest P., Verlaan I., van der Wall E., Kopan R., Vooijs M. (2009) J. Biol. Chem. 284, 31018–31027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bozkulak E. C., Weinmaster G. (2009) Mol. Cell Biol. 29, 5679–5695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kovall R. A., Blacklow S. C. (2010) Curr. Top. Dev. Biol. 92, 31–71 [DOI] [PubMed] [Google Scholar]
- 31. Kovall R. A. (2007) Curr. Opin. Struct. Biol. 17, 117–127 [DOI] [PubMed] [Google Scholar]
- 32. Dyczynska E., Sun D., Yi H., Sehara-Fujisawa A., Blobel C. P., Zolkiewska A. (2007) J. Biol. Chem. 282, 436–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ohtsuka T., Ishibashi M., Gradwohl G., Nakanishi S., Guillemot F., Kageyama R. (1999) EMBO J. 18, 2196–2207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cao Y., Kang Q., Zhao Z., Zolkiewska A. (2002) J. Biol. Chem. 277, 26403–26411 [DOI] [PubMed] [Google Scholar]
- 35. Le Pabic H., Bonnier D., Wewer U. M., Coutand A., Musso O., Baffet G., Clément B., Théret N. (2003) Hepatology 37, 1056–1066 [DOI] [PubMed] [Google Scholar]
- 36. Le Pabic H., L'Helgoualc'h A., Coutant A., Wewer U. M., Baffet G., Clément B., Théret N. (2005) J. Hepatol. 43, 1038–1044 [DOI] [PubMed] [Google Scholar]
- 37. Solomon E., Li H., Duhachek Muggy S., Syta E., Zolkiewska A. (2010) J. Biol. Chem. 285, 21969–21977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Barter M. J., Pybus L., Litherland G. J., Rowan A. D., Clark I. M., Edwards D. R., Cawston T. E., Young D. A. (2010) Matrix Biol. 29, 602–612 [DOI] [PubMed] [Google Scholar]
- 39. Ray A., Dhar S., Ray B. K. (2010) Mol. Cancer Res. 8, 1261–1270 [DOI] [PubMed] [Google Scholar]
- 40. Blokzijl A., Dahlqvist C., Reissmann E., Falk A., Moliner A., Lendahl U., Ibáñez C. F. (2003) J. Cell Biol. 163, 723–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sun Y., Lowther W., Kato K., Bianco C., Kenney N., Strizzi L., Raafat D., Hirota M., Khan N. I., Bargo S., Jones B., Salomon D., Callahan R. (2005) Oncogene 24, 5365–5374 [DOI] [PubMed] [Google Scholar]
- 42. Fu Y., Chang A., Chang L., Niessen K., Eapen S., Setiadi A., Karsan A. (2009) J. Biol. Chem. 284, 19452–19462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tang Y., Urs S., Boucher J., Bernaiche T., Venkatesh D., Spicer D. B., Vary C. P., Liaw L. (2010) J. Biol. Chem. 285, 17556–17563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Oeckinghaus A., Ghosh S. (2009) Cold Spring Harb. Perspect. Biol. 1, a000034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. O'Dea E., Hoffmann A. (2010) Cold Spring Harb. Perspect. Biol. 2, a000216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Shin H. M., Minter L. M., Cho O. H., Gottipati S., Fauq A. H., Golde T. E., Sonenshein G. E., Osborne B. A. (2006) EMBO J. 25, 129–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Vilimas T., Mascarenhas J., Palomero T., Mandal M., Buonamici S., Meng F., Thompson B., Spaulding C., Macaroun S., Alegre M. L., Kee B. L., Ferrando A., Miele L., Aifantis I. (2007) Nat. Med. 13, 70–77 [DOI] [PubMed] [Google Scholar]
- 48. Song L. L., Peng Y., Yun J., Rizzo P., Chaturvedi V., Weijzen S., Kast W. M., Stone P. J., Santos L., Loredo A., Lendahl U., Sonenshein G., Osborne B., Qin J. Z., Pannuti A., Nickoloff B. J., Miele L. (2008) Oncogene 27, 5833–5844 [DOI] [PubMed] [Google Scholar]
- 49. Hao L., Rizzo P., Osipo C., Pannuti A., Wyatt D., Cheung L. W., Sonenshein G., Osborne B. A., Miele L. (2010) Oncogene 29, 201–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Espinosa L., Cathelin S., D'Altri T., Trimarchi T., Statnikov A., Guiu J., Rodilla V., Inglés-Esteve J., Nomdedeu J., Bellosillo B., Besses C., Abdel-Wahab O., Kucine N., Sun S. C., Song G., Mullighan C. C., Levine R. L., Rajewsky K., Aifantis I., Bigas A. (2010) Cancer Cell 18, 268–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Aifantis I., Vilimas T., Buonamici S. (2007) Cell Cycle 6, 403–406 [DOI] [PubMed] [Google Scholar]
- 52. Osipo C., Golde T. E., Osborne B. A., Miele L. A. (2008) Lab. Invest. 88, 11–17 [DOI] [PubMed] [Google Scholar]
- 53. Pomerantz J. L., Denny E. M., Baltimore D. (2002) EMBO J. 21, 5184–5194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Fabian M. R., Sonenberg N., Filipowicz W. (2010) Annu. Rev. Biochem. 79, 351–379 [DOI] [PubMed] [Google Scholar]
- 55. Luna C., Li G., Qiu J., Epstein D. L., Gonzalez P. (2009) Mol. Vis. 15, 2488–2497 [PMC free article] [PubMed] [Google Scholar]
- 56. Houbaviy H. B., Murray M. F., Sharp P. A. (2003) Dev. Cell 5, 351–358 [DOI] [PubMed] [Google Scholar]
- 57. Wang H., Garzon R., Sun H., Ladner K. J., Singh R., Dahlman J., Cheng A., Hall B. M., Qualman S. J., Chandler D. S., Croce C. M., Guttridge D. C. (2008) Cancer Cell 14, 369–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Mott J. L., Kurita S., Cazanave S. C., Bronk S. F., Werneburg N. W., Fernandez-Zapico M. E. (2010) J. Cell Biochem. 110, 1155–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Liu S., Wu L. C., Pang J., Santhanam R., Schwind S., Wu Y. Z., Hickey C. J., Yu J., Becker H., Maharry K., Radmacher M. D., Li C., Whitman S. P., Mishra A., Stauffer N., Eiring A. M., Briesewitz R., Baiocchi R. A., Chan K. K., Paschka P., Caligiuri M. A., Byrd J. C., Croce C. M., Bloomfield C. D., Perrotti D., Garzon R., Marcucci G. (2010) Cancer Cell 17, 333–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Skaug B., Jiang X., Chen Z. J. (2009) Annu. Rev. Biochem. 78, 769–796 [DOI] [PubMed] [Google Scholar]
- 61. Wertz I. E., Dixit V. M. (2010) Cold Spring Harb. Perspect. Biol. 2, a003350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Thatcher E. J., Flynt A. S., Li N., Patton J. R., Patton J. G. (2007) Dev. Dyn. 236, 2172–2180 [DOI] [PubMed] [Google Scholar]
- 63. Lu Y., Thomson J. M., Wong H. Y., Hammond S. M., Hogan B. L. (2007) Dev. Biol. 310, 442–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Yu J., Wang F., Yang G. H., Wang F. L., Ma Y. N., Du Z. W., Zhang J. W. (2006) Biochem. Biophys. Res. Commun. 349, 59–68 [DOI] [PubMed] [Google Scholar]
- 65. Hwang H. W., Wentzel E. A., Mendell J. T. (2007) Science 315, 97–100 [DOI] [PubMed] [Google Scholar]
- 66. Ray B. K., Dhar S., Shakya A., Ray A. (2011) Proc. Natl. Acad. Sci. U.S.A. 108, 103–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. He A., Zhu L., Gupta N., Chang Y., Fang F. (2007) Mol. Endocrinol. 21, 2785–2794 [DOI] [PubMed] [Google Scholar]
- 68. Keating D. T., Sadlier D. M., Patricelli A., Smith S. M., Walls D., Egan J. J., Doran P. P. (2006) Respir. Res. 7, 114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. van Rooij E., Sutherland L. B., Thatcher J. E., DiMaio J. M., Naseem R. H., Marshall W. S., Hill J. A., Olson E. N. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 13027–13032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Fabbri M., Garzon R., Cimmino A., Liu Z., Zanesi N., Callegari E., Liu S., Alder H., Costinean S., Fernandez-Cymering C., Volinia S., Guler G., Morrison C. D., Chan K. K., Marcucci G., Calin G. A., Huebner K., Croce C. M. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 15805–15810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Garzon R., Liu S., Fabbri M., Liu Z., Heaphy C. E., Callegari E., Schwind S., Pang J., Yu J., Muthusamy N., Havelange V., Volinia S., Blum W., Rush L. J., Perrotti D., Andreeff M., Bloomfield C. D., Byrd J. C., Chan K., Wu L. C., Croce C. M., Marcucci G. (2009) Blood 113, 6411–6418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Jiao J., Herl L. D., Farese R. V., Gao F. B. (2010) PLoS One 5, e10551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Xiong Y., Fang J. H., Yun J. P., Yang J., Zhang Y., Jia W. H., Zhuang S. M. (2010) Hepatology 51, 836–845 [DOI] [PubMed] [Google Scholar]
- 74. Eyholzer M., Schmid S., Wilkens L., Mueller B. U., Pabst T. (2010) Br. J. Cancer 103, 275–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Iorio M. V., Ferracin M., Liu C. G., Veronese A., Spizzo R., Sabbioni S., Magri E., Pedriali M., Fabbri M., Campiglio M., Ménard S., Palazzo J. P., Rosenberg A., Musiani P., Volinia S., Nenci I., Calin G. A., Querzoli P., Negrini M., Croce C. M. (2005) Cancer Res. 65, 7065–7070 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.