Abstract
Tristetraprolin (TTP) is an AU-rich element-binding protein that regulates mRNA stability. We previously showed that TTP acts as a negative regulator of VEGF gene expression in colon cancer cells. The p38 MAPK pathway is known to suppress the TTP activity. However, until now the signaling pathway to enhance TTP function is not well known. Here, we show that casein kinase 2 (CK2) enhances the TTP function in the regulation of the VEGF expression in colon cancer cells. CK2 increased TTP protein levels and enhanced VEGF mRNA decaying activity of TTP. TTP was not a direct target of CK2. Instead, CK2 increased the phosphorylation of MKP-1, which led to a decrease in the phosphorylation of p38 MAPK. Inhibition of MKP-1 by siRNA attenuated the increase in TTP function and the decrease of p38 phosphorylation induced by CK2α overexpression. TGF-β1 increased the expressions of CK2 and TTP and the TTP function. The siRNA against CK2α or TTP reversed TGF-β1-induced increases in the expression of CK2 and TTP and the TTP function. Our data suggest that CK2 enhances the protein level and activity of TTP via the modulation of the MKP-1-p38 MAPK signaling pathway and that TGF-β1 enhances the activity of CK2.
Keywords: Colon Cancer, Gene Regulation, Protein Stability, RNA-binding Protein, RNA Turnover, CK2, MKP-1, TGF-beta, VEGF, Tristetraprolin
Introduction
Tristetraprolin (TTP)3 is an AU-rich element (ARE)-binding protein that mediates the decay of ARE-containing mRNAs such as those encoding cytokines and proto-oncogenes (1). TTP also regulates the stability of VEGF transcripts (2), indicating a possible role for TTP in angiogenesis and tumor growth (3, 4). Up-regulation of TTP mRNA can be induced by 12-O-tetradecanoylphorbol-13-acetate (5), insulin (6), lipopolysaccharide (LPS) (7, 8), GM-CSF (9), TGF-β (10), IL-10 (11), and glucocorticoid (12). TTP mRNA contains AREs, and its stability is regulated by TTP itself (8).
TTP is highly phosphorylated both in vitro and in vivo, and its phosphorylation is regulated by p42 MAPK (13), p38 MAPK (13, 14), c-Jun N-terminal kinase (JNK) (13), and MAPK-activated protein kinase 2 (MAPKAP kinase 2 or MK2) (7, 15, 16). Among these protein kinases, the p38 MAPK/MK-2 pathway has been reported to be a crucial regulator of the expression, stability, and function of TTP (7, 17, 18). Phosphorylation of TTP by MK2 increases the binding of 14-3-3 proteins, thereby excluding TTP from stress granules, inactivating TTP, and increasing the stabilities of target mRNAs (15, 16, 19). The TTP protein is unstable and is rapidly degraded by proteasomes (17, 20); however, phosphorylation of TTP by p38 MAPK protects TTP from proteasomal degradation (17). Recent evidence suggests that phosphorylation-induced inhibition of TTP can be reversed by protein phosphatase 2A (PP2A) through the inactivation of p38 MAPK and MK2 (21). Despite these studies on TTP phosphorylation, the signaling pathways responsible for the induction of TTP activity are not well known.
Casein kinase 2 (CK2), a highly conserved and ubiquitously expressed serine/threonine kinase, is a tetramer composed of two catalytic subunits (α and α′) and two regulatory subunits (β) in an α2β2, αα′β2, or α′2β2 configuration (22). CK2 phosphorylates a large number of substrates, many of which are involved in gene expression and cell growth (23–28). CK2 is up-regulated in a variety of human cancers (29) and creates a cellular environment favorable to neoplasia by regulating a variety of biological processes (26, 30–32). In contrast to the functions of CK2 that favor the neoplastic growth of cells, several investigations have demonstrated an opposing function of this enzyme. Exogenous expression of CK2α deactivates MEK, suppresses cell growth, and inhibits the foci formation induced by activated Ras (33). In addition, CK2 phosphorylates p53 (34), which enhances the sequence-specific DNA binding function of p53 in vitro (35) and ensures the maintenance of G2 arrest and apoptosis following spindle damage (36).
We previously reported that TTP down-regulates the expression of VEGF and inhibits the growth of human colon cancer cells in vitro and in vivo (37). In this study, we demonstrate that CK2 increases the VEGF mRNA decaying activity of TTP in human colon cancer cells by protecting the TTP protein from phosphorylation and proteasomal degradation. CK2 decreased phosphorylation of p38 MAPK, and the effects of CK2 were attenuated either by treatment with an inhibitor of MAPK phosphatase 1 (MKP-1) or an siRNA against MKP-1, suggesting that CK2 function is mediated by MKP-1, which is known to dephosphorylate p38 MAPK (38). Moreover, we show that transforming growth factor β (TGF-β) increases CK2 activity, resulting in an increase in the VEGF mRNA decaying activity of TTP. Finally, TGF-β suppressed the growth of colon cancer cells, and this event was mediated by CK2 and TTP, indicating that TGF-β acts as a tumor suppressor via the activation of CK2 and TTP in Colo320 cells. Collectively, our results show that CK2 increases the VEGF mRNA decaying activity of TTP through activation of MKP-1 and that TGF-β plays a role in the activation of the CK2-MKP-1-TTP signaling pathway.
EXPERIMENTAL PROCEDURES
Cells
Human colon cancer cells (Colo320) were purchased from Korean Cell Line Bank (KCLB, Seoul, Korea). Colo320 cells were cultured in RPMI 1640 media, supplemented with 10% FBS (heat-inactivated fetal bovine serum) (Invitrogen), and maintained at 37 °C in a humidified atmosphere of 5% CO2. For the MTS cell proliferation assay, cells were plated in RPMI 1640 medium in triplicate at 1.2 × 104 cells/well in 96-well culture plates. At the indicated times, CellTiter 96® Aqueous One Solution Reagent (Promega, Madison, WI) was added to each well according to the manufacturer's protocols, and absorbance at 490 nm (A490) was determined for each well using a Wallac Vector 1420 multilabel counter (EG&G Wallac, Turku, Finland).
Semi-quantitative RT-PCR
Five micrograms of DNase I-treated total RNA was reverse-transcribed using oligo(dT) and Superscript II reverse transcriptase (Invitrogen) according to the manufacturer's instructions. Semi-quantitative RT-PCR was performed using Taq polymerase (Sun Genetics, Daejeon, Korea) and the appropriate primers as follows: TTP, 5′-CGCTACAAGACTGAGCTAT-3′ and 5′-GAGGTAGAACTTGTGACAGA-3′; GAPDH, 5′-ACATCAAGAAGGTGGTGAAG-3′ and 5′-CTGTTGCTGTAGCCAAATTC-3′.
Quantitative Real Time PCR
For RNA kinetic analysis, we used actinomycin D and assessed VEGF mRNA expression using quantitative real time PCR, performed using the ABI Prism 7900 HT to monitor real time increases in the fluorescence of SYBR Green dye (Qiagen, Hilden, Germany). Specificities of each primer pair were confirmed via melting curve analysis and agarose gel electrophoresis. The PCR primer pairs were as follows: qVEGF, 5′-ATCTTCAAGCCATCCTGTGTGC-3′ and 5′-TGCGCTTGTCACATTTTTTCTTG-3′; qTTP, 5′-CCCCAAATACAAGACGGAACTC-3′ and 5′-GGGCCGCCAGGTCTTC-3′; and qGAPDH, 5′-ACATCAAGAAGGTGGTGAAG-3′ and 5′-CTGTTGCTGTAGCCAAATTC-3′.
Plasmid, siRNAs, Transfection, and Dual-Luciferase Assay
The pcDNA6/V5-TTP plasmid construct has been described previously (37). Plasmid constructs containing full-length cDNA of human CK2α or human ubiquitin were purchased from Addgene (Cambridge, UK). Full-length cDNA of human CK2α and human ubiquitin were amplified from these plasmids, and full-length cDNA of human MKP-1 was amplified from the cDNA of Colo320 cells using PCR. PCR products were ligated into BamHI/XhoI sites of pcDNA3-HA or pcDNA3.1-FLAG vectors (Invitrogen) to generate pcDNA3/HA-CK2α, pcDNA3/FLAG-Ub, and pcDNA3/FLAG-MKP-1. Site-directed mutants of TTP with single (S21A, S169A, S279A, or S325A), double (S60A/S186A), or quadruple amino acid substitutions (S21A/S169A/S279A/S325A) and MKP-1 with double amino acid substitutions (S131A/S235A) were generated using pcDNA6/V5-TTP and pcDNA3/FLAG-MKP-1 as a template, respectively, using a QuikChange site-directed mutagenesis kit (Stratagene, San Diego) according to the manufacturer's instructions. Mutagenic primers used for generation of site-directed mutants of TTP or MKP-1 were as follows: TTP-S21A, 5′-AGTGCCCGTGCCAGCCGACCATGGAGGG-3′ and 5′-CCTCCATGGTCGGCTGGCACGGGCACTG-3′; TTP-S60A, 5′-CTGGCCGCTCCACCGCCCTAGTGGAGGGC-3′ and 5′-GCCCTCCACTAGGGCGGTGGAGCGGCCAG-3′; TTP-S169A, 5′-CATCCACAACCCTGCCGAAGACCTGGCGG-3′ and 5′-CCGCCAGGTCTTCGGCAGGGTTGTGGATG-3′; TTP-S186A, 5′-CTTCGCCAGAGCATCGCCTTCTCCGGCCTGC-3′ and 5′-GCAGGCCGGAGAAGGCGATGCTCTGGCGAAG-3′; TTP-S279A, 5′-GTACAGTCCCTGGGAGCCGACCCTGATGAATATG-3′ and 5′-CATATTCATCAGGGTCGGCTCCCAGGGACTGTAC-3′;TTP-S325A, 5′-CGCATCTCTGTTGCCGAGCTCGAGTCTAG-3′ and 5′-CTAGACTCGAGCTCGGCAACAGAGATGCG-3′; MKP-1-S131A, 5′-GAAGCGTTTTCGGCTGCCTGCCCGGAGCTG-3′ and 5′-CAGCTCCGGGCAGGCAGCCGAAAACGCTTC-3′; and MKP-1-S235A, 5′-GGCAGACATCAGCGCCTGGTTCAACGAGGC-3′ and 5′-GCCTCGTTGAACCAGGCGCTGATGTCTGCC-3′. Cells were transfected with plasmid constructs using TurboFectTM in vitro transfection reagent (Fermentas, Hanover, Germany).
Small interfering RNAs (siRNA) against human CK2α (siCK2α) (sc-38963), human MKP-1 (siMKP-1) (sc-35937), human TTP (siTTP) (sc-36761), and control siRNA (scRNA) (sc-37007) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Colo320 cells (1.5 or 3 × 105) were transfected with each siRNA using LipofectamineTM RNAiMAX (Invitrogen). VEGF 3′UTR was PCR-amplified from the cDNA of Colo320 cells using Taq polymerase (Solgent, Daejeon, Korea) and the following primers: VEGF-3′UTR, 5′-CTAGAGGTACTTATTTAATAGCCCTTTTTAATTAGAAATTAAAACAGTTAATTTAATTAAAGAGTAGGGTTTTTTCAGTAT-3′ and 5′-CTAGATACTGAAAAAACCCTACTCTTTAATTAAATTAACTGTTTTAATTTCTAATTAAAAAGGGCTATTAAATAAGTACCT-3′. PCR products were inserted into the XhoI/NotI sites of a psiCHECK2 Renilla/firefly Dual-Luciferase expression vector (Promega).
For luciferase assays, Colo320 cells were cotransfected with pcDNA6/V5-TTP and psiCHECK2-VEGF 3′UTR or psiCHECK2-Bcl2 3′UTR construct using the TurboFectTM in vitro transfection reagent. Transfected cells were lysed with lysis buffer (Promega) and mixed with luciferase assay reagent (Promega), and the chemiluminescent signal was measured in a Wallac Victor 1420 Multilabel Counter (EG&G Wallac). The activity of Renilla luciferase was normalized to that of firefly luciferase in each sample. All luciferase assay results reported here represent at least three independent experiments, each consisting of three wells per transfection.
SDS-PAGE Analysis and Immunoblotting
Proteins were resolved using SDS-PAGE, transferred onto Hybond-P membranes (GE Healthcare), and probed with appropriate dilutions of antibodies as follows: rabbit anti-human TTP antibody (ab36558, Abcam); anti-human CK2α antibody (sc-12738, Santa Cruz Biotechnology); anti-p38 MAPK antibody (9212, Cell Signaling Technology, Danvers); anti-phospho-p38 MAPK antibody (9215, Cell Signaling); anti-MK2 antibody (3042, Cell Signaling); anti-phospho-MK2 antibody (sc-101729, Santa Cruz Biotechnology); anti-MKP-1 antibody (sc-370, Santa Cruz Biotechnology); anti-phospho-MKP-1 antibody (2857, Cell Signaling Technology); anti-ubiquitin antibody (sc-9133, Santa Cruz Biotechnology); anti-V5 antibody (Genentech, San Francisco); anti-HA antibody (H9658, Sigma); and anti-FLAG antibody (F1804, Sigma). Immunoreactivity was detected using the ECL detection system (GE Healthcare). Films were exposed at multiple time points to ensure that the images were not saturated.
Phosphorylation Site Prediction
The potential for phosphorylation sites in TTP protein was predicted using NetPhos. The amino acid sequences of human TTP protein were obtained from GenBankTM (accession number NP_003398).
Effects of Protein Kinase Inhibitors on the mRNA Destabilizing Activity of TTP
Inhibitors against the protein kinases CK2 (5,6-dichlorobenzimidazole riboside (DRB) and 1,3,5-tribromobenzene (TBB), Sigma), phosphatidylinositol 3-kinase (PI3K) (LY294002, Calbiochem), mammalian target of rapamycin (rapamycin, Calbiochem), MEK1 (PD98059, Calbiochem), PKC (Ro-32-0432, Calbiochem), p38 MAPK (SB203580, Calbiochem), JNK (SP600125, Calbiochem), and MKP-1 (Triptolide, Calbiochem) were dissolved in dimethyl sulfoxide (DMSO) and used at the concentrations indicated in the figure legends. Colo320 cells were cotransfected with pcDNA6/V6-TTP and psiCHECK2-VEGF 3′UTR using the TurboFectTM in vitro transfection reagent. At 24 h post-transfection, cells were treated with protein kinase inhibitors for 12 h, and the chemiluminescent signal in the cell lysate was measured in a Wallac Victor 1420 multilabel counter. The activity of Renilla luciferase was normalized to that of firefly luciferase in each sample. All luciferase assays reported here represent at least three independent experiments, each consisting of three wells per transfection.
Detection of Ubiquitinylation
Cells were cotransfected with pcDNA6/V5-TTP and pcDNA3/FLAG-Ub using TurboFectTM in vitro transfection reagent. At 24 h post-transfection, cells were treated with 30 μm DRB for 12 h in the presence or absence of 50 μm MG-132 (Calbiochem) and lysed with RIPA buffer (50 mm Tris-HCl, pH 8.0, 150 mm NaCl, 5 mm EDTA, 0.5% Nonidet P-40, 0.1% SDS). Proteins in the cell lysates were immunoprecipitated with anti-V5 antibodies for 3 h at 4 °C, followed by incubation with protein A-agarose overnight at 4 °C. Proteins in the immunoprecipitates were resolved using SDS-PAGE, followed by Western blotting with anti-FLAG antibody.
Phosphatase Treatment
Cells were transfected with pcDNA6/V5-TTP using TurboFectTM in vitro transfection reagent. At 24 h post-transfection, cells were treated with 30 μm DRB for 12 h in the presence or absence of 50 μm MG-132 (Calbiochem) and were resuspended in modified RIPA buffer (50 mm Tris, pH 7.5, 1 mm MgCl2, 0.1% Triton X-100, 0.2 mm PMSF, 5 μg/ml leupeptin, 0.25% aprotinin). Fifty microgram cell lysates were incubated with 20 units of calf intestinal phosphatase (New England Biolabs) in a reaction buffer (500 mm potassium acetate, 200 mm Tris acetate, 100 mm magnesium acetate, 10 mm dithiothreitol, pH 7.9). After incubation at 37 °C for 30 min, the proteins were resolved using SDS-PAGE, followed by immunoblotting with anti-V5 antibody.
Cycloheximide-based Protein Chase Experiment
Colo320 cells were transfected with pcDNA6/V5-TTP using the TurboFectTM in vitro transfection reagent. At 24 h post-transfection, cells were incubated with 10 μg/ml cycloheximide (CHX, Calbiochem) to stop protein synthesis in the presence or absence of DRB. Cells were harvested at 0, 1, 3, 5, and 10 h after addition of CHX, and total cell lysates were analyzed using immunoblotting with anti-V5 antibody.
Exposure to TGF-β
Colo320 cells were cotransfected with psiCHECK2-VEGF 3′UTR and pcDNA6/V5-TTP, TTP siRNA, or CK2 siRNA using the TurboFectTM in vitro transfection reagent. After 24 h of incubation, cells were incubated with 2 ng/ml human TGF-β in the presence of or absence of 30 μm DRB for 12 h, and chemiluminescent signals in the cell lysate were measured in a Wallac Victor 1420 Multilabel Counter. The activity of Renilla luciferase was normalized to that of firefly luciferase in each sample. All luciferase assays reported here represent at least three independent experiments, each consisting of three wells per transfection.
In Vitro CK2 Kinase Assay
CK2 activity was assayed using a CK2-sensitive synthetic p53 peptide (RRRDDDSDDD) (Cyclex Co. Ltd., Nagano, Japan), which is phosphorylated at serine 46 by CK2. Briefly, Colo320 cells were incubated with 2 ng/ml human TGF-β in the presence or absence of 30 μm DRB for 12 h. Cell lysates were incubated with the synthetic p53 peptide coated on a 96-well plate with the kinase reaction buffer. The amount of phosphorylated substrate was measured by using a horseradish peroxidase-conjugated anti-phospho-p53 serine 46-specific antibody. Kinase activity was calculated by subtracting the mean of the background control samples without enzyme from the mean of samples with enzyme.
Statistical Analysis
For statistical comparisons, p values were determined using Student's t test or one-way analysis of variance.
RESULTS
CK2 Inhibition via CK2 Inhibitors or siRNA Decreases the VEGF mRNA Decaying Activity of TTP
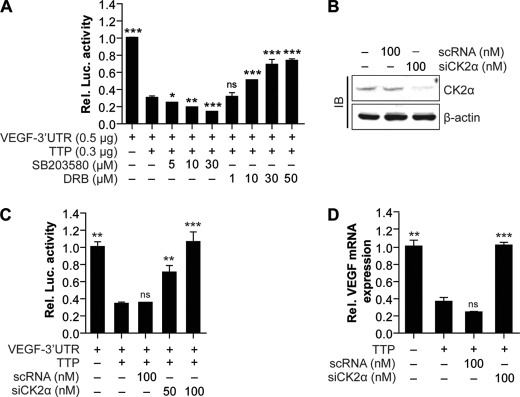
TTP has been reported to be phosphorylated by various kinds of protein kinases (39). Phosphorylation of TTP by p38 MAPK/MK2 signals inhibits TTP activity (40), but specific protein kinases that can increase TTP activity are not known. We previously reported that TTP reduced VEGF expression and inhibited the growth of colon cancer cells (37). Here, we aimed to identify protein kinases that can increase the VEGF mRNA decaying activity of TTP. To screen protein kinases that increase the VEGF mRNA decaying activity of TTP, we studied the effects of several protein kinase inhibitors on the VEGF mRNA decaying activity of TTP. We have successfully used the VEGF ARE-containing reporter mRNA to study VEGF mRNA turnover by TTP in human colon cancer cells (37); we used a transient transfection system in which the ARE-dependent turnover of the reporter transcripts and its sensitivity to protein kinase inhibitors could be evaluated. To characterize the effects of protein kinase inhibitors on the VEGF decaying activity of TTP, Colo320 cells were cotransfected with a TTP expression construct (pcDNA/V5-TTP) and a luciferase reporter construct that incorporated the 3′UTR of VEGF mRNA (psiCHECK2-VEGF 3′UTR). Transfected cells were treated with various kinds of protein kinase inhibitors, and luciferase activity was measured at 12 h post-treatment. Consistent with the previous report (41), overexpression of TTP reduced luciferase activity (Fig. 1A). Treatment with SP600125 (Jun N-terminal protein kinase inhibitor), Ro-32-0432 (protein kinase C inhibitor), LY294002 (phosphatidylinositol 3-kinase inhibitor), PD98059 (MEK1 inhibitor), or rapamycin (mammalian target of rapamycin inhibitor) did not affect the TTP-mediated reduction of luciferase activity (supplemental Fig. 1). However, SB203580 (p38 MAPK inhibitor) enhanced the TTP inhibitory activity in a dose-dependent manner (Fig. 1A). Conversely, treatment with the CK2 inhibitor DRB attenuates the TTP-mediated reduction of luciferase activity in a dose-dependent manner (Fig. 1A). The effect of SB203580 on TTP activity is consistent with those of previous reports that have suggested that the p38 MAPK/MK-2 pathway inhibits the mRNA decaying activity of TTP (7, 17, 18). It is also possible that SB203580 enhanced TTP activity by inhibiting other kinases than p38 MAPK as SB203580 is not selective and is known to inhibit a number of other kinases, including CK1 (42). However, our results showing that the CK2 inhibitor DRB increases luciferase activity suggests that the CK2 signal activates the mRNA decaying activity of TTP. Similar results were obtained by another CK2 inhibitor, TBB (supplemental Fig. 2).
FIGURE 1.
Inhibition of CK2 decreases the VEGF mRNA decaying activity of TTP. A, CK2 inhibitor DRB decreases the mRNA decaying activity of TTP. Colo320 cells were cotransfected with psiCHECK2-VEGF 3′UTR and pcDNA6/V5-TTP. At 24 h post-transfection, cells were treated with CK2 inhibitor DRB or p38 MAPK inhibitor SB203580 at indicated concentrations for 12 h (A), and luciferase (Luc.) activity was determined. Renilla luciferase activity was normalized to firefly activity. The luciferase activity obtained from psiCHECK2-VEGF 3′UTR-transfected cells was set to 1. Each bar represents the mean ± S.D. of three independent experiments (*, p < 0.05; **, p < 0.01; ***, p < 0.001). ns, not significant. B and C, inhibition of CK2α by siRNA (siCK2α) decreases the mRNA decaying activity of TTP. Colo320 cells were cotransfected with psiCHECK2-VEGF 3′UTR, pcDNA6/V5-TTP, and siCK2α or scRNA. At 24 h post-transfection, expression level of CK2α was determined by immunoblotting (IB) (B) and luciferase activity was determined (C). Renilla luciferase activity was normalized to firefly activity. The luciferase activity obtained from psiCHECK2-VEGF 3′UTR-transfected cells was set to 1. Each bar represents the mean ± S.D. of three independent experiments (**, p < 0.01; ***, p < 0.001). D, TTP-mediated down-regulation of endogenous VEGF mRNA is attenuated by siCK2α. Colo320 cells were cotransfected with pcDNA6/V5-TTP and siCK2α or scRNA. At 24 h post-transfection, VEGF mRNA was determined by quantitative real time PCR. The expression level obtained from mock-transfected cells was set to 1. Each bar represents the mean ± S.D. of three independent experiments (**,p < 0.01; ***, p < 0.001).
To confirm whether inhibition of CK2 increases luciferase activity, we analyzed the effect of siRNA against CK2α (siCK2α) on the reporter mRNA decaying activity of TTP. Cells were cotransfected with pcDNA6/V5-TTP, psiCHECK2-VEGF 3′UTR, and siCK2, and luciferase activity was measured 24 h after transfection. Knockdown of endogenous CK2α by siRNA (Fig. 1B) significantly decreased the TTP inhibitory effect (Fig. 1C). Taken together, these results indicate that inhibition of CK2 decreases the mRNA decaying activity of TTP. All of these data were obtained by analyzing the expression levels of reporter transcripts containing VEGF 3′UTR.
Next, we wished to determine whether CK2 inhibition also affects the TTP inhibitory effects on endogenous VEGF mRNA. Colo320 cells were cotransfected with pcDNA6/V5-TTP and siCK2α or control siRNA (scRNA), and the expression level of VEGF mRNA was analyzed using real time PCR. Overexpression of TTP decreased the expression level of VEGF mRNA (Fig. 1D). Although control siRNA treatment did not affect the TTP inhibitory effect, down-regulation of CK2α by siRNA attenuated it. The expression level of endogenous TTP in Colo320 cells was extremely low (37), and in the absence of transfected TTP, neither SB203580 nor DRB affected the reporter mRNA decaying activity of TTP and the expression level of endogenous VEGF mRNA level (supplemental Fig. 3). Collectively, our results suggest that inhibition of CK2 decreases the VEGF mRNA decaying activity of TTP.
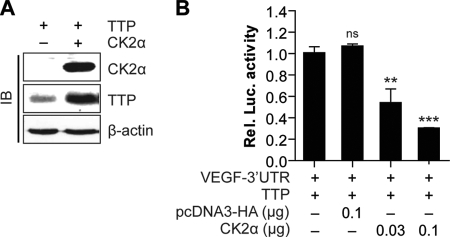
CK2 Overexpression Increases the mRNA Decaying Activity of TTP
Overexpression studies were performed to further analyze the role of CK2 in the regulation of the TTP inhibitory effect. Cells were transiently cotransfected with pcDNA/V5-TTP, psiCHECK2-VEGF 3′UTR, and a CK2α expression construct (pcDNA3/HA-CK2α). Overexpressions of TTP and CK2α were confirmed using an immunoblotting assay (Fig. 2A). At 24 h post-transfection, luciferase activity was measured. Transfection of the control vector pcDNA3/HA did not affect the TTP inhibitory effect. However, overexpression of CK2α enhanced the TTP inhibitory effect in a dose-dependent manner (Fig. 2B). The results suggest that CK2α is involved in the enhancement of the TTP inhibitory effect.
FIGURE 2.
Overexpression of CK2α increases VEGF mRNA decaying activity of TTP. Colo320 cells were cotransfected with psiCHECK2-VEGF 3′UTR and pcDNA6/V5-TTP pcDNA3/HA-CK2α. At 24 h post-transfection, the expression levels of CK2α and TTP were determined by immune blots (IB) (A) and luciferase activity was determined (B). Renilla luciferase activity was normalized to firefly activity. The luciferase activity obtained from cells cotransfected with psiCHECK2-VEGF 3′UTR and pcDNA6/V5-TTP was set to 1. Each bar represents the mean ± S.D. of three independent experiments (**, p < 0.01; ***, p < 0.001). ns, not significant.
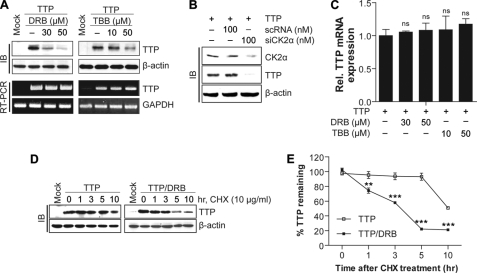
CK2 Inhibition by CK2 Inhibitors or siRNA Decreases TTP Protein Stability
To determine whether CK2 inhibition affects the stability of the TTP protein, Colo320 cells were treated with DRB, TBB, or siCK2, and the expression level of TTP protein was analyzed using an immunoblotting assay. CK2 inhibition by DRB, TBB (Fig. 3A), or siCK2α (Fig. 3B) significantly decreased TTP protein levels, indicating that CK2 affects the expression level of TTP protein. CK2 is known to phosphorylate and regulate several transcription factors (30, 43); thus, it is possible that CK2 controls TTP expression through the regulation of TTP transcription. To determine whether CK2 inhibition affects the expression level of TTP transcripts, Colo320 cells were treated with DRB or TBB, and the expression levels of TTP transcript were analyzed using RT-PCR and real time PCR. Neither treatment with DRB nor TBB affected the expression level of TTP transcript in Colo320 cells (Fig. 3, A and C). We next determined whether CK2 inhibition affected the stability of TTP protein. Colo320 cells transfected with pcDNA6/V5-TTP were treated with 100 μg/ml CHX to block protein synthesis in the presence or absence of DRB. At the indicated time points after CHX treatment, cells were collected, and the expression level of TTP protein was analyzed using immunoblotting with anti-V5 antibody. In the absence of DRB, the half-life of the TTP protein was longer than 10 h. However, in the presence of DRB, its half-life was reduced, and at 5 h after CHX treatment, about 80% of TTP protein was degraded (Fig. 3, D and E). These data demonstrate that CK2 functions to enhance the stabilization of TTP protein.
FIGURE 3.
Inhibition of CK2 by DRB decreases the instability of TTP protein. A–C, inhibition of CK2α by CK2 inhibitors or siRNA decreases the expression level of the TTP protein but does not affect the expression level of the TTP transcript. Colo320 cells were treated with CK2 inhibitor DRB or TBB at the indicated concentrations for 12 h or transfected with siCK2α or scRNA. TTP protein levels were determined by immunoblotting (IB) (A, top panel, and B) and TTP mRNA levels were determined by (A, bottom panel) semi-quantitative PCR, and real time PCR (C). D and E, DRB treatment decreases the stability of TTP protein. Colo320 cells were transfected with pcDNA6/V5-TTP, and at 24 h post-transfection cells were incubated with 10 μg/ml cycloheximide (CHX) in the presence or absence of DRB. D, cells were harvested at indicated times. and TTP protein levels were determined by immunoblotting with anti-V5 antibody. E, graph showing the expression levels of TTP protein. The expression levels of TTP protein in CHX-treated cells at 0 h were set to 1. The results are presented as the means ± S.D. of three independent experiments. (**, p < 0.01; ***, p < 0.001). ns, not significant.
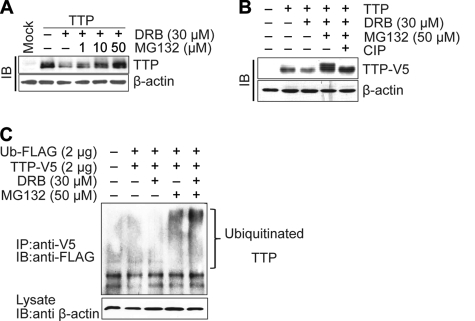
CK2 Decreases the Proteasomal Degradation and Phosphorylation of TTP
To determine whether the proteasome pathway is involved in the degradation of the TTP protein, Colo320 cells transfected with pcDNA6/V5-TTP were treated with DRB in the presence or absence of the specific proteasome inhibitor MG132. At 24 h post-treatment, the expression level of TTP protein was examined using an immunoblotting assay. MG132 blocked the DRB-induced degradation of TTP protein in a dose-dependent manner (Fig. 4A). These findings suggest that DRB-induced degradation of TTP protein is dependent on proteasomal activity. Interestingly, along with blocking the degradation of TTP, MG132 treatment led to the appearance of higher molecular weight TTP proteins (Fig. 4A). The higher molecular weight TTP proteins appear to be the result of phosphorylation, because this form could be completely converted to the lower molecular weight TTP protein by in vitro incubation with calf intestinal alkaline phosphatase (CIP, Fig. 4B). Treatment with DRB decreased the level of lower molecular weight TTP but increased the level of higher molecular weight TTP, indicating that DRB treatment induced the phosphorylation of TTP (supplemental Fig. 4).
FIGURE 4.
CK2 inhibitor DRB enhances phosphorylation, ubiquitination, and proteasomal degradation of the TTP protein. Colo320 cells were transfected with pcDNA6/V5-TTP, and at 24 h post-transfection cells were treated with DRB in the presence or absence of MG132 at the indicated concentrations. A, proteasome inhibitor MG132 abolishes the decrease of TTP protein induced by DRB. TTP protein levels were determined by immunoblotting (IB) with anti-V5 antibody. B, treatment of calf intestinal alkaline phosphatase (CIP) converts slow migrating TTP to fast migrating TTP. Cell lysates were subjected to in vitro calf intestinal alkaline phosphatase treatment (20 units) for 30 min at 35 °C (5th lane). The samples were then analyzed for TTP protein by immunoblotting with anti-V5 antibody. C, CK2 inhibitor DRB increases the ubiquitination of TTP. Colo320 cells were cotransfected with pcDNA6/V5-TTP and pcDNA3/FLAG-UB. At 24 h post-transfection, cells were treated with DRB in the presence or absence of MG132. Immunoprecipitation (IP) was performed by incubating the cell lysates with anti-V5 antibody and then blotting with anti-FLAG antibody.
To assess whether TTP protein is ubiquitinated prior to its degradation by DRB, in vivo ubiquitination assays were performed. Colo320 cells were cotransfected with pcDNA6/V5-TTP and pcDNA3/FLAG-Ub. At 24 h post-transfection, cells were treated with MG132 in the presence or absence of DRB and incubated for an additional 2 h. TTP proteins were then immunoprecipitated with anti-V5 antibody. When the proteins were probed with anti-FLAG antibody, a high molecular mass protein ladder was observed in the presence of MG132 but not in the absence of MG132, suggesting that TTP protein was ubiquitinated (Fig. 4C). Ubiquitinated TTP was increased by DRB treatment (Fig. 4C), suggesting that CK2 inhibition increased TTP ubiquitinylation. Together, our data indicate that the CK2 can decrease the phosphorylation and ubiquitinylation of TTP, resulting in protection of TTP from proteasomal degradation.
TTP Is Not a Direct Target of CK2
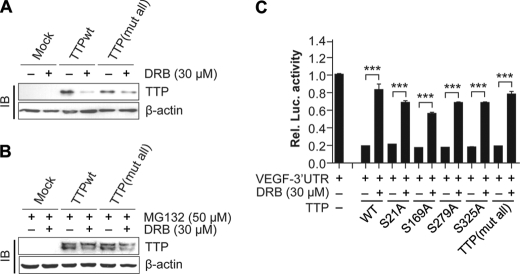
Because CK2 functions to decrease the phosphorylation of TTP (Fig. 4), it is possible to speculate that TTP is not a direct target of CK2. To confirm this hypothesis, we tested whether DRB affects the phosphorylation, degradation, and mRNA decaying activity of TTP with mutation at CK2-specific phosphorylation sites. Scanning the amino acid sequence of TTP for potential CK2 phosphorylation sites revealed four different sites: Ser-21, Ser-169, Ser-279, and Ser-325. We generated four point mutants by replacing the coding sequence for Ser at amino acid residues 21, 169, 279, or 325 with that for Ala (pcDNA6/V5-TTP(S21A); pcDNA6/V5-TTP(S169A); pcDNA6/V5-TTP(S279A); pcDNA6/V5-TTP(S325A)). In addition, we generated one point mutant by replacing the coding sequences for all four Ser sites with Ala (pcDNA6/V5-TTP(mut all)). To determine the effects of these point mutations on the degradation and phosphorylation of TTP protein induced by DRB, cells were transfected with wild-type TTP or TTP point mutation constructs. At 24 h post-transfection, cells were treated with DRB in the presence or absence of MG132, and the expression levels of TTP proteins were analyzed using immunoblotting. Site-directed mutant of TTP with quadruple amino acid substitutions (S21A/S169A/S279A/ S325A) (Fig. 5, A and B) did not change the degradation and phosphorylation of TTP proteins induced by DRB treatment. Next, cells were cotransfected with psiCHECK2-VEGF 3′UTR and with wild-type TTP or TTP point mutation constructs, and we tested the effects of these mutations on the decrease in the mRNA decaying activity of TTP induced by DRB. Consistent with the effects of these mutations on the degradation and phosphorylation of TTP, any mutations of TTP did not affect the decrease in the DRB-induced mRNA decaying activity of TTP (Fig. 5C). These data suggest that the predicted CK2-specific phosphorylation sites in the TTP protein are not responsible for the effects of CK2 on the degradation, phosphorylation, and mRNA decaying activity of TTP protein. Thus, TTP may not be the direct target of CK2 in Colo320 cells.
FIGURE 5.
Mutations of putative CK2 phosphorylation sites of TTP do not disrupt the effects of CK2 on TTP. A, mutation at the four CK2 phosphorylation sites does not affect the decrease of TTP protein levels induced by CK2 inhibitor DRB. Colo320 cells were transfected with pcDNA6/V5-TTP or pcDNA6/V5-TTP(mut all) and were treated with DRB for 12 h. The expression level of the TTP protein was determined by immunoblotting (IB) with anti-V5 antibody. B, mutation at the four CK2 phosphorylation sites does not change the phosphorylation of TTP induced by DRB. Colo320 cells transfected with pcDNA6/V5-TTP or pcDNA6/V5-TTP(mut all) were treated with MG132 in the presence or absence of DRB for 12 h. The expression level of TTP was determined by immunoblotting with anti-V5 antibody. C, mutation of the CK2 phosphorylation sites does not affect the decrease of mRNA decaying activity of TTP induced by DRB. Colo320 cells were cotransfected with psiCHECK2-VEGF 3′UTR and wild-type TTP (pcDNA6/V5-TTP) or point mutant TTP: pcDNA6/V5-TTP(S26A), pcDNA6/V5-TTP(S169A), pcDNA6/V5-TTP(S279A), pcDNA6/V5-TTP(S325A), or pcDNA6/V5-TTP(mut all). At 24 h post-transfection, cells were treated with DRB for 12 h, and luciferase activity was determined. Renilla luciferase activity was normalized to firefly activity. The luciferase activity obtained from cells transfected with psiCHECK2-VEGF 3′UTR was set to 1. Each bar represents the mean ± S.D. of three independent experiments (***, p < 0.001).
CK2 Decreases the Phosphorylations of p38 MAPK and MK2
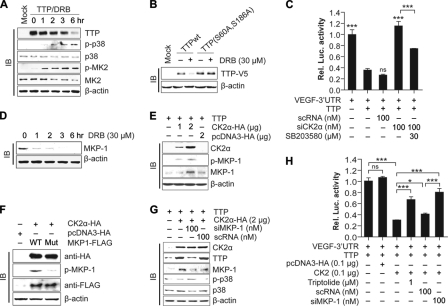
The TTP protein is phosphorylated by MK2 (7, 15), and its stability has been reported to be regulated by the p38 MAPK/MK2 pathway (7, 17, 18). To determine whether CK2 affects the p38 MAPK/MK2 pathway, cells were transfected with pcDNA6/V5-TTP. At 24 h post-transfection, cells were treated with DRB, and cell lysates were analyzed for phospho-p38 MAPK and phospho-MK2 using an immunoblotting assay. DRB treatment resulted in a slight decrease in the expression levels of total p38 MAPK and MK2. However, it increased the phosphorylations of both p38 MAPK and MK2 (Fig. 6A). Human TTP is phosphorylated by MK2 at serine residues Ser-60 and Ser-186 (44). We generated a double mutant of TTP (S60A/S186A) and tested whether these sites function in DRB-induced degradation of TTP. Although DRB treatment decreased the protein level of wild-type TTP, it did not affect that of the double mutant TTP (Fig. 6B), suggesting that the effect of CK2 on the TTP protein degradation was mediated by MK2. To determine whether the effect of CK2 inhibition on the mRNA decaying activity of TTP was mediated via the p38 MAPK pathway, cells were cotransfected with pcDNA6/V5-TTP, psiCHECK2-VEGF 3′UTR, and siRNA against CK2α (siCK2α) or control siRNA (scRNA). At 24 h post-transfection, cells were treated with the p38 MAPK inhibitor SB203580. Results showed that the increase in luciferase activity induced by siCK2α was significantly attenuated by SB203580 (Fig. 6C). These data suggest that the CK2 enhances the TTP protein stability by suppression of the p38 MAPK pathway.
FIGURE 6.
Effects of CK2 on TTP is mediated by the MKP-1/p38 MAPK pathways. A, CK2 inhibition by DRB increases phosphorylation of p38 MAPK and MK2. Colo320 cells were transfected with pcDNA6/V5-TTP, and the cells were treated with 30 μm DRB at 24 h post-transfection. Cells were harvested at indicated times after DRB treatment, and cell lysates were analyzed for TTP, p38 MAPK, phosphorylated p38 MAPK, MK2, and phosphorylated MK2 by immunoblotting (IB). B, mutation of MK2 phosphorylation sites of TTP blocks the DRB-induced degradation of TTP. Colo320 cells were transfected with pcDNA6/V5-TTP or pcDNA6/V5-TTP(S60A and S186A) and were treated with DRB for 12 h. The expression level of the TTP protein was determined by immunoblotting with anti-V5 antibody. C, p38 MAPK inhibitor SB203580 attenuates the decrease mRNA decaying activity of TTP induced by siRNA against CK2α. Colo320 cells were cotransfected with psiCHECK2-VEGF 3′UTR, pcDNA6/V5-TTP, and siCK2α or scRNA. Cells were treated with SB203580 for 12 h, and luciferase activity was determined. Renilla luciferase activity was normalized to firefly activity. The luciferase activity obtained from cells transfected with psiCHECK2-VEGF 3′UTR was set to 1. Each bar represents the mean ± S.D. of three independent experiments (***, p < 0.001). D, inhibition of CK2α by DRB decreases the expression of MKP-1. Colo320 cells were treated with DRB, and cells were harvested at the indicated times after DRB treatment. Cell lysates were analyzed for MKP-1 by immunoblotting with anti-MKP-1 antibody. E, overexpression of CK2α increases the expression of MKP-1. Colo320 cells were cotransfected with pcDNA6/V5-TTP and pcDNA3/HA-CK2α. At 24 h post-transfection, expression of CK2α and MKP-1 was determined by immunoblotting. F, mutation of CK2 phosphorylation sites of MKP-1 blocks the CK2α-induced phosphorylation of MKP-1. Colo320 cells were cotransfected with pcDNA6/V5-TTP, pcDNA3/HA-CK2α, and pcDNA3.1/FLAG-MKP-1 or pcDNA3.1/FLAG-MKP-1 (S131A and S235A). At 24 h post-transfection, cell lysates were analyzed for CK2α, MKP-1, and phosphorylated MKP-1 by immunoblotting. G, inhibition of MKP-1 by siRNA abolishes the effects of CK2 on TTP expression and p38 MAPK phosphorylation. Colo320 cells were cotransfected with pcDNA6/V5-TTP, pcDNA3/HA-CK2α, and siRNA against MKP-1 (siMKP-1) or scRNA. At 24 h post-transfection, cell lysates were analyzed for CK2α, TTP, MKP-1, p38 MAPK, and phosphorylated p38 MAPK by immunoblotting. H, inhibition of MKP-1 by MKP-1 inhibitor triptolide or siRNA abolishes the effects of CK2α on mRNA decaying activity of TTP. Colo320 cells were cotransfected with psiCHECK2-VEGF 3′UTR, pcDNA6/V5-TTP, pcDNA3/HA-CK2α, and siMKP-1 or scRNA. Cells were treated with triptolide for 12 h, and the luciferase activity was determined. Renilla luciferase activity was normalized to firefly activity. The luciferase activity obtained from cells cotransfected with psiCHECK2-VEGF 3′UTR and pcDNA6/V5-TTP was set to 1. Each bar represents the mean ± S.D. of three independent experiments (*, p < 0.05; ***, p < 0.001). ns, not significant.
MKP-1 Mediates the Effects of CK2 on the Phosphorylations of p38 MAPK and TTP Activity
Inhibition of CK2 by DRB increased the phosphorylation of p38 MAPK (Fig. 6A), indicating that CK2 affects the phosphorylation of p38 MAPK via activation of protein phosphatase. MAPK phosphatases are a subclass of protein-tyrosine phosphatases that can dephosphorylate both phosphotyrosine and phosphothreonine residues on MAPKs (45). Of the known MAPK phosphatases, MKP-1 was shown to be responsible for the dephosphorylation of p38 MAPK (38). We therefore determined a potential effect of CK2 on MKP-1 expression. Inhibition of CK2 by DRB (Fig. 6D) or TBB (supplement Fig. 5) significantly decreased MKP-1 protein, whereas overexpression of CK2α increased the MKP-1 protein (Fig. 6E). Our data clearly show that CK2 acts as a positive regulator of MKP-1 expression. CK2 seems to control proteasomal degradation of MKP-1 because MG132 treatment significantly attenuated the decrease in MKP-1 induced by the CK2 inhibitor DRB (supplemental Fig. 6). Our next goal was to determine whether CK2-specific phosphorylation sites in MKP-1 protein are responsible for the effects of CK2 on the MKP-1 protein. MKP-1 protein contains two potential CK2 phosphorylation sites at serine residues Ser-131 and Ser-235. We generated a double mutant (S131A/S235A) and tested whether these mutations affect the phosphorylation of MKP-1 protein induced by CK2α overexpression. Although overexpression of CK2α increased the phosphorylation of wild-type MKP-1 protein, it did not induce that of mutant MKP-1 (Fig. 6F), indicating that CK2α directly phosphorylate MKP-1.
We tested whether MKP-1 is necessary for the effects of CK2 on the phosphorylation of p38 MAPK and degradation and mRNA decaying activity of the TTP protein. Cells were cotransfected with pcDNA6/V5-TTP, pcDNA3/HA-CK2α, and siRNA against MKP-1 (siMKP-1) or control siRNA (scRNA). At 24 h post-transfection, cells were analyzed for the expressions of CK2α, TTP, MKP-1, p38 MAPK, and phospho-p38 MAPK. Inhibition of MKP-1 by siRNA increased the phosphorylation of p38 MAPK and attenuated the increase of TTP protein induced by CK2α (Fig. 6G). These data confirmed the involvement of MKP-1 in the CK2-mediated regulation of p38 MAPK phosphorylation and TTP protein degradation. To determine whether MKP-1 is involved in the CK2-mediated regulation of the mRNA decaying activity of TTP, cells were cotransfected with pcDNA6/V5-TTP, pcDNA3/HA-CK2α, and psiCHECK2-VEGF 3′UTR. Transfected cells were treated with siRNA against MKP-1 (siMKP-1) or triptolide, a MKP-1 inhibitor, and luciferase activity was measured. Inhibition of MKP-1 by siMKP-1 or triptolide significantly attenuated the decrease in luciferase activity induced by CK2α overexpression (Fig. 6H). These data suggest that CK2 regulates TTP activity via the MKP-1 pathway.
TGF-β Increases the CK2 Activity and mRNA Decaying Activity of TTP
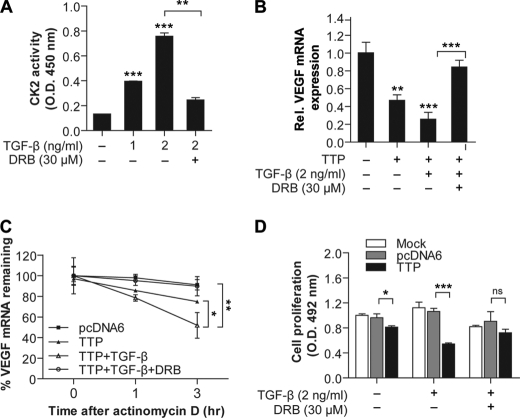
CK2 has been reported to be activated by TGF-β stimulation (46–48). To determine whether TGF-β stimulation increases CK2 activity in Colo320 cells, cells were stimulated with TGF-β for 4 h, and CK2 activity in the cell lysates was analyzed. TGF-β1 markedly enhanced CK2 activity in a dose-dependent manner in cells (Fig. 7A).
FIGURE 7.
TGF-β increases mRNA decaying activity of TTP through activation of CK2/MKP-1 pathway. A, TGF-β increases CK2 activity. Colo320 cells were treated with TGF-β at the indicated concentrations in the presence or absence of DRB for 4 h. CK2 activity in the cell lysates was determined as described under “Experimental Procedures.” The CK2 activity obtained from untreated cells was set to 1. Each bar represents the mean ± S.D. of three independent experiments (**, < 0.01; ***, p < 0.001). B, TTP-mediated down-regulation of endogenous VEGF mRNA is enhanced by TGF-β, and this effect is mediated by CK2. Colo320 cells were transfected with pcDNA6/V5-TTP. At 24 h post-transfection, cells were treated with 2 ng/ml of TGF-β in the presence or absence of DRB. VEGF mRNA was determined by quantitative real time PCR. The expression level obtained from mock-transfected and untreated cells was set to 1. Each bar represents the mean ± S.D. of three independent experiments (**, < 0.01; ***, p < 0.001). C, TTP-mediated destabilization of VEGF mRNA is enhanced by TGF-β, and this effect is mediated by CK2. Colo320 cells were transfected with pcDNA6/V5-TTP. At 24 h post-transfection, cells were treated with 2 ng/ml of TGF-β in the presence or absence of DRB for 6 h. Expression of VEGF mRNA was determined by quantitative real time PCR at indicated times after the addition of 5 μg/ml actinomycin D. Results shown on the graph represent means ± S.E. of three independent experiments (*, < 0.05; **, p < 0.01). D, TGF-β significantly enhances the growth inhibitory effect of TTP, and DRB attenuates the TGF-β effect. Colo320 cells were transfected with pcDNA6/V5-TTP or empty vector. At 24 h post-transfection, cells were treated with 2 ng/ml of TGF-β in the presence or absence of DRB. Cell viability was determined at 48 h using a 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium cell proliferation assay. The data represent means ± S.D. of three independent experiments (*, p < 0.05; ***, p < 0.001).
We next determined whether TGF-β1 enhances the mRNA decaying activity of TTP. TGF-β significantly enhanced the TTP-mediated decay of endogenous VEGF transcripts, and the CK2 inhibitor DRB reversed the effects of TGF-β on TTP activity (Fig. 7B). In addition, TGF-β decreased the half-life of VEGF mRNA, and this effect was blocked by DRB (Fig. 7C). TGF-β1 was reported to activate TTP promoter activity, thereby increasing the TTP transcription levels in the human T cell line HuT78 (10). However, in Colo320 cells, TGF-β1 did not activate TTP promoter activity (supplemental Fig. 7), indicating that the TGF-β stimulation did not increase transcription of TTP. Our results suggest that TGF-β-stimulated TTP activity is mediated by CK2.
Previously, we reported that overexpression of TTP promotes VEGF mRNA degradation and inhibits the growth of Colo320 cells (37). Thus, it is possible that TGF-β treatment inhibits the growth of Colo320 cells via the CK2-TTP pathway. To test this possibility, cells were transfected with pcDNA6/V5-TTP, and their growth was analyzed after TGF-β treatment in the presence or absence of DRB. Consistent with our previous report (37), TTP overexpression inhibited the growth of Colo320 cells (Fig. 7D). The treatment of TGF-β significantly enhanced the inhibitory effect of TTP, and DRB attenuated the TGF-β effect (Fig. 7F), indicating that TGF-β can suppress the growth of Colo320 cells through activation of the CK2-TTP pathway.
DISCUSSION
In our previous work, we showed that TTP enhances the decay of VEGF and suppresses the growth of Colo320 human colon cancer cells (37). Here, we provide evidence that CK2 increases the VEGF mRNA decaying activity of TTP through regulation of proteasomal degradation of TTP protein in Colo320 cells. (i) Inhibition of CK2 by CK2 inhibitors or siRNA treatment suppressed the TTP-inhibitory effects on the expression of a luciferase reporter gene containing VEGF mRNA 3′UTR and endogenous VEGF mRNA. (ii) Overexpression of CK2α enhanced the TTP-inhibitory effects on the expression of a luciferase reporter gene containing VEGF mRNA 3′UTR. (iii) Inhibition of CK2 by CK2 inhibitors or siRNA treatment decreased the expression level of TTP protein, and overexpression of CK2α increased it. (iv) In the cycloheximide chase experiments, a CK2 inhibitor destabilized the TTP protein, reducing its half-life from more than 5 h to less than 3 h. (v) Treatment with the proteasome inhibitor MG132 prevented the TTP protein from degradation induced by the CK2 inhibitor and led to the appearance of a phosphorylated TTP protein. (vi) Treatment with a CK2 inhibitor enhanced the phosphorylation and ubiquitination of the TTP protein.
In this study, we found that phosphorylation of TTP induced by a CK2 inhibitor led to degradation of TTP through the ubiquitin/proteasome pathway. This is in contrast to previous reports demonstrating that phosphorylation of TTP by p38 MAPK/MK2 pathways prevents TTP protein from decay by proteasome (17, 20). One possible explanation of this discrepancy is that the enhanced stability of phosphorylated TTP protein could be based on the increased binding to 14-3-3 protein (15, 16). 14-3-3 proteins influence the subcellular localization of target proteins (49, 50) and protect target proteins from proteolysis by acting as chaperones (51, 52). Consistent with these functions, binding of 14-3-3 protein alters subcellular localization of TTP (16, 17); this may affect the ubiquitination and proteasomal degradation of TTP. However, in Colo320 cells, CK2 inhibitor decreased the expression level of 14-3-3 protein (supplemental Fig. 8). Thus, it is possible to speculate that the expression level of 14-3-3 would not be enough to protect the phosphorylated TTP from degradation by the ubiquitin/proteasome pathway. A more detailed analysis of the role of 14-3-3 binding in TTP protein stability may provide insights into the relationship between phosphorylation and degradation of TTP protein.
CK2 has been thought to function as a survival factor by increasing cell proliferation (53), malignant transformation (54), and decreasing apoptosis (26). However, there are several reports that support an anti-proliferative role for CK2 (33, 35, 36). Regarding the role of CK2 in the control of VEGF expression, CK2 has been reported to be involved in the expression of VEGF (55) and the angiogenesis (56). However, even though the molecular mechanism is presently unknown, in Colo320 cells, CK2 increased the VEGF mRNA decaying activity of TTP and acted as a negative regulator of cell growth.
CK2 is able to enhance the mRNA decaying activity of TTP through phosphorylation of TTP because it is a protein kinase that phosphorylates a variety of proteins and regulates their activity (23–28). Scanning the amino acid sequence of TTP revealed four potential CK2 phosphorylation sites. Thus, it is possible that CK2 regulates TTP through phosphorylation of these CK2 sites. However, mutation of serines to alanines within the CK2 phosphorylation sites of the TTP protein did not change the effect of CK2 on the VEGF mRNA decaying activity, phosphorylation, or degradation of TTP. In addition, CK2 inhibitor increased the phosphorylation of mutant TTP proteins. Although we cannot exclude that there are other CK2 sites within TTP that can be phosphorylated by CK2, our data suggest that TTP is not the direct target of CK2.
Our data showed that CK2 can also affect the phosphorylation of p38 MAPK and MK2. Inhibition of CK2 by CK2 inhibitor increased the phosphorylation of p38 MAPK and MK2. p38 MAPK is a well known kinase that is involved in TTP phosphorylation and inhibition of mRNA decaying activity of TTP (16–18). Thus, we hypothesized that CK2 influences the mRNA decaying activity of TTP through the control of p38 MAPK phosphorylation. Consistent with this hypothesis, the p38 MAPK inhibitor SB203580 attenuated the decrease of mRNA decaying activity of TTP induced by the CK2 inhibitor. However, because the CK2 inhibitor increased the phosphorylation of p38 MAPK, p38 MAPK does not seem to be a direct target of CK2. The phosphorylation and the activity of p38 MAPK are controlled by a balance between activities of upstream activators, kinases (57), and negative regulators, protein phosphatases (58). Previously, it was reported that protein phosphatase 2a (PP2A) regulates phosphorylation of p38 MAPK and MK2 and, in turn, TTP phosphorylation (21). Here, we provide evidence that CK2 affects the phosphorylation of p38 MAPK via MAPK phosphatase-1 (MKP-1), which was shown to dephosphorylate p38 MAPK (21). Inhibition of CK2 decreased the expression level of MKP-1; overexpression of CK2α increased the MKP-1 expression level, and inhibition of MKP-1 by siRNA increased the phosphorylation of p38 MAPK. Inhibition of MKP-1 by MKP-1 inhibitor triptolide or siRNA attenuated the increase of mRNA decaying activity of TTP induced by CK2α overexpression. Collectively, these results suggest that CK2 enhances the mRNA decaying activity of TTP via MKP-1/p38 MAPK signal pathways.
We next sought to determine the factors responsible for the stimulation of CK2 activity and the increase of mRNA decaying activity of TTP. CK2 activity is modulated by various cytokines and hormones (59–61). We found that TGF-β increases the expression level of TTP protein and enhances the VEGF mRNA decaying activity of TTP. A previous report suggested that TGF-β increases the expression level of TTP by enhancing TTP promoter activity (10). However, in Colo320 cells, TGF-β did not affect TTP promoter activity, indicating that the TGF-β-induced increase in the TTP protein and the mRNA decaying activity of TTP were not due to the increase in the TTP promoter activity. Instead, we found that TGF-β enhanced the mRNA decaying activity of TTP through activation of CK2. TGF-β increased CK2 activity, and inhibition of CK2 by inhibitor DRB or siRNA abrogated the increase of mRNA decaying activity of TTP induced by TGF-β.
TGF-β is considered the most potent and widespread inhibitor of cell growth known in mammals (62). However, in contrast, TGF-β can also act as a tumor promoter in a context-dependent manner (63). Regarding angiogenesis, TGF-β was shown to promote VEGF synthesis and to exert angiogenic effects (64, 65). However, several studies have also reported that TGF-β has antiangiogenic properties (66–69). In our study, TGF-β inhibited VEGF synthesis and cell growth, indicating that TGF-β acts as a tumor suppressor in Colo320 cells. The inhibitory effect of TGF-β was abrogated by siRNA against TTP or CK2, suggesting that it was mediated by CK2/TTP pathways. The mechanisms responsible for the in vitro biphasic effect of TGF-β are not known. Recently, it was suggested that changes in the expression pattern of specific genes are responsible for the switch of TGF-β from tumor suppressor to tumor promoter (63, 70, 71). In this regard, Disabled homolog 2 (DAB2) is a good candidate gene responsible for this change. DAB2 is a multifunctional adapter protein, which is involved in TGF-β-mediated Smad activation (72) and acts as a negative regulator of multiple signaling pathways, including the ERK/MAPK (73), Src (74), and Wnt pathways (75). Hannigan et al. (70) found that down-regulation of DAB2 switches TGF-β from a tumor suppressor to a tumor promoter both in vitro and in vivo. We found that Colo320 cells express DAB2, and the expression level is slightly increased by TGF-β treatment (supplemental Fig. 9). Even though we did not analyze the effect of DAB2 expression on TGF-β functions, it is possible that DAB2 is one of the factors responsible for the inhibitory function of TGF-β in Colo320 cells.
Previously it was reported that the p38 MAPK pathway is required for TGF-β-stimulated VEGF synthesis (76, 77). Activation of p38 MAPK increases VEGF mRNA stabilization (78). This action is presumably mediated through an AU-rich region of the 3′UTR of VEGF mRNA. TTP is known to be a direct target for p38 MAPK, and phosphorylation of TTP by p38 MAPK suppresses mRNA decaying activity of TTP (7, 15–19). Thus, it is conceivable that if TGF-β signal activates p38 MAPK pathways, it can suppress the VEGF mRNA decaying activity of TTP and promote VEGF synthesis. However, in cells that express DAB2, such as Colo320 cells, TGF-β blocks the p38 MAPK pathways via a DAB2 adapter molecule and, instead, activates CK2 leading to an increase in the VEGF mRNA decaying activity of TTP via the MPK-1 pathway. Further studies are needed to address whether expression of DAB2 changes the effect of TGF-β on the VEGF mRNA decaying activity of TTP and the cell growth in Colo320 cells.
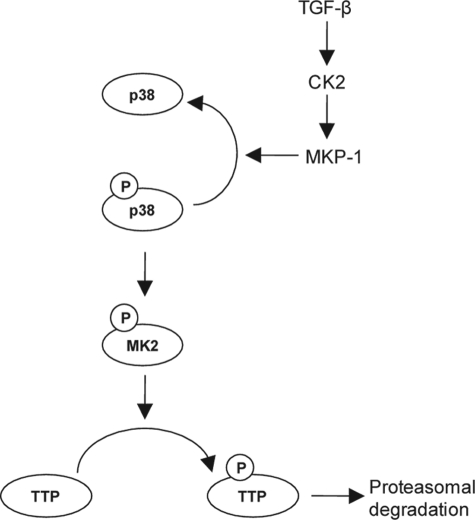
In this study, we have shown that CK2 suppresses phosphorylation of TTP through MKP-1-dependent inactivation of the p38 MAPK pathway (Fig. 8) and that these events increase the stability and the VEGF mRNA decaying activity of TTP. In contrast to our results, several studies have reported that phosphorylation of TTP by p38 MAPK protects the TTP protein from proteasomal degradation (17, 20). Ubiquitination targets the proteins for degradation by the proteasome (79), and our results indicate that CK2 influences the ubiquitination of TTP proteins. Thus, further studies to understand how CK2 controls ubiquitination of TTP protein may provide important information to explain this discrepancy. Our results also demonstrate that TGF-β suppresses VEGF synthesis and cell growth through activation of the CK2/MKP-1/TTP pathway. TGF-β and CK2 have been known to promote malignant progression and metastasis and have been considered as attractive pharmacological targets (80, 81). However, from our results, it appears that TGF-β and CK2 can negatively regulate cell proliferation. Thus, before the clinical use of inhibitors of TGF-β or CK2 is tried, further studies are required to unravel the mechanisms involved in the regulation of their functions in cell growth and to develop biomarkers predictive of cellular responses to them. Collectively, our results offer evidence of a role for the TGF-β/CK2/MKP-1 pathway (Fig. 8) in the regulation of the mRNA decaying activity of TTP.
FIGURE 8.
Model of regulation of TTP expression by CK2. The p38 MAPK/MK2 signaling pathway can induce phosphorylation (P) of TTP, which leads to proteasomal degradation of TTP protein. However, when CK2 is activated by TGF-β stimulation, it can activate MKP-1, which in turn inhibits the p38 MAPK/MK2 pathway by dephosphorylation of p38 MAPK, thereby protecting the TTP protein from proteasomal degradation.
Supplementary Material
This work was supported by Korea Research Foundation Grants KRF-2005-070-C00088, BRL2009-0087350, 2009-0094050, and 2009-0070260 funded by the Korean government (MOEHRD) and by Korea Health 21 R&D Project Grant A101086 from the Ministry of Health and Welfare.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–9.
- TTP
- tristetraprolin
- ARE
- AU-rich element
- DRB
- 5,6-dichlorobenzimidazole riboside
- TBB
- 1,3,5-tribromobenzene
- CHX
- cycloheximide.
REFERENCES
- 1. Baou M., Jewell A., Murphy J. J. (2009) J. Biomed. Biotechnol. 2009, 634520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ciais D., Cherradi N., Bailly S., Grenier E., Berra E., Pouyssegur J., Lamarre J., Feige J. J. (2004) Oncogene 23, 8673–8680 [DOI] [PubMed] [Google Scholar]
- 3. Essafi-Benkhadir K., Onesto C., Stebe E., Moroni C., Pagès G. (2007) Mol. Biol. Cell 18, 4648–4658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Suswam E., Li Y., Zhang X., Gillespie G. Y., Li X., Shacka J. J., Lu L., Zheng L., King P. H. (2008) Cancer Res. 68, 674–682 [DOI] [PubMed] [Google Scholar]
- 5. Varnum B. C., Lim R. W., Sukhatme V. P., Herschman H. R. (1989) Oncogene 4, 119–120 [PubMed] [Google Scholar]
- 6. Cao H., Urban J. F., Jr., Anderson R. A. (2008) Obesity 16, 1208–1218 [DOI] [PubMed] [Google Scholar]
- 7. Mahtani K. R., Brook M., Dean J. L., Sully G., Saklatvala J., Clark A. R. (2001) Mol. Cell. Biol. 21, 6461–6469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tchen C. R., Brook M., Saklatvala J., Clark A. R. (2004) J. Biol. Chem. 279, 32393–32400 [DOI] [PubMed] [Google Scholar]
- 9. Varnum B. C., Lim R. W., Kujubu D. A., Luner S. J., Kaufman S. E., Greenberger J. S., Gasson J. C., Herschman H. R. (1989) Mol. Cell. Biol. 9, 3580–3583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ogawa K., Chen F., Kim Y. J., Chen Y. (2003) J. Biol. Chem. 278, 30373–30381 [DOI] [PubMed] [Google Scholar]
- 11. Schaljo B., Kratochvill F., Gratz N., Sadzak I., Sauer I., Hammer M., Vogl C., Strobl B., Müller M., Blackshear P. J., Poli V., Lang R., Murray P. J., Kovarik P. (2009) J. Immunol. 183, 1197–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Smoak K., Cidlowski J. A. (2006) Mol. Cell. Biol. 26, 9126–9135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cao H., Dzineku F., Blackshear P. J. (2003) Arch. Biochem. Biophys. 412, 106–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Carballo E., Cao H., Lai W. S., Kennington E. A., Campbell D., Blackshear P. J. (2001) J. Biol. Chem. 276, 42580–42587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chrestensen C. A., Schroeder M. J., Shabanowitz J., Hunt D. F., Pelo J. W., Worthington M. T., Sturgill T. W. (2004) J. Biol. Chem. 279, 10176–10184 [DOI] [PubMed] [Google Scholar]
- 16. Stoecklin G., Stubbs T., Kedersha N., Wax S., Rigby W. F., Blackwell T. K., Anderson P. (2004) EMBO J. 23, 1313–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Brook M., Tchen C. R., Santalucia T., McIlrath J., Arthur J. S., Saklatvala J., Clark A. R. (2006) Mol. Cell. Biol. 26, 2408–2418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hitti E., Iakovleva T., Brook M., Deppenmeier S., Gruber A. D., Radzioch D., Clark A. R., Blackshear P. J., Kotlyarov A., Gaestel M. (2006) Mol. Cell. Biol. 26, 2399–2407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Johnson B. A., Stehn J. R., Yaffe M. B., Blackwell T. K. (2002) J. Biol. Chem. 277, 18029–18036 [DOI] [PubMed] [Google Scholar]
- 20. Deleault K. M., Skinner S. J., Brooks S. A. (2008) Mol. Immunol. 45, 13–24 [DOI] [PubMed] [Google Scholar]
- 21. Sun L., Stoecklin G., Van Way S., Hinkovska-Galcheva V., Guo R. F., Anderson P., Shanley T. P. (2007) J. Biol. Chem. 282, 3766–3777 [DOI] [PubMed] [Google Scholar]
- 22. Graham K. C., Litchfield D. W. (2000) J. Biol. Chem. 275, 5003–5010 [DOI] [PubMed] [Google Scholar]
- 23. Ahmed K. (1999) Crit. Rev. Eukaryot. Gene Expr. 9, 329–336 [DOI] [PubMed] [Google Scholar]
- 24. Guerra B., Boldyreff B., Sarno S., Cesaro L., Issinger O. G., Pinna L. A. (1999) Pharmacol. Ther. 82, 303–313 [DOI] [PubMed] [Google Scholar]
- 25. Pinna L. A. (2002) J. Cell Sci. 115, 3873–3878 [DOI] [PubMed] [Google Scholar]
- 26. Litchfield D. W. (2003) Biochem. J. 369, 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tawfic S., Yu S., Wang H., Faust R., Davis A., Ahmed K. (2001) Histol. Histopathol. 16, 573–582 [DOI] [PubMed] [Google Scholar]
- 28. Ahmed K., Davis A. T., Wang H., Faust R. A., Yu S., Tawfic S. (2000) J. Cell Biochem. Suppl. 35, 130–135 [DOI] [PubMed] [Google Scholar]
- 29. Guerra B., Issinger O. G. (1999) Electrophoresis 20, 391–408 [DOI] [PubMed] [Google Scholar]
- 30. Meggio F., Pinna L. A. (2003) FASEB J. 17, 349–368 [DOI] [PubMed] [Google Scholar]
- 31. Ahmad K. A., Wang G., Slaton J., Unger G., Ahmed K. (2005) Anticancer Drugs 16, 1037–1043 [DOI] [PubMed] [Google Scholar]
- 32. Mehra A., Shi M., Baker C. L., Colot H. V., Loros J. J., Dunlap J. C. (2009) Cell 137, 749–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hériché J. K., Lebrin F., Rabilloud T., Leroy D., Chambaz E. M., Goldberg Y. (1997) Science 276, 952–955 [DOI] [PubMed] [Google Scholar]
- 34. Meek D. W., Simon S., Kikkawa U., Eckhart W. (1990) EMBO J. 9, 3253–3260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hupp T. R., Meek D. W., Midgley C. A., Lane D. P. (1992) Cell 71, 875–886 [DOI] [PubMed] [Google Scholar]
- 36. Sayed M., Pelech S., Wong C., Marotta A., Salh B. (2001) Oncogene 20, 6994–7005 [DOI] [PubMed] [Google Scholar]
- 37. Lee H. H., Son Y. J., Lee W. H., Park Y. W., Chae S. W., Cho W. J., Kim Y. M., Choi H. J., Choi D. H., Jung S. W., Min Y. J., Park S. E., Lee B. J., Cha H. J., Park J. W. (2010) Int. J. Cancer 126, 1817–1827 [DOI] [PubMed] [Google Scholar]
- 38. Chen P., Hutter D., Yang X., Gorospe M., Davis R. J., Liu Y. (2001) J. Biol. Chem. 276, 29440–29449 [DOI] [PubMed] [Google Scholar]
- 39. Cao H., Lin R. (2008) Protein J. 27, 163–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sandler H., Stoecklin G. (2008) Biochem. Soc. Trans. 36, 491–496 [DOI] [PubMed] [Google Scholar]
- 41. Lee H. H., Vo M. T., Kim H. J., Lee U. H., Kim C. W., Kim H. K., Ko M. S., Lee W. H., Cha S. J., Min Y. J., Choi D. H., Suh H. S., Lee B. J., Park J. W., Cho W. J. (2010) J. Biol. Chem. 285, 17329–17337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Godl K., Wissing J., Kurtenbach A., Habenberger P., Blencke S., Gutbrod H., Salassidis K., Stein-Gerlach M., Missio A., Cotten M., Daub H. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 15434–15439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. St-Denis N. A., Litchfield D. W. (2009) Cell. Mol. Life Sci. 66, 1817–1829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rigby W. F., Roy K., Collins J., Rigby S., Connolly J. E., Bloch D. B., Brooks S. A. (2005) J. Immunol. 174, 7883–7893 [DOI] [PubMed] [Google Scholar]
- 45. Owens D. M., Keyse S. M. (2007) Oncogene 26, 3203–3213 [DOI] [PubMed] [Google Scholar]
- 46. Zdunek M., Silbiger S., Lei J., Neugarten J. (2001) Kidney Int. 60, 2097–2108 [DOI] [PubMed] [Google Scholar]
- 47. Negulescu O., Bognar I., Lei J., Devarajan P., Silbiger S., Neugarten J. (2002) Kidney Int. 62, 1989–1998 [DOI] [PubMed] [Google Scholar]
- 48. Singh N. N., Ramji D. P. (2006) Arterioscler. Thromb. Vasc. Biol. 26, 1323–1329 [DOI] [PubMed] [Google Scholar]
- 49. Lopez-Girona A., Furnari B., Mondesert O., Russell P. (1999) Nature 397, 172–175 [DOI] [PubMed] [Google Scholar]
- 50. Seimiya H., Sawada H., Muramatsu Y., Shimizu M., Ohko K., Yamane K., Tsuruo T. (2000) EMBO J. 19, 2652–2661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Jeanclos E. M., Lin L., Treuil M. W., Rao J., DeCoster M. A., Anand R. (2001) J. Biol. Chem. 276, 28281–28290 [DOI] [PubMed] [Google Scholar]
- 52. Vincenz C., Dixit V. M. (1996) J. Biol. Chem. 271, 20029–20034 [DOI] [PubMed] [Google Scholar]
- 53. Litchfield D. W., Slominski E., Lewenza S., Narvey M., Bosc D. G., Gietz R. D. (1996) Biochem. Cell Biol. 74, 541–547 [DOI] [PubMed] [Google Scholar]
- 54. Faust R. A., Gapany M., Tristani P., Davis A., Adams G. L., Ahmed K. (1996) Cancer Lett. 101, 31–35 [DOI] [PubMed] [Google Scholar]
- 55. Brown M. S., Diallo O. T., Hu M., Ehsanian R., Yang X., Arun P., Lu H., Korman V., Unger G., Ahmed K., Van Waes C., Chen Z. (2010) Clin. Cancer Res. 16, 2295–2307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kramerov A. A., Saghizadeh M., Caballero S., Shaw L. C., Li Calzi S., Bretner M., Montenarh M., Pinna L. A., Grant M. B., Ljubimov A. V. (2008) Mol. Cell. Biochem. 316, 177–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Avruch J. (2007) Biochim. Biophys. Acta 1773, 1150–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Keyse S. M. (2000) Curr. Opin. Cell Biol. 12, 186–192 [DOI] [PubMed] [Google Scholar]
- 59. Ackerman P., Glover C. V., Osheroff N. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 821–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Orlandini M., Semplici F., Ferruzzi R., Meggio F., Pinna L. A., Oliviero S. (1998) J. Biol. Chem. 273, 21291–21297 [DOI] [PubMed] [Google Scholar]
- 61. Sommercorn J., Mulligan J. A., Lozeman F. J., Krebs E. G. (1987) Proc. Natl. Acad. Sci. U.S.A. 84, 8834–8838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Siegel P. M., Massagué J. (2003) Nat. Rev. Cancer 3, 807–821 [DOI] [PubMed] [Google Scholar]
- 63. Roberts A. B., Wakefield L. M. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 8621–8623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Brogi E., Wu T., Namiki A., Isner J. M. (1994) Circulation 90, 649–652 [DOI] [PubMed] [Google Scholar]
- 65. Stavri G. T., Zachary I. C., Baskerville P. A., Martin J. F., Erusalimsky J. D. (1995) Circulation 92, 11–14 [DOI] [PubMed] [Google Scholar]
- 66. Pepper M. S. (1997) Cytokine Growth Factor Rev. 8, 21–43 [DOI] [PubMed] [Google Scholar]
- 67. Zheng W., Seftor E. A., Meininger C. J., Hendrix M. J., Tomanek R. J. (2001) Am. J. Physiol. Heart Circ. Physiol. 280, H909–H917 [DOI] [PubMed] [Google Scholar]
- 68. van Royen N., Hoefer I., Buschmann I., Heil M., Kostin S., Deindl E., Vogel S., Korff T., Augustin H., Bode C., Piek J. J., Schaper W. (2002) FASEB J. 16, 432–434 [DOI] [PubMed] [Google Scholar]
- 69. Mallet C., Vittet D., Feige J. J., Bailly S. (2006) Stem Cells 24, 2420–2427 [DOI] [PubMed] [Google Scholar]
- 70. Hannigan A., Smith P., Kalna G., Lo Nigro C., Orange C., O'Brien D. I., Shah R., Syed N., Spender L. C., Herrera B., Thurlow J. K., Lattanzio L., Monteverde M., Maurer M. E., Buffa F. M., Mann J., Chu D. C., West C. M., Patridge M., Oien K. A., Cooper J. A., Frame M. C., Harris A. L., Hiller L., Nicholson L. J., Gasco M., Crook T., Inman G. J. (2010) J. Clin. Invest. 120, 2842–2857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Tang B., Vu M., Booker T., Santner S. J., Miller F. R., Anver M. R., Wakefield L. M. (2003) J. Clin. Invest. 112, 1116–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Hocevar B. A., Smine A., Xu X. X., Howe P. H. (2001) EMBO J. 20, 2789–2801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Zhou J., Hsieh J. T. (2001) J. Biol. Chem. 276, 27793–27798 [DOI] [PubMed] [Google Scholar]
- 74. Zhou J., Scholes J., Hsieh J. T. (2003) J. Biol. Chem. 278, 6936–6941 [DOI] [PubMed] [Google Scholar]
- 75. Hocevar B. A., Mou F., Rennolds J. L., Morris S. M., Cooper J. A., Howe P. H. (2003) EMBO J. 22, 3084–3094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Yamamoto T., Kozawa O., Tanabe K., Akamatsu S., Matsuno H., Dohi S., Uematsu T. (2001) J. Cell. Biochem. 82, 591–598 [DOI] [PubMed] [Google Scholar]
- 77. Bian Z. M., Elner S. G., Elner V. M. (2007) Exp. Eye Res. 84, 812–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Pagès G., Berra E., Milanini J., Levy A. P., Pouysségur J. (2000) J. Biol. Chem. 275, 26484–26491 [DOI] [PubMed] [Google Scholar]
- 79. Hershko A., Ciechanover A. (1998) Annu. Rev. Biochem. 67, 425–479 [DOI] [PubMed] [Google Scholar]
- 80. Saunier E. F., Akhurst R. J. (2006) Curr. Cancer Drug Targets 6, 565–578 [DOI] [PubMed] [Google Scholar]
- 81. Sarno S., Pinna L. A. (2008) Mol. Biosyst. 4, 889–894 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.