Abstract
The tumor necrosis factor (TNF) superfamily member TNF-like weak inducer of apoptosis (TNFSF12, CD255) (TWEAK) can stimulate apoptosis in certain cancer cells. Previous studies suggest that TWEAK activates cell death indirectly, by inducing TNFα-mediated autocrine signals. However, the underlying death-signaling mechanism has not been directly defined. Consistent with earlier work, TWEAK assembled a proximal signaling complex containing its cognate receptor FN14, the adaptor TRAF2, and cellular inhibitor of apoptosis protein 1 (cIAP1). Neither the death domain adaptor Fas-associated death domain nor the apoptosis-initiating protease caspase-8 associated with this primary complex. Rather, TWEAK induced TNFα secretion and TNF receptor 1-dependent assembly of a death-signaling complex containing receptor-interacting protein 1 (RIP1), FADD, and caspase-8. Knockdown of RIP1 by siRNA prevented TWEAK-induced association of FADD with caspase-8 but not formation of the FN14-TRAF2-cIAP1 complex and inhibited apoptosis activation. Depletion of the RIP1 E3 ubiquitin ligase cIAP1 enhanced assembly of the RIP1-FADD-caspase-8 complex and augmented cell death. Conversely, knockdown of the RIP1 deubiquitinase CYLD inhibited these functions. Depletion of FADD, caspase-8, BID, or BAX and BAK but not RIP3 attenuated TWEAK-induced cell death. Pharmacologic inhibition of the NF-κB pathway or siRNA knockdown of RelA attenuated TWEAK induction of TNFα and association of RIP1 with FADD and caspase-8. These results suggest that TWEAK triggers apoptosis by promoting assembly of a RIP1-FADD-caspse-8 complex via autocrine TNFα-TNFR1 signaling. The proapoptotic activity of TWEAK is modulated by cIAP1 and CYLD and engages both the extrinsic and intrinsic signaling pathways.
Keywords: Apoptosis, Caspase, Receptors, TRAF, Tumor Necrosis Factor (TNF)
Introduction
TWEAK (tumor necrosis factor-like weak inducer of apoptosis, TNFSF12, CD255)2 was initially identified as a novel member of the tumor necrosis factor (TNF) superfamily and has been reported to be capable of inducing cell death in certain cancer cell lines (1, 2). Subsequent studies revealed that TWEAK activates a number of additional cellular functions including proliferation, differentiation, and migration (3–7). TWEAK is expressed in a variety of tissues and by certain types of immune cells and cancer cell lines. The human TWEAK gene encodes a 249-amino acid type II transmembrane protein that is proteolytically processed into a 156-amino acid soluble cytokine. The secreted form is biologically active and contains the receptor-binding domain. The physiological receptor of TWEAK, FN14 (TNFRSF12A, TWEAKR, CD266) (8, 9), was discovered initially as a fibroblast growth factor-induced gene (10). Similarly to TWEAK, FN14 is expressed on a variety of cell types. FN14 is the smallest member of the TNF receptor (TNFR) superfamily. The mature cell surface human FN14 polypeptide is 102 amino acids long, consisting of a single extracellular cysteine-rich domain, a transmembrane region, and a short cytoplasmic tail that contains a TNFR-associated factor (TRAF) binding motif but lacks a canonical death domain. Yeast two-hybrid analysis suggests that TRAF1, 2, 3, and 5 can interact with FN14 (11). Furthermore, mouse embryo fibroblasts with TRAF2 and TRAF5 gene knockouts are deficient in FN14 signaling, demonstrating the importance of these adaptors for TWEAK activity (12). The signaling functions of TWEAK include activation of the canonical and non-canonical NF-κB pathways (13–15), likely via TRAF2 and 5. In vivo, the TWEAK-FN14 system has been implicated in a number of biological functions, including immunomodulation (16), tissue homeostasis and regeneration (17, 18), angiogenesis (19), autoimmune disease (4, 20, 21), control of bone metabolism (22), and skeletal muscle wasting (23).
Two key apoptosis signaling pathways have been identified to date (24–26). The cell-extrinsic pathway is activated upon binding of proapoptotic ligands such as Fas/CD95L (TNFSF6) or Apo2L/TRAIL (TNFSF10) to their cognate death domain-containing receptors on the surface of target cells. This leads to receptor clustering and recruitment FADD and caspase-8 to form the death-inducing signaling complex. FADD binds to death receptors such as Fas/CD95 (TNFRSF6) or DR4 (TNFRSF10A) and DR5 (TNFRSF10B) via death-domain interactions and recruits caspase-8 to the death-inducing signaling complex through death effector domain interactions. In turn, caspase-8 is activated and autocatalytically processed, an event that is augmented by Cullin-3-mediated caspase-8 ubiquitination and aggregation (27). Caspase-8 then activates downstream executioners caspase-3 and caspase-7, triggering the apoptotic demise of the cell. Binding of TNFα (TNFSF2) to TNFR1 (TNFRSF1A) leads to association of a signaling complex in which the TNFR1 death domain recruits the adaptor protein TNFR-associated protein with death domain, TRADD (28–39). TRADD subsequently assembles a signaling complex (complex I) at the plasma membrane, which includes RIP1, TRAF2 or 5, and cellular inhibitor of apoptosis proteins cIAP1 and 2. This complex can activate downstream pathways, including the NF-κB, JNK, and p38 MAPK cascades. Following internalization of TNFR1, posttranslational modifications and conformational changes in RIP1 and TRAF2 lead to their dissociation from TRADD and in turn to association of two additional cytoplasmic complexes (complexes IIA and IIB). TRADD can recruit FADD and caspase-8 (complex IIA) to initiate apoptosis. Dissociated RIP1 also can interact with FADD and caspase-8 (complex IIB), further activating apoptosis.
The intrinsic apoptosis pathway is initiated when BH3-only proteins are activated transcriptionally or posttranslationally in response to intracellular stresses (e.g. DNA damage). BH3 proteins neutralize antiapoptotic Bcl-2 family proteins and/or activate the proapoptotic family members BAX and BAK to induce mitochondrial outer membrane permeabilization (MOMP). Cytochrome C is then released, promoting apoptosome formation with the adaptor Apaf-1 and the initiator protease caspase-9. The apoptosome drives activation of caspase-9 and, consequently, of the executioner proteases caspase-3 and caspase-7. An additional proapoptotic protein released from mitochondria is SMAC/DIABLO, which competes with inhibitor of apoptosis proteins such as XIAP for binding to caspases, reversing their caspase-inhibitory activity. In various cell types, commonly referred to as type II cells, apoptosis commitment in response to cell-extrinsic stimuli requires cross-talk between the extrinsic and intrinsic pathways (25, 38). This cross-talk occurs through caspase-8-mediated processing of the BH3-only protein BID. Truncated BID then engages BAX and BAK to induce MOMP and release cytochrome C and SMAC, further augmenting caspase activation.
The ability of TWEAK to induce apoptosis in certain cells as a single agent or in combination with other cytokines such as IFNγ is intriguing, given the absence of a death domain in FN14. Previous work has revealed that in Kym1 rhabdomyosarcoma cells, the cytotoxic activity of TWEAK is exerted indirectly via induction of TNFα and consequent autocrine action (40). More recent studies suggest that a similar mechanism underlies TWEAK-induced cell death in SKOV3 and OVCAR4 ovarian carcinoma cells (41). While biochemical evidence exists for the assembly of a proximal TWEAK signaling complex consisting of FN14, TRAF2, and cIAP1 (41), the specific intracellular signaling complexes and pathways that trigger apoptosis activation in response to TWEAK have not been directly identified. In this study, we demonstrate that TWEAK stimulates apoptosis via a TNFα-TNFR1-dependent complex containing RIP1, the adaptor FADD, and the apoptosis-initiating protease caspase-8. Furthermore, we show that TWEAK relies on both the extrinsic and intrinsic apoptosis pathways to trigger cell death. These results provide direct molecular insight into the key intracellular biochemical events that mediate apoptosis activation by TWEAK.
EXPERIMENTAL PROCEDURES
Cell Culture and Experimental Reagents
HSC3 (Japanese Collection of Research Bioresources (JCRB) Cell Bank) cells were cultured in DMEM. Culture media were supplemented with 10% FBS (Sigma Aldrich, catalog no. F2442), 2 mm l-glutamine, penicillin (100 units/ml), and streptomycin (100 units/ml) (PenStrep Invitrogen, catalog no. 15140). Antibodies for IB analysis of caspase-8 (9746), caspase-3 (9668), caspase-7 (9494), FN14 (4403), β-actin (4970), TNFR1 (3736S), BAX (2772), BAK (3814), and RelA/p65 (4764S) were from Cell Signaling Technology (Danvers, MA). Antibodies for IB analysis of caspase-8 (804–429-C050) and CYLD (210–910-C050) were from Enzo Life Sciences (Plymouth Meeting, PA). Antibody for IB analysis of caspsase-8 (06–775) was from Millipore (Billerica, MA). Antibodies for IB analysis of TRAF2 (sc-7181 clone H-249) were from Santa Cruz Biotechnology (Santa Cruz, CA). Antibodies for IB analysis of caspase-9 (5511246), TRAF2 (558890), FADD (610399), and TRADD (610573) were from BD Biosciences. Antibody for IB analysis of FLAG (anti-M2 F3165–1MG) was from Sigma Aldrich. Antibodies for IB analysis of cIAP1 (AF8181), RIP1 (MAB3585), and BID (AF846) were from R&D Systems (Minneapolis, MN). Antibodies for IP analysis of caspase-8 (sc-6136 clone C20) and RIP1 (sc-7887 clone H-207) were from Santa Cruz Biotechnology. Antibody for IP of FN14 (4403) was from Cell Signaling Technology. Antibody for IB analysis of RIP3 (IMG-5846A) was from Imgenex Corp. (San Diego, CA). Antibody for IP of TRAF2 (sc-7181 clone H-249) was from Santa Cruz Biotechnology. Control antibody for IP against FGFR3 (sc-123) was from Santa Cruz Biotechnology. Horseradish peroxidase-conjugated anti-mouse IgG heavy chain (115–035-205), anti-mouse IgG light chain (115–035-174), anti-rabbit IgG light chain (211–032-171), and anti-goat (705–035-003) were from Jackson ImmunoResearch Laboratories, Inc. (West Grove, PA). Horseradish peroxidase-conjugated anti-rabbit IgG heavy chain (A1949–1VL) was from Sigma Aldrich. BMS-345541 (B9935), from Sigma-Aldrich, was diluted to 30 mm in dimethyl sulfoxide. FLAG-TWEAK was generated at Genentech (South San Francisco, CA), in which the FLAG sequence (DYKDDDDK) was added to TWEAK (amino acids 98–249) at the N terminus. Recombinant TNFR1:Fc was from Genentech, Inc. Pan-caspase inhibitor zVAD-FMK (FMK001) was from R&D Systems.
siRNA Transfection
The siRNAs were from Dharmacon Research (Lafayette, CO) or Invitrogen. Non-targeting siRNA control (Stealth RNAi Negative Universal Control LO #522887A and HI #435991) and Lipofectamine RNAiMAX (13778–075) were from Invitrogen. RIP1 (A = D-004445–03 and B = D-004445–04), FADD (A = D-003800–01, B = D-003800–02, C = D-003800–04, and D = D-003800–05), TNFR1 (A = D-005197–02 and B = D-005197–03), RelA/p65 (A = J-003533–06, B = J-003533–067, C = J-003533–08, and D = J-003533–09), FN14 (1299003 Invitrogen), cIAP1 (A = D-004390–06 and B = D-004390–07), CYLD (A = D-004609–01 and B = D-004609–04), caspase-8 (A = D-003466–02, B = D-003466–04, and C = D-003466–07), BID (A = D-004387–01, B = D-004387–02, C = D-004387–03, and E = D-004387–05), BAX (A = D-003308–01 and B = D-003308–02), BAK (A = D-003305–01 and B = D-003305–03), TRADD (A = D-004452–01, B = D-004452–02, C = D-004452–03, and D = D-004452–04), caspase-3 (A = D-004307–01, B = D-004307–02, C = D-004307–04, and D = D-004307–05), and caspase-7 (A = D-004407–01, B = D-004407–02, C = D-004407–04, and D = D-004407–05), RIP3 (A = J-003534–05, B = J-003534–06, C = J-003534–07, and D = J-003534–08). FADD (A = D-003800–01 and B = D-003800–05), BID (A = D-004387–01, B = D-004387–02, C = D-004387–03, and D = D-004387–04), BAX (A = D-003308–01), BAK (A = D-003305–01), and FN14 (1299003) were used for IP experiments. siRNAs (10 pmol) were transfected using Lipofectamine RNAiMAX according to the manufacturer's protocol. siRNAs were incubated with HSC3 for 48 h followed by replating into 96-well white assay plates (cell viability or caspase-3/7 assays), 6-well dishes (IB analysis), or 10-cm dishes (IP). For cell viability or caspase-3/7 experiments, cells were treated with TWEAK for 48 h. Assays were performed in triplicate and are reported as mean ± S.E.
Cell Viability and Caspase-3/7 Assays
10,000 HSC3 cells/well were seeded in 96-well white assay plates in 100 ml of DMEM and treated with TWEAK for 48 h. Cell viability was measured using Promega (Madison, WI) CellTiter Glo (G7570). Caspase-3/7 was measured using Promega Caspase-Glo 3/7 assay systems (G8091). Assays were performed in triplicate and are reported as mean ± S.E. Experiments with RIP1, TNFR1, or cIAP1 used a pool of two siRNAs: RIP1 (A and B), TNFR1 (B and C), and cIAP1 (C and D).
MOMP Analysis
The MitoProbe DilC1(5) assay kit for flow cytometry (M34151) was from Invitrogen. HSC3 cells were seeded in 6-well dishes and treated with TWEAK (3 μg/ml) for the indicated times. Cells were removed using cell dissociation buffer (C1419, Sigma Aldrich). Experiments with RIP1 or TNFR1 used a pool of two siRNAs: RIP1 (A and B) and TNFR1 (B and C). MOMP was analyzed according to the manufacturer's protocol. A total of 10,000 events were captured using FACS.
Immunoprecipitation and Immunoblot Analysis
HSC3 cells were seeded overnight in 10-cm dishes and treated with TWEAK (3 μg/ml) for specific amounts of time. When applicable 20 μm zVAD-FMK was added 1 h prior to TWEAK treatment. Cells were collected by centrifugation and lysed at 4 °C in 20 mm TRIS-HCl buffer (pH 7.5) containing 150 mm NaCl, 2 mm EDTA, 10% glycerol, 1% Triton X-100, 0.57 mm PMSF, and 1× complete EDTA-free protease inhibitor mixture tablet (11873580001, Roche). Cells lysates (2 mg of protein) were incubated with specific IP antibodies as listed above overnight at 4 °C. The following day, 30 μl of bead volume of protein A/G-agarose (Thermo Scientific, catalog no. 20421) was added for 2 h at room temperature, and the beads were washed four times with lysis buffer. Novex Tris-glycine SDS sample buffer (1×) (Invitrogen) was added, and the beads were boiled for 5 min at 95 °C. Lysates for siRNA experiments, caspase-8, caspase-9, caspase-3, caspase-7, TRAF2, and cIAP1 time-course experiments were generated using 1× radioimmune precipitation assay lysis buffer (20–188, Millipore, Billerica, MA) according to the manufacturer's protocol. Novex Tris-glycine SDS sample buffer (1×) was added, and the lysates were boiled for 5 min at 95 °C. Samples were analyzed by SDS-PAGE and detected by IB with specific antibodies as listed above. Experiments were repeated at least twice. IP experiments with siRNA used a pool of two oligos: FADD (A and D), RIP1 (A and B), TNFR1 (B and C), cIAP1 (C and D), CYLD (A and D), BID (A and B), and Bax (A)/Bak (B) following the transfection protocol as stated above.
Caspase-8 Activity
Cells seeded in 10-cm dishes were treated with TWEAK (3 μg/ml) for 6 h in the absence or presence of zVAD-FMK (20 μm). Caspase-8 or RIP1 was immunoprecipitated as described in the previous section. After washing, the protein A/G beads were resuspended in 90 μl of PBS and added to 96-well white plates. Then 90 μl of Caspase-8 Glo Promega (G8201) was added to measure activity.
ELISA
Cells were seeded in 10-cm dishes and treated with TWEAK (3 μg/ml). Conditioned medium was collected for the indicated times and stored at −80°C. TNFα ELISA (DTA00C) from R&D Systems was performed according to the manufacturer's protocol. Assays were performed in duplicate and are reported as mean ± S.D.
TNFα mRNA Analysis
Cells were seeded in 10-cm dishes and treated with TWEAK (3 μg/ml) for the indicated times. RNA was purified using the RNAeasy kit from Qiagen (74104). RT-PCR was performed using the cDNA RT kit (Applied Biosystems, Carlsbad, CA). Transcript levels of TNFα were measured (HS00174128_m1, Applied Biosystems), and the comparative CT method was used to determine the fold change in expression.
RESULTS
TWEAK Induces Caspase-dependent Apoptosis in HSC3 and HEC1A Cells
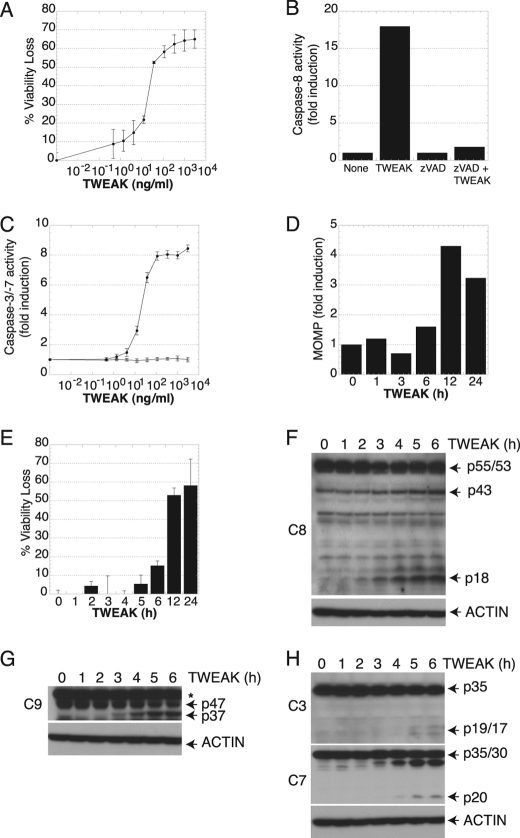
To define the molecular events that mediate cell death induction by TWEAK, we purposely selected cell lines that were previously shown to be sensitive to TWEAK in the absence of any added sensitizing agents such as IFNγ or cycloheximide, namely, human oral squamous carcinoma HSC3 and human endometrial carcinoma HEC1A cells (42–44). Treatment of HSC3 and HEC1A cells with TWEAK induced a substantial loss of cell viability in a dose-dependent fashion, with half-maximal activity at 10–100 ng/ml (Fig. 1A and supplemental Fig. S1). HSC3 cell death was accompanied by activation of the initiator caspase-8 and executioners caspase-3 and caspase-7, and caspase activation could be blocked by the pan-caspase inhibitor zVAD-FMK (Fig. 1, B and C), consistent with induction of apoptosis. Kinetic analysis showed that TWEAK caused a significant loss of mitochondrial membrane integrity by 6–12 h (Fig. 1D), further confirming the apoptotic nature of the effect of TWEAK. Kinetic analysis of cell death indicated substantial loss of cell viability by 6–12 h after TWEAK addition (Fig. 1E), in keeping with the time frame of MOMP induction.
FIGURE 1.
TWEAK activates apoptosis in HSC3 cells. A, cells were treated with the indicated concentrations of TWEAK for 48 h and assayed for viability. B, after cells were treated with TWEAK (3 μg/ml, 6 h), caspase-8 was subjected to IP and assayed for activity. C, cells were treated with the indicated concentrations of TWEAK for 48 h in the absence (●) or presence (○) of the pan-caspase inhibitor zVAD-FMK (20 μm) and assayed for caspase-3/7 activity. D, cells were treated with TWEAK (3 μg/ml) for the indicated number of hours and assayed for MOMP or viability (E). F–H, cells were treated with TWEAK (10 μg/ml) for the indicated number of hours and subjected to IB analysis with antibodies specific to caspase-8 (F), caspase-9 (G), or caspase-3 and caspase-7 (H). The asterisk indicates a nonspecific band.
To interrogate the activation of caspases in response to TWEAK, we examined processing events by IB analysis with caspase-specific antibodies. Caspase-8 is the key initiator protease in the cell-extrinsic pathway (24, 45). TWEAK augmented processing of caspase-8 and generation of its active p18 fragment by 2–3 h (Fig. 1F). Caspase-8 processing was associated with depletion of the caspase-8 substrate BID, which was detectable by 3 h (supplemental Fig. S2), confirming caspase-8 activation. Caspase-9 is the key initiator protease in the cell-intrinsic apoptosis pathway (24, 45). TWEAK stimulated detectable processing of caspase-9, indicated by generation of the active p37 fragment by 3–4 h (Fig. 1G) (46). Furthermore, TWEAK induced processing of the effectors caspase-3 and caspase-7 by 5 h, evident by appearance of the respective p19/17 and p18 fragments of these enzymes (Fig. 1H). These results suggested that TWEAK engages key initiator caspases of both the extrinsic and intrinsic pathways, leading to downstream activation of executioner caspases.
TWEAK Assembles a Proximal Signaling Complex Consisting of FN14, TRAF2, and cIAP1, Leading to Depletion of TRAF2 and cIAP1
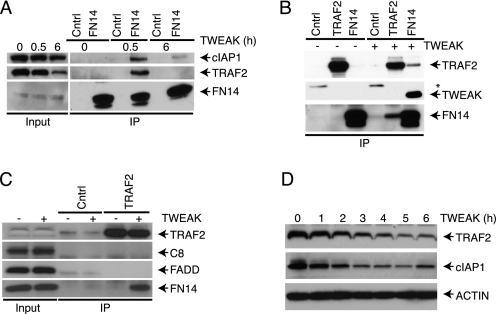
Recent work has suggested that TRAF2 and cIAP1 may play a role in regulating apoptosis stimulation by TNFα, SMAC mimetics, and perhaps TWEAK (39, 47, 48). To confirm that TWEAK engages TRAF2 and cIAP1, we tested for FN14 association with specific signaling components in response to TWEAK by FN14 IP followed by IB analysis. IP of FN14 revealed a TWEAK-dependent, specific interaction with TRAF2 and cIAP1 by 30 min (Fig. 2A). Reciprocal IP of FN14 or TRAF2 confirmed this TWEAK-induced association between FN14 and TRAF2 (Fig. 2B). TWEAK similarly induced the interaction of FN14 with TRAF2 and cIAP1 in HEC1A cells (supplemental Fig. S3). In contrast to TRAF2 and cIAP1, neither FADD nor caspase-8 showed significant association with FN14 in response to TWEAK (Fig. 2C). In keeping with previous data (41), TWEAK stimulation also led to depletion of cIAP1 and TRAF2 over a period of 3–6 h (Fig. 2D), suggesting degradation of these proteins.
FIGURE 2.
TWEAK assembles a proximal signaling complex containing FN14, TRAF2, and cIAP1. A, cells were treated with TWEAK (3 μg/ml) for the indicated amount of time and subjected to IP of FN14 followed by IB with antibodies to FN14, TRAF2, and cIAP1. B, cells were treated with TWEAK (3 μg/ml, 0.5 h) and subjected to IP of FN14 or TRAF2, followed by IB analysis of FN14 and TRAF2. C, cells were treated with TWEAK (3 μg/ml, 0.5 h) and subjected to IP of TRAF2 followed by IB of FN14, FADD, caspase-8, or TRAF2. D, cells were treated with TWEAK (3 μg/ml) for the indicated time and subjected to IB analysis of TRAF2 and cIAP1. The asterisk indicates a nonspecific band.
TWEAK Induces Assembly of a Death-signaling Complex Containing RIP1, FADD, and Caspase-8
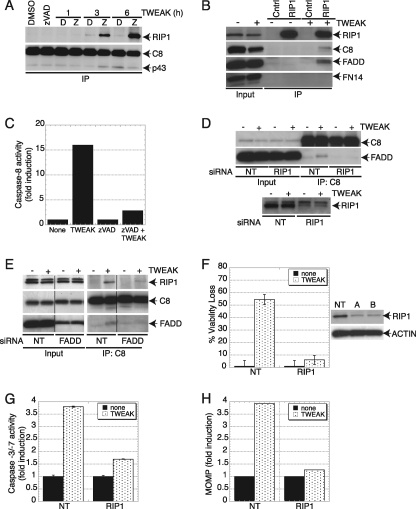
RIP1 plays a key role in NF-κB activation by TNFα (49), although recent work with mouse embryonic fibroblasts suggests that the RIP1 gene is not essential for TNFα-induced NF-κB activation in this latter cell type (50). RIP1 also has been implicated in apoptosis activation by TNFα, particularly in combination with cycloheximide or SMAC mimetics (39, 48). Caspase-8 IP revealed TWEAK-driven association of RIP1 with caspase-8, which was detected more clearly by addition of zVAD-FMK during TWEAK stimulation (Fig. 3A). Reciprocal IP through RIP1 confirmed the association of RIP1 with caspase-8 in response to TWEAK and further revealed the presence of FADD in this complex (Fig. 3B). Importantly, we detected substantial caspase-8 activity in RIP1 IPs from TWEAK-stimulated cells (Fig. 3C), demonstrating that caspase-8 association with RIP1 leads to its activation. We observed a similar TWEAK-induced interaction of RIP1 and caspase-8 in HEC1A cells (supplemental Fig. S4). Knockdown of FN14 by siRNA prevented the association of RIP1 and caspase-8 (supplemental Fig. S5), verifying that TWEAK induces this complex by acting through FN14. Moreover, depletion of RIP1 by siRNA disrupted the interaction of caspase-8 with FADD (Fig. 3D), indicating that RIP1 acts upstream of this association. Similarly, siRNA knockdown of FADD also inhibited the interaction of RIP1 with caspase-8 (Fig. 3E), suggesting that FADD acts as the adaptor that links RIP1 and caspase-8, likely by binding to RIP1 through death domain interactions and to caspase-8 via death effector-domain association. Knockdown of RIP1 inhibited TWEAK-induced cell death to a similar extent as depletion of FN14 (Fig. 3F and data not shown), suggesting that RIP1 plays a critical role in transmission of the apoptotic signal. Indeed, RIP1 depletion also inhibited activation of caspases-3/7 and induction of MOMP in response to TWEAK (Fig. 3, G and H), confirming an important role for RIP1 in TWEAK-induced apoptosis. The adaptor TRADD also can recruit FADD and caspase-8 to enhance TNFR1-mediated apoptosis (37–39). However, siRNA knockdown of TRADD, which led to substantial TRADD protein depletion, did not inhibit TWEAK-induced apoptosis (supplemental Fig. S6), nor was TRADD detected in the proximal or secondary complexes induced by TWEAK (data not shown), indicating that TRADD is dispensable for apoptosis activation by TWEAK. This finding is in concert with reports suggesting that RIP1 rather than TRADD is the primary mediator of apoptosis induction by TNFα plus SMAC mimetics (39, 48).
FIGURE 3.
TWEAK induces association of a death-signaling complex containing RIP1, FADD, and caspase-8, leading to caspase-8 activation. A, cells were treated with TWEAK (3 μg/ml) for the indicated time in the absence (D-dimethyl sulfoxide) or presence of zVAD-FMK and subjected to IP of caspase-8 followed by IB analysis of caspase-8 and RIP1. B, cells were treated with TWEAK (3 μg/ml, 6 h) and subjected to IP of RIP1 followed by IB analysis of FN14, FADD, caspase-8, or RIP1. C, cells were treated with TWEAK (3 μg/ml, 6 h), RIP1 was immunoprecipitated from cell lysates, and IPs were assayed for caspase-8 activity. D, cells were transfected with two siRNA oligonucleotides against RIP1 for 48 h, treated with TWEAK (3 μg/ml, 6 h) and subjected to IP of caspase-8 followed by IB analysis of FADD or caspase-8. E, cells were transfected with siRNA against FADD for 48 h, treated with TWEAK (3 μg/ml, 6 h), and subjected to IP of caspase-8 followed by IB analysis of FADD, caspase-8, or RIP1. F–H, cells were transfected with RIP1 siRNA as in D, treated with TWEAK for 48 h, and then analyzed for viability (F), caspase-3/7 activity (G), or MOMP (H). In F, IB analysis shows siRNA knockdown levels. For the IP experiments, zVAD-FMK (20 μm) was added 1 h prior to incubation with TWEAK. A vertical line denotes lane removal from the original image.
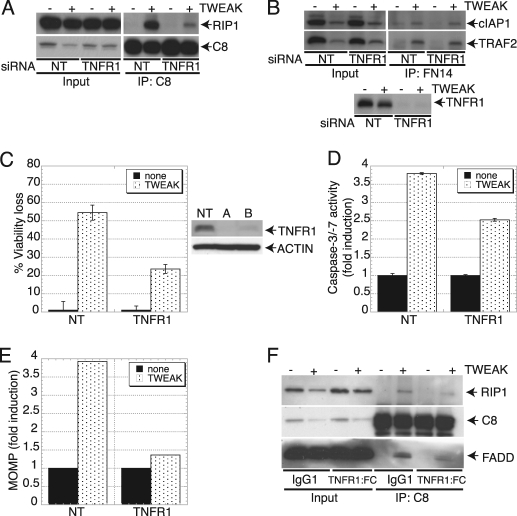
Assembly of the Death-signaling Complex by TWEAK Requires TNFR1
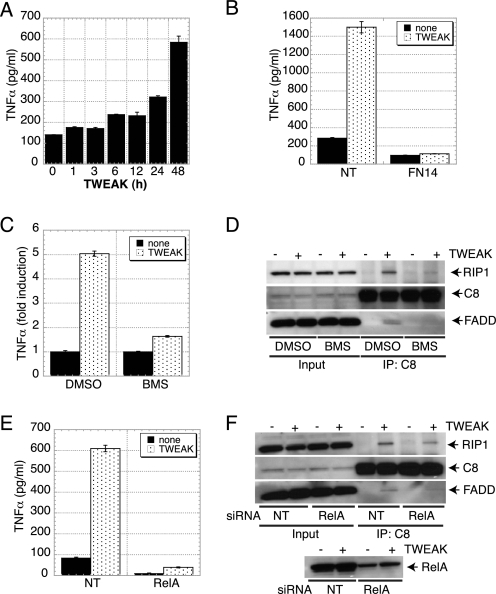
Given the role of RIP1 in TNFα signaling (28–33) and our evidence that RIP1 is important for apoptosis activation by TWEAK (Fig. 3, F–H), we reasoned that TWEAK might engage RIP1 via a TNFα-TNFR1 signaling mechanism. Consistent with this notion, TWEAK induced secretion of TNFα protein, which was detectable as early as 6 h and continued to increase through 48 h (Fig. 4A). Additionally, TWEAK stimulated a measurable increase in TNFα mRNA by 3 h (supplemental Fig. S8). Knockdown of FN14 by siRNA blocked the induction of TNFα (Fig. 4B), confirming that this effect was based on a specific TWEAK interaction with FN14. Furthermore, treatment of HSC3 cells with the NF-κB pathway inhibitor BMS-345541 or siRNA knockdown of the NF-κB subunit RelA attenuated TNFα production and association of the RIP1-FADD-caspase-8 complex (Fig. 4, C–F). These results indicate that TWEAK begins to induce TNFα mRNA transcription and protein production within a time frame that precedes or overlaps caspase activation and apoptosis (Fig. 1, D and E).
FIGURE 4.
TWEAK induces TNFα and inhibiting NF-κB abrogates formation of the proapoptotic complex between RIP1-FADD-C8. A, cells were incubated with TWEAK (3 μg/ml) for the indicated amount of time, and cell supernatants were analyzed with a TNFα-specific ELISA. B, cells were transfected with FN14 siRNA for 48 h, incubated with TWEAK (3 μg/ml) for 48 h, and analyzed for TNFα secretion as in A. C, HSC3 cells were treated with BMS-345541 (30 μm) in the absence or presence of TWEAK (3 μg/ml) for 24 h. Cell supernatants were analyzed with a TNFα-specific ELISA. D, cells were treated with TWEAK (3 μg/ml) for 6 h in the presence of zVAD-FMK and BMS-345541. Subsequently, caspase-8 was immunoprecipitated from whole cell lysates followed by IP of RIP1, FADD, and caspase-8. E, HSC3 cells were transfected with two siRNA oligonucleotides against RelA for 48 h and then treated with TWEAK (3 μg/ml, 24 h). Cell supernatants were analyzed with a TNFα-specific ELISA. F, cells transfected with RelA siRNA were treated with TWEAK (3 μg/ml) for 6 h in presence of zVAD-FMK (20 μm), and caspase-8 was immunoprecipitated followed by IB analysis of RIP1, FADD, caspase-8, and RelA.
Upon recruitment by TNFR1 in response to TNFα stimulation, RIP1 undergoes ubiquitination-based modification events that promote NF-κB activation (31–33). A key E3 ubiquitin ligase involved in the ubiquitination of RIP1 is cIAP1 (51). Conversely, further evidence indicates that deubiquitination of RIP1 by the ubiquitin hydrolase CYLD (cylindromatosis) promotes dissociation of RIP1 from TNFR1, allowing RIP1 to recruit and activate caspase-8 via FADD and thereby trigger apoptosis (39). To examine the importance of TNFR1 for TWEAK-induced apoptosis, we first tested the effect of TNFR1 siRNA. TNFR1 knockdown inhibited the association of RIP1 with caspase-8 in response to TWEAK, as indicated by caspase-8 IP (Fig. 5A). In contrast, TNFR1 depletion did not affect the assembly of the proximal FN14-TRAF2-cIAP1 TWEAK signaling complex, as shown by FN14 IP (Fig. 5B). Importantly, siRNA silencing of TNFR1 protected cells from TWEAK-induced death (Fig. 5C) in conjunction with inhibiting caspase-3/7 activation and MOMP (D and E). Furthermore, addition of a TNFR1:Fc fusion protein to block TNFα activity disrupted the ability of TWEAK to stimulate interaction between RIP1, FADD, and caspase-8 (Fig. 5F). These results indicate that TWEAK induces the assembly of a death-inducing RIP1-FADD-caspase-8 complex via autocrine TNFα-TNFR1 signaling.
FIGURE 5.
TWEAK induces TNFα and drives TNFR1-dependent apoptosis signaling. A, cells were transfected with TNFR1 siRNA for 48 h, stimulated with TWEAK (3 μg/ml, 6 h), and association of RIP1 and caspase-8 was analyzed by caspase-8 IP followed by immunoblot analysis as indicated. B, cells were transfected with TNFR1 siRNA and treated with TWEAK as in A, and assembly of the FN14-TRAF2-cIAP1 complex was examined by FN14 IP followed by immunoblot analysis as indicated. C–E, cells were transfected with TNFR1 siRNA, treated with TWEAK as in A, and analyzed for viability (C), caspase-3/7 activity (D), and MOMP (E). In B, IB analysis shows siRNA knockdown levels. For the IP experiments, zVAD-FMK (20 μm) was added 1 h prior to incubation with TWEAK. F, HSC3 cells were treated with TWEAK (3 μg/ml) for 6 h in the presence of zVAD-FMK (20 μm) and TNFR1:FC (100 μg/ml). Cell lysates were immunoprecipitated with caspase-8 followed by IB analysis of RIP1, FADD, and caspase-8.
Recent evidence suggests an essential role for RIP1 and its related kinase RIP3 in the induction of programmed necrosis or necroptosis in response to TNFα (52). Knockdown of RIP3 by siRNA did not attenuate TWEAK-induced cell death (supplemental Fig. S9), suggesting that the cytotoxicity of TWEAK was not mediated by necroptosis.
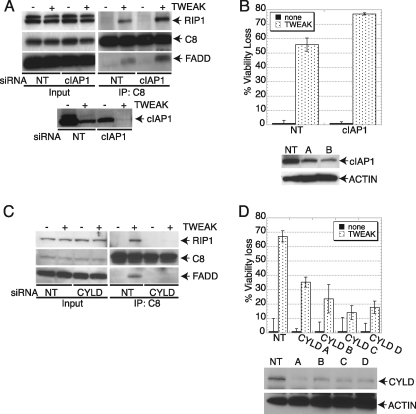
Next, we tested whether cIAP1 and CYLD may impact the ability of RIP1 to interact with FADD and caspase-8 in response to TWEAK. Depletion of cIAP1 by siRNA enhanced the association of RIP1 with FADD and caspase-8 in response to TWEAK (Fig. 6A), augmenting apoptosis stimulation (B). In contrast, siRNA knockdown of CYLD disrupted TWEAK-induced assembly of the RIP1-FADD-caspase-8 complex (Fig. 6C), attenuated apoptosis stimulation (D), and decreased TNFα production (supplemental Fig. S10). Taken together, these results suggest that TWEAK induces apoptosis by driving TNFα-TNFR1-mediated engagement of RIP1 to recruit FADD and caspase-8 in a manner that is negatively controlled by cIAP1 and positively modulated by CYLD.
FIGURE 6.
Modulation of TWEAK-induced apoptosis by cIAP1 and CYLD. A, cells were transfected with cIAP1 siRNA for 48 h and stimulated with TWEAK (3 μg/ml, 6 h), and association of RIP1 and caspase-8 was analyzed by caspase-8 IP followed by IB analysis of FADD, caspase-8, cIAP1, or RIP1. B, cells were transfected with cIAP1 siRNA for 48 h, stimulated with TWEAK (3 μg/ml, 48 h), and then analyzed for viability. IB analysis shows siRNA knockdown levels. C, cells were transfected with CYLD siRNA for 48 h, stimulated with TWEAK (3 μg/ml, 6 h), and then association of RIP1 and caspase-8 was analyzed by caspase-8 IP followed by IB analysis of FADD, caspase-8, or RIP1. D, cells were transfected with CYLD siRNA for 48 h, stimulated with TWEAK (3 μg/ml, 48 h), and then analyzed for viability. IB analysis shows siRNA knockdown levels.
TWEAK Engages Both the Extrinsic and Intrinsic Apoptosis Pathways
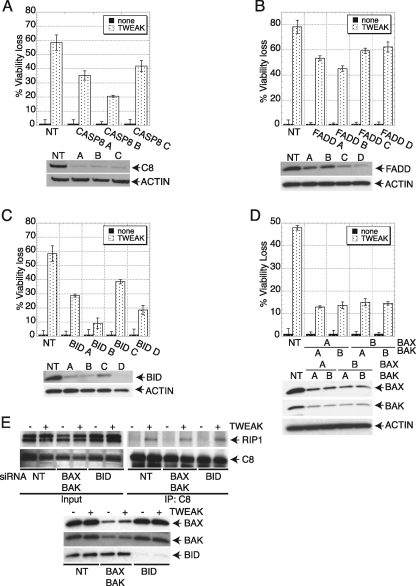
Although our findings indicated that TWEAK activates caspase-8, a key initiator caspase in the extrinsic pathway, the significant induction of MOMP in response to TWEAK (Figs. 1D, 3H, and 5G), together with the evidence for caspase-9 processing (Fig. 1G) and the well documented evidence for cross-talk between extrinsic and intrinsic apoptosis signals, suggested that TWEAK may engage both pathways. Knockdown of caspase-8 or FADD afforded partial protection against TWEAK-induced cell death (Fig. 7, A and B), confirming extrinsic pathway involvement. Moreover, depletion of BID by siRNA also gave partial protection against TWEAK-induced death (Fig. 7C), suggesting that cross-talk to the intrinsic pathway contributed to this function. Indeed, siRNA knockdown of BAX and BAK also inhibited TWEAK-induced cell death (Fig. 7D), further supporting the latter conclusion. Finally, siRNA depletion of caspase-3 or caspase-7 also provided protection against TWEAK-induced cell death (supplemental Fig. S7, A and B), confirming the apoptotic nature of this response. Although siRNA knockdown of both extrinsic and intrinsic pathway components inhibited TWEAK-induced cell death, knockdown of BAX and BAK or BID did not prevent assembly of the RIPK1-FADD-caspase-8 complex (Fig. 7E), supporting the notion that this complex acts upstream of intrinsic pathway engagement via BID processing by caspase-8. These data suggest that, at least in HSC3 cells, TWEAK relies on both the extrinsic and intrinsic pathways to induce apoptosis via type II signaling.
FIGURE 7.
TWEAK requires both the extrinsic and intrinsic apoptosis signaling pathways to induce cell death. A–D, cells were transfected with siRNAs oligonucleotides against caspase-8 (A), FADD (B), BID (C), or BAX and BAK (D) for 48 h, stimulated with TWEAK (3 μg/ml, 6 h), and then analyzed for cell viability. In A–D, IB analysis shows siRNA knockdown levels. E, cells were transfected with siRNA against BAX and BAK or BID for 48 h, stimulated with TWEAK (3 μg/ml, 6 h), and subjected to caspase-8 IP followed by IB analysis of BID, BAX, BAK, caspase-8, or RIP1.
DISCUSSION
TWEAK is capable of inducing a number of diverse cell functions, including proliferation, differentiation, migration, and cell death, depending on cellular context (3, 5, 7). The ability of TWEAK to induce apoptosis in certain cells has been perplexing, given that its cognate receptor, FN14, lacks a cytoplasmic death domain. Studies in Kym1 cells (40) and subsequent work with the SKOV3 and OVCAR4 cell lines (41) indicated that the cell death-inducing activity of TWEAK is exerted indirectly via autocrine TNFα signaling. However, the specific signaling complexes and pathways that transmit the death signal in response to TWEAK have not been previously defined. To further elucidate the mechanisms underlying the cytotoxic action of TWEAK, we chose as our main model a cell line that is sensitive to TWEAK as a single agent, without requirement for exogenous sensitizing factors such as IFNγ or cycloheximide, namely human squamous cell carcinoma HSC3 cells (42, 43). We further verified some of our key observations in human endometrial carcinoma HEC1A cells. TWEAK stimulation led to substantial activation of apoptotic cell death, characterized by increased processing of the initiator proteases caspase-8 and caspase-9, induction of MOMP, and activation of the executioner proteases caspase-3 and caspase-7. Biochemical studies showed that TWEAK stimulated the assembly of a proximal signaling complex at the level of its cognate receptor FN14, which recruited the adaptor TRAF2 and the E3 ubiquitin ligase cIAP1, previously implicated in ubiquitination of RIP1 (51). Neither FADD nor caspase-8 were recruited to this proximal complex. However, both TRAF2 and cIAP1 levels decreased within a few hours after TWEAK addition, suggesting degradation of these proteins, which is consistent with earlier data (41). TWEAK stimulation led to secretion of TNFα, likely through activation of NF-κB signaling via the proximal FN14-TRAF2-cIAP1 complex (see below), in keeping with earlier observations in other cell types (41). Indeed, pharmacologic inhibition of the NF-κB pathway or siRNA depletion of the NF-κB subunit RelA attenuated TWEAK-induced TNFα production and consequent cell death signaling. TWEAK induced TNFα-TNFR1-mediated assembly of a death-signaling complex containing RIP1, FADD, and caspase-8, leading to caspase-8 activation and subsequent apoptosis. Association of the second complex was dependent on RIP1, as well as on FADD. This complex appeared to be critical for apoptosis activation in response to TWEAK because its disruption by siRNA knockdown of TNFR1 or RIP1 blocked apoptosis activation. In contrast, TRADD was not involved in association with FADD and caspase-8 in response to TWEAK, nor was it required for TWEAK-induced cell death. RIP3 also was dispensable for the latter response, consistent with apoptotic rather than necroptotic cell death in response to TWEAK.
TWEAK stimulation led to processing of initiator caspase-8 as well as caspase-9, suggesting that both the extrinsic and intrinsic apoptosis pathways participate in a “type II” signaling in response to TWEAK. Indeed, siRNA knockdown of BID, which plays a critical role in mediating cross-talk between the extrinsic and intrinsic pathways, attenuated TWEAK-induced cell death, as did depletion of BAX and BAK, which operate downstream of BID.
The consequences of TWEAK-dependent activation of the proximal FN14-TRAF2-cIAP1 complex may be mechanistically similar to the effect of pharmacological IAP antagonists, which lead to depletion of cIAPs by inducing their autoubiquitination and consequent proteasomal degradation (39, 48, 53). Recent work with small molecule IAP antagonists demonstrates that cIAPs are critical modulators of NF-κB activation by TNFR superfamily members such as TNFR1 (39) or FN14 (41). These receptors recruit cIAP1/2 via TRAF2 to activate the classical NF-κB pathway or inhibit the alternative NF-κB pathway. In the classical pathway, p50-RelA NF-κB heterodimers are kept in the cytoplasm through interaction with IκB. Upon stimulation (e.g. binding of TNFα to TNFR1), TRAF2 and cIAP1/2 are recruited to the receptor. In turn, the cIAPs, which possess RING-based E3 ubiquitin ligase activity, ubiquitinate RIP1, augmenting its recruitment to the receptor complex (51). Ubiquitinated RIP1 then engages the IκB kinase complex by binding to the transforming growth factor-β-activated kinase 1 (TAK1) via TAK1-binding proteins 1 and 2 (TAB1/2) (54). IκB kinase phosphorylates IκB, targeting this protein for ubiquitination and consequent proteasomal degradation (29, 41, 55). Once IκB is depleted, NF-κB is free to translocate to the nucleus to activate the transcription of target genes. Simultaneously, TRAF2 and the cIAPs repress the alternative pathway by ubiquitinating NF-κB-inducing kinase and targeting it for degradation (56). However, loss of cIAPs (e.g. as induced by SMAC mimetics) stabilizes NF-κB-inducing kinase, increasing alternative NF-κB pathway activation and transcription of various target genes including TNFα. Although the driving signal is yet to be identified, CYLD deubiquitinates RIP1, releasing it from TNFR1 and allowing it to recruit and activate caspase-8 via FADD. CYLD can indirectly associate with its targets through adaptor proteins such as p62 or NEMO (57). Our data suggest that TWEAK may use a similar mechanism, causing degradation of cIAPs via FN14 and promoting TNFα secretion, thus enhancing apoptosis activation via TNFR1. Consistent with this hypothesis, there is published evidence that TWEAK can activate the NF-κB pathway (14). Tweak-induced assembly of the RIP1-FADD-caspase-8 complex, which is augmented in the absence of cIAP1 and attenuated by depletion of CYLD, plays a critical role in transmitting the apoptotic signal; and cross-talk between the extrinsic and intrinsic pathways through BID processing reinforces stimulation of caspase activity by driving cell death. Our present results support the conclusion that TWEAK triggers apoptosis via TNFα-TNFR1 signaling and elucidate the key death-signaling molecular events mediating this activity. Small molecule IAP antagonists are currently being developed as novel anticancer therapeutics (48, 58, 59). Recombinant soluble TWEAK or agonistic FN14 antibodies may provide protein-based alternatives or complements to these agents, with similar mechanisms of action yet distinct pharmacology and target specificity (60).
Supplementary Material
Acknowledgments
We thank Dylan Daniel, Becky Yang, and Rena Wang for helping with initiation of these studies, Scot Marsters for generation of recombinant soluble TWEAK, and members of the Ashkenazi and Vucic laboratories for useful discussions.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S10.
- TWEAK
- tumor necrosis factor-like weak inducer of apoptosis
- TNF
- tumor necrosis factor
- TNFR
- tumor necrosis factor receptor
- TRAF
- tumor necrosis factor receptor-associated factor
- cIAP
- cellular inhibitor of apoptosis protein
- MOMP
- mitochondrial outer membrane permeabilization
- IB
- immunoblot
- IP
- immunoprecipitation
- RIP1
- receptor-interacting protein 1
- FADD
- Fas-associated death domain
- CYLD
- cylindromatosis
- BID
- BH3 interacting domain death agonist
- Apo2L/TRAIL
- Apo2 ligand or TNF-related apoptosis-inducing ligand
- TRADD
- TNFR-associated protein with death domain
- SMAC/DIABLO
- second mitochondria-derived activator of caspase or direct IAP binding protein with low pI
- XIAP
- X-linked inhibitor of apoptosis
- zVAD-FMK
- carbobenzoxy-valyl-alanyl-aspartyl-[O-methyl]-fluoromethylketone
- NEMO
- NF-κB essential modulator.
REFERENCES
- 1. Chicheportiche Y., Bourdon P. R., Xu H., Hsu Y. M., Scott H., Hession C., Garcia I., Browning J. L. (1997) J. Biol. Chem. 272, 32401–32410 [DOI] [PubMed] [Google Scholar]
- 2. Marsters S. A., Sheridan J. P., Pitti R. M., Brush J., Goddard A., Ashkenazi A. (1998) Curr. Biol. 8, 525–528 [DOI] [PubMed] [Google Scholar]
- 3. Vince J. E., Silke J. (2006) Cell Death Differ. 13, 1842–1844 [DOI] [PubMed] [Google Scholar]
- 4. Gao H. X., Campbell S. R., Burkly L. C., Jakubowski A., Jarchum I., Banas B., Saleem M. A., Mathieson P. W., Berman J. W., Michaelson J. S., Putterman C. (2009) Cytokine 46, 24–35 [DOI] [PubMed] [Google Scholar]
- 5. Burkly L. C., Michaelson J. S., Hahm K., Jakubowski A., Zheng T. S. (2007) Cytokine 40, 1–16 [DOI] [PubMed] [Google Scholar]
- 6. Winkles J. A., Tran N. L., Berens M. E. (2006) Cancer Lett. 235, 11–17 [DOI] [PubMed] [Google Scholar]
- 7. Winkles J. A. (2008) Nat. Rev. Drug Discov. 7, 411–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wiley S. R., Cassiano L., Lofton T., Davis-Smith T., Winkles J. A., Lindner V., Liu H., Daniel T. O., Smith C. A., Fanslow W. C. (2001) Immunity 15, 837–846 [DOI] [PubMed] [Google Scholar]
- 9. Brown S. A., Hanscom H. N., Vu H., Brew S. A., Winkles J. A. (2006) Biochem. J. 397, 297–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Meighan-Mantha R. L., Hsu D. K., Guo Y., Brown S. A., Feng S. L., Peifley K. A., Alberts G. F., Copeland N. G., Gilbert D. J., Jenkins N. A., Richards C. M., Winkles J. A. (1999) J. Biol. Chem. 274, 33166–33176 [DOI] [PubMed] [Google Scholar]
- 11. Brown S. A., Richards C. M., Hanscom H. N., Feng S. L., Winkles J. A. (2003) Biochem. J. 371, 395–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Han S., Yoon K., Lee K., Kim K., Jang H., Lee N. K., Hwang K., Young Lee S. (2003) Biochem. Biophys. Res. Commun. 305, 789–796 [DOI] [PubMed] [Google Scholar]
- 13. Ashwell J. D. (2008) J. Cell Biol. 182, 15–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Saitoh T., Nakayama M., Nakano H., Yagita H., Yamamoto N., Yamaoka S. (2003) J. Biol. Chem. 278, 36005–36012 [DOI] [PubMed] [Google Scholar]
- 15. Sanz A. B., Justo P., Sanchez-Niño M. D., Blanco-Colio L. M., Winkles J. A., Kreztler M., Jakubowski A., Blanco J., Egido J., Ruiz-Ortega M., Ortiz A. (2008) J. Am. Soc. Nephrol. 19, 695–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Maecker H., Varfolomeev E., Kischkel F., Lawrence D., LeBlanc H., Lee W., Hurst S., Danilenko D., Li J., Filvaroff E., Yang B., Daniel D., Ashkenazi A. (2005) Cell 123, 931–944 [DOI] [PubMed] [Google Scholar]
- 17. Ortiz A., Sanz A. B., Muñoz-García B., Moreno J. A., Sánchez-Niño M. D., Martín-Ventura J. L., Egido J., Blanco-Colio L. M. (2009) Cytokine Growth Factor Rev. 20, 251–258 [DOI] [PubMed] [Google Scholar]
- 18. Feng S. L., Guo Y., Factor V. M., Thorgeirsson S. S., Bell D. W., Testa J. R., Peifley K. A., Winkles J. A. (2000) Am. J. Pathol. 156, 1253–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lynch C. N., Wang Y. C., Lund J. K., Chen Y. W., Leal J. A., Wiley S. R. (1999) J. Biol. Chem. 274, 8455–8459 [DOI] [PubMed] [Google Scholar]
- 20. Kaplan M. J., Lewis E. E., Shelden E. A., Somers E., Pavlic R., McCune W. J., Richardson B. C. (2002) J. Immunol. 169, 6020–6029 [DOI] [PubMed] [Google Scholar]
- 21. Zhao Z., Burkly L. C., Campbell S., Schwartz N., Molano A., Choudhury A., Eisenberg R. A., Michaelson J. S., Putterman C. (2007) J. Immunol. 179, 7949–7958 [DOI] [PubMed] [Google Scholar]
- 22. Perper S. J., Browning B., Burkly L. C., Weng S., Gao C., Giza K., Su L., Tarilonte L., Crowell T., Rajman L., Runkel L., Scott M., Atkins G. J., Findlay D. M., Zheng T. S., Hess H. (2006) J. Immunol. 177, 2610–2620 [DOI] [PubMed] [Google Scholar]
- 23. Mittal A., Bhatnagar S., Kumar A., Lach-Trifilieff E., Wauters S., Li H., Makonchuk D. Y., Glass D. J., Kumar A. (2010) J. Cell Biol. 188, 833–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ashkenazi A., Dixit V. M. (1998) Science 281, 1305–1308 [DOI] [PubMed] [Google Scholar]
- 25. Peter M. E., Krammer P. H. (2003) Cell Death Differ. 10, 26–35 [DOI] [PubMed] [Google Scholar]
- 26. Danial N. N., Korsmeyer S. J. (2004) Cell 116, 205–219 [DOI] [PubMed] [Google Scholar]
- 27. Jin Z., Li Y., Pitti R., Lawrence D., Pham V. C., Lill J. R., Ashkenazi A. (2009) Cell 137, 721–735 [DOI] [PubMed] [Google Scholar]
- 28. Chen G., Goeddel D. V. (2002) Science 296, 1634–1635 [DOI] [PubMed] [Google Scholar]
- 29. Karin M. (2006) Nature 441, 431–436 [DOI] [PubMed] [Google Scholar]
- 30. Muppidi J. R., Tschopp J., Siegel R. M. (2004) Immunity 21, 461–465 [DOI] [PubMed] [Google Scholar]
- 31. Ea C. K., Deng L., Xia Z. P., Pineda G., Chen Z. J. (2006) Mol. Cell 22, 245–257 [DOI] [PubMed] [Google Scholar]
- 32. Li H., Kobayashi M., Blonska M., You Y., Lin X. (2006) J. Biol. Chem. 281, 13636–13643 [DOI] [PubMed] [Google Scholar]
- 33. Wu C. J., Conze D. B., Li T., Srinivasula S. M., Ashwell J. D. (2006) Nat. Cell Biol. 8, 398–406 [DOI] [PubMed] [Google Scholar]
- 34. Chu Z. L., McKinsey T. A., Liu L., Gentry J. J., Malim M. H., Ballard D. W. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 10057–10062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kreuz S., Siegmund D., Scheurich P., Wajant H. (2001) Mol. Cell. Biol. 21, 3964–3973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang C. Y., Mayo M. W., Korneluk R. G., Goeddel D. V., Baldwin A. S., Jr. (1998) Science 281, 1680–1683 [DOI] [PubMed] [Google Scholar]
- 37. Micheau O., Tschopp J. R. (2003) Cell 114, 181–190 [DOI] [PubMed] [Google Scholar]
- 38. Wilson N. S., Dixit V., Ashkenazi A. (2009) Nat. Immunol. 10, 348–355 [DOI] [PubMed] [Google Scholar]
- 39. Wang L., Du F., Wang X. (2008) Cell 133, 693–703 [DOI] [PubMed] [Google Scholar]
- 40. Schneider P., Schwenzer R., Haas E., Mühlenbeck F., Schubert G., Scheurich P., Tschopp J., Wajant H. (1999) Eur. J. Immunol. 29, 1785–1792 [DOI] [PubMed] [Google Scholar]
- 41. Vince J. E., Chau D., Callus B., Wong W. W., Hawkins C. J., Schneider P., McKinlay M., Benetatos C. A., Condon S. M., Chunduru S. K., Yeoh G., Brink R., Vaux D. L., Silke J. (2008) J. Cell Biol. 182, 171–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nakayama M., Kayagaki N., Yamaguchi N., Okumura K., Yagita H. (2000) J. Exp. Med. 192, 1373–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nakayama M., Ishidoh K., Kayagaki N., Kojima Y., Yamaguchi N., Nakano H., Kominami E., Okumura K., Yagita H. (2002) J. Immunol. 168, 734–743 [DOI] [PubMed] [Google Scholar]
- 44. Wang D., Fung J. N., Tuo Y., Hu L., Chen C. (2010) Cancer Lett. 294, 91–100 [DOI] [PubMed] [Google Scholar]
- 45. Boatright K. M., Salvesen G. S. (2003) Curr. Opin. Cell Biol. 15, 725–731 [DOI] [PubMed] [Google Scholar]
- 46. Zou H., Li Y., Liu X., Wang X. (1999) J. Biol. Chem. 274, 11549–11556 [DOI] [PubMed] [Google Scholar]
- 47. Silke J., Brink R. (2010) Cell Death Differ. 17, 35–45 [DOI] [PubMed] [Google Scholar]
- 48. Petersen S. L., Wang L., Yalcin-Chin A., Li L., Peyton M., Minna J., Harran P., Wang X. (2007) Cancer Cell 12, 445–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wertz I. E., Dixit V. M. (2008) Cytokine Growth Factor Rev. 19, 313–324 [DOI] [PubMed] [Google Scholar]
- 50. Wong W. W., Gentle I. E., Nachbur U., Anderton H., Vaux D. L., Silke J. (2010) Cell Death Differ. 17, 482–487 [DOI] [PubMed] [Google Scholar]
- 51. Park S. M., Yoon J. B., Lee T. H. (2004) FEBS Lett. 566, 151–156 [DOI] [PubMed] [Google Scholar]
- 52. Cho Y. S., Challa S., Moquin D., Genga R., Ray T. D., Guildford M., Chan F. K. (2009) Cell 137, 1112–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Varfolomeev E., Goncharov T., Fedorova A. V., Dynek J. N., Zobel K., Deshayes K., Fairbrother W. J., Vucic D. (2008) J. Biol. Chem. 283, 24295–24299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hayden M. S., Ghosh S. (2008) Cell 132, 344–362 [DOI] [PubMed] [Google Scholar]
- 55. Karin M., Lin A. (2002) Nature Immunol. 3, 221–227 [DOI] [PubMed] [Google Scholar]
- 56. Vallabhapurapu S., Matsuzawa A., Zhang W., Tseng P. H., Keats J. J., Wang H., Vignali D. A., Bergsagel P. L., Karin M. (2008) Nat. Immunol. 9, 1364–1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sun S. C. (2010) Cell Death Differ. 17, 25–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Varfolomeev E., Blankenship J. W., Wayson S. M., Fedorova A. V., Kayagaki N., Garg P., Zobel K., Dynek J. N., Elliott L. O., Wallweber H. J., Flygare J. A., Fairbrother W. J., Deshayes K., Dixit V. M., Vucic D. (2007) Cell 131, 669–681 [DOI] [PubMed] [Google Scholar]
- 59. Wu H., Tschopp J., Lin S. C. (2007) Cell 131, 655–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Culp P. A., Choi D., Zhang Y., Yin J., Seto P., Ybarra S. E., Su M., Sho M., Steinle R., Wong M. H., Evangelista F., Grove J., Cardenas M., James M., Hsi E. D., Chao D. T., Powers D. B., Ramakrishnan V., Dubridge R. (2010) Clinical Cancer Research 16, 497–508 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.