Abstract
Sesquiterpene lactones (STLs) are terpenoid natural products possessing the γ-lactone, well known for their diverse biological and medicinal activities. The occurrence of STLs is sporadic in nature, but most STLs have been isolated from plants in the Asteraceae family. Despite the implication of the γ-lactone group in many reported bioactivities of STLs, the biosynthetic origins of the γ-lactone ring remains elusive. Germacrene A acid (GAA) has been suggested as a central precursor of diverse STLs. The regioselective (C6 or C8) and stereoselective (α or β) hydroxylation on a carbon of GAA adjacent to its carboxylic acid at C12 is responsible for the γ-lactone formation. Here, we report two cytochrome P450 monooxygenases (P450s) capable of catalyzing 6α- and 8β-hydroxylation of GAA from lettuce and sunflower, respectively. To identify these P450s, sunflower trichomes were isolated to generate a trichome-specific transcript library, from which 10 P450 clones were retrieved. Expression of these clones in a yeast strain metabolically engineered to synthesize substrate GAA identified a P450 catalyzing 8β-hydroxylation of GAA, but the STL was not formed by spontaneous lactonization. Subsequently, we identified the closest homolog of the GAA 8β-hydroxylase from lettuce and discovered 6α-hydroxylation of GAA by the recombinant enzyme. The resulting 6α-hydroxy-GAA spontaneously undergoes a lactonization to yield the simplest form of STL, costunolide. Furthermore, we demonstrate the milligram per liter scale de novo synthesis of costunolide using the lettuce P450 in an engineered yeast strain, an important advance that will enable exploitation of STLs. Evolution and homology models of these two P450s are discussed.
Keywords: Cytochrome P450, Enzyme Catalysis, Enzymes, Metabolism, Plant, Genomics
Introduction
Sesquiterpenoids are structurally diverse isoprenoid natural products derived from C15 farnesyl diphosphate. Among them, one major subclass is represented by sesquiterpene lactone (STL),2 which has a characteristic γ-lactone moiety on its C15 backbone. Many STLs are known to have strong bioactivity partly due to their lactone ring moiety. STLs of known pharmaceutical activities include artemisinin for malarial treatment (1), parthenolide for a migraine medication (2), santonin as an anthelmintic drug (3), and thapsigargin for a sarco-endoplasmic reticulum calcium-ATPase inhibitor in the treatment of certain cancers (4). In addition, many other STLs also have ecophysiological roles as allelochemicals, insect repellents, animal allergens, and poisons (5, 6).
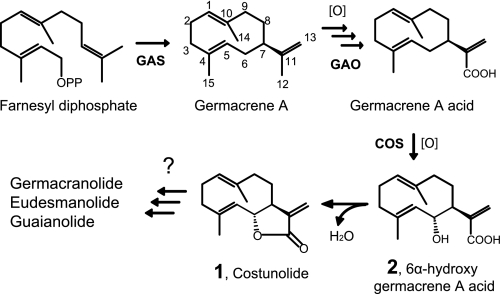
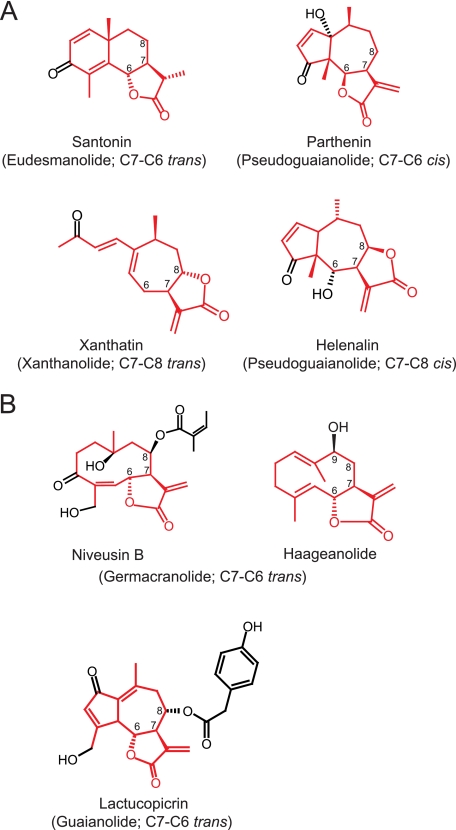
The simplest STL, costunolide, is derived from germacrene A backbone (see Fig. 1), but several variations of sesquiterpene backbones also occur in nature such as germacranolide, eudesmanolide, guaianolide, pseudo-guaianolide, xanthanolide, and bakkenolide (5–7). Some of these structures are depicted in Fig. 2. In addition to their sesquiterpene backbones, STLs have structural variations on their lactone rings. In nature, a majority of STLs possess the typical α-methylene γ-lactone moiety, but the methylene group is occasionally reduced to a single bond (see santonin in Fig. 2A and artemisinin). Lactone rings can also form either in cis- or trans-configuration with C6-C7 or C7-C8 regiospecificity (see Fig. 2A) (5, 7), but the biochemical basis of this lactone ring formation is largely unknown.
FIGURE 1.
Costunolide biosynthetic pathway in Asteraceae. COS, costunolide synthase.
FIGURE 2.
Structures of STLs found in various Asteraceae plants. A, four representative structures of STLs with distinct regio- and stereo-characteristics of γ-lactone rings are shown, and their sesquiterpene backbones are labeled in red. B, structures of STLs found in sunflower cv. HA300 and lettuce. Santonin is from Artemisia maritima (Anthemideae, Asteroideae); parthenin is from Parthenium hysterophorus (Heliantheae, Asteroideae); xanthatin is from Xanthium strumarium (Heliantheae, Asteroideae); helenalin is from Arnica montana (Heliantheae, Asteroideae); niveusin B and haageanolide are from H. annuus (Heliantheae, Asteroideae); and lactucopicrin is from L. sativa (Cichorieae, Cichorioideae). Tribe and subfamily are given in parentheses.
STLs are present sporadically in several plant families such as Magnoliaceae (8), Lauraceae (9), Cupressaceae (10), and Apiaceae (11), but they are most commonly found in Asteraceae (or Compositae) (12). Asteraceae first appeared in South America ∼50 million years ago (13, 14) and has since radiated on all continents to become one of the largest families of land plants (15). Throughout the diversification of this family, most Asteraceae plants retain STLs as a major secondary metabolic constituents, suggesting a positive selection of STL biosynthesis in the adaptive evolution. Therefore, studies of STL in Asteraceae through the 50-million year evolutionary time frame presents an ideal model to understand the adaptive enzyme evolution and chemical diversifications in distinct ecological niches. Considering the likely evolutionary significance of STL biosynthesis in Asteraceae, comparative metabolic and genomics studies from various medicinal Asteraceae plants will facilitate elucidation of the STL biosynthetic pathways in other plants.
Sunflower (Helianthus annuus) and lettuce (Lactuca sativa) are the two representative crop species of Asteraceae, from which extensive genomics resources have been generated. Phytochemical analyses showed that they synthesize structurally related yet distinct STLs (see Fig. 2B) (16, 17). For sunflower and lettuce STL biosynthesis, it has been shown that farnesyl diphosphate is cyclized to form C15 hydrocarbon germacrene A by germacrene A synthase (GAS) (Fig. 1) (18–20). Subsequently, the C12 methyl group of germacrene A is oxidized three times by a single cytochrome P450 monooxygenase (P450), germacrene A oxidase (GAO), to yield germacrene A acid (germacra-1(10),4,11(13)-trien-12-oic acid; GAA) (21). GAO are evolutionarily conserved across the Asteraceae family whereas the homolog, amorphadiene oxidase, uniquely diverged in a single species of Artemisia annua (21, 22). The biochemical data from these studies suggested that the three sequential oxidative reactions by GAO are conserved at the basal species, Barnadesia spinosa, as well as three major subfamilies of Asteraceae (Asteroideae, Cichorioideae, and Carduoideae) (21). Minor mutations on GAO may give rise to amorphadiene oxidase to utilize the new substrate amorphadiene in A. annua by an unknown co-evolutionary mechanism (21).
Despite our comprehensive knowledge of GAA biosynthesis in various Asteraceae plants, it remains unknown how the central γ-lactone ring is synthesized from GAA. Cell-free enzyme assays using chicory root extract hinted that 6α-hydroxylation of GAA is catalyzed by a P450-type reaction, followed by a spontaneous lactone ring formation to yield costunolide (Fig. 1) (23). It has been proposed that costunolide can be converted to eudesmanolide and guaianolide, and the chicory cell-free extract could transform costunolide to a guaianolide compound, leucodin (23).
The germacranolide-type STL, costunolide, is accepted as a central precursor of diverse STLs. Costunolide was first isolated from costus roots (Saussurea lappa) (24) and is now found in several plant species, including Magnolia spp. and lettuce (25, 26). It is known that costunolide is not only the precursor of STLs but also displays several bioactivities. Reported pharmaceutical activities of costunolide include anti-inflammatory (25), anti-carcinogenic (27), anti-diabetic (28), anti-fungal (29), and anti-viral activities (30). Hence, costunolide can be a valuable chemical feedstock as the precursor of various bioactive STLs. However, it only accumulates as a metabolic end product in a limited number of plant species such as an endangered Himalayan medicinal plant costus (S. lappa). Therefore, research aimed at the elucidation of costunolide biosynthesis is important not only as the first step to the understanding of STL biosynthesis but also for the sustainable supply of costunolide without perturbing delicate ecosystems.
In this report, we present the cloning and functional characterization of P450 genes from lettuce and sunflower and identified GAA hydroxylation activities through use of a metabolically engineered yeast strain. The coding regions of these P450s show 65% sequence identity, whereas the enzymes catalyze distinct 6α- or 8β-hydroxylation of GAA. We further found that the 6α-hydroxy-GAA can undergo a spontaneous lactonization to form costunolide. Using this regio- and stereoselective P450, we synthesized several milligrams of costunolide from a liter culture of the engineered yeast strain. This is the first report to use a regio- and stereoselective P450 to microbially manufacture the simplest STL costunolide and a major step toward the exploitation of this family of bioactive terpene natural products.
EXPERIMENTAL PROCEDURES
Plant Materials
H. annuus L. cv. HA300 and Lactuca sativa cv. Mariska were grown under greenhouse conditions with 16 h of light (330 μmol s−1 m−2) and 8 h of dark.
Costunolide Standard and Structural Confirmation
The authentic costunolide standard was purchased from AvaChem Scientific (San Antonio, TX). The structure of the purchased costunolide was confirmed by one-dimensional and two-dimensional NMR analyses. Experiments performed were one-dimensional proton, one-dimensional 13C with proton decoupling, 13C attached proton test with proton decoupling, COSY, Total Correlation Spectroscopy, Heteronuclear Single Quantum Coherence, and Heteronuclear Multiple Bond Coherence.
RNA Isolation and cDNA Library Construction
Trichomes found in the anther appendages of sunflower florets were used to generate a trichome-specific library. Trichomes in the secretory stage were manually isolated as described (16). The trichomes were collected in 200 μl ice-cold RNA extraction buffer (Aurum Total RNA Isolation kit, Bio-Rad). When trichomes from 200 florets (∼40,000 trichomes) were collected, the vial was frozen at −80 °C. Altogether, trichomes from 5,000 florets were isolated. For total RNA isolation, the frozen aliquots were thawed on ice. Two ceramic beads (2.8-mm diameter, Precellys, Peqlab) were added to each vial and mixer mill (MM20, Retsch) was used for cell disruption (16 Hz, 1.5 min). Total RNA was isolated by the Aurum total RNA isolation kit according to the manufacturer's instructions. RNA quantity and integrity was evaluated on a Bioanalyzer 2100 using a RNA 6000 Pico Chip (Agilent). The SMART cDNA library synthesis kit was used to generate the cDNA library in the pDNR-lib vector (Clontech). 5′-Single pass sequencings of 1,130 clones were performed by the Quintara Biosciences, and the sequences were assembled by the FIESTA system at the National Research Council-Plant Biotechnology Institute (Saskatoon, Canada).
cDNA Isolation and Plasmid Construction
Oligonucleotides used in this work are given in supplemental Table 1. 5′- or 3′-RACE was performed using a SMART RACE cDNA Amplification kit (Clontech), following the recommended protocol. 5′-RACE was conducted for C49, C63, C113, C12, C7, C100, C28, S1, S2, and S3. 3′-RACE was conducted for LsCOS using lettuce cDNA. The open reading frames (ORFs) of these P450s were subcloned into pYES-DEST52 or pESC-Ura vector for expression. The ORFs of C49, C113, C63, and C7 were amplified using primers (1a–4b). C49 and C113 were re-amplified by a pair of primers, 5a and 5b, and then cloned into pDONR 221 plasmid by the gateway BP reaction. C63 and C7 were cloned into pENTR/D-TOPO vector according to the provided protocol (Invitrogen). The gateway LR reactions were performed for C49, C113, C63, and C7 to generate the respective yeast expression plasmids in pYES-DEST52 according to the provided protocol (Invitrogen). These ORFs were cloned in translational fusions with the V5 epitope in pYES-DEST52 vector. Independently, the ORFs of C12, C100, C28, S1, S2, and S3 were amplified using primers (6a–11b). Amplified fragments were digested with NheI or XbaI and subcloned into the SpeI site of pESC-Ura vector to make translational fusions to the FLAG epitope. Expression of the cloned P450 genes were assessed by immunoblots using commercially available anti-V5 or anti-FLAG antibodies. For functional in vivo screening, these plasmids and substrate-supplying plasmid, pESC-Leu2d::GAS/LsGAO/CPR, were co-transformed in the EPY300 strain (21, 31).
For in vitro enzyme assay, sunflower C12 (HaG8H) and LsCOS were co-expressed with Artemisia annua CPR in pESC-Ura vector. To make this dual expression plasmid, A. annua CPR from the pESC-Ura::CPR plasmid was digested by BamHI and SalI, and the digested fragment was ligated to the corresponding sites in pESC-Ura::C12, resulting in pESC-Ura::C12/CPR. Partial sequences at the start codon of LsCOS were obtained from the Compositae Genome Project Database of the University of California Davis. After determining the sequences at the stop codon by 3′-RACE, the ORF of LsCOS was amplified from the cDNA templates from lettuce leaf with a primer pair, 12a and 12b, followed by the digestion with XbaI and ligation into the SpeI site of pESC-Ura::CPR plasmid, resulting in pESC-Ura::CPR/LsCOS.
For de novo synthesis of costunolide in yeast, a quadruple expression plasmid, pESC-Leu2d::GAS/LsGAO/CPR/LsCOS, was constructed as follows. The plasmid, pESC-Leu2d::GAS/LsGAO/CPR (21), was digested with ScaI and BspEI. The digested product containing partial sequence of GAS and full-length sequences of LsGAO and CPR was ligated to the corresponding sites of pESC-Leu2d::GAS. This cloning created a plasmid, pESC-Leu2d::GAS/LsGAO/CPR-Gal10_Cassette, which contains GAS/LsGAO/CPR and a newly introduced empty cloning site (i.e. Gal10 promoter-multiple cloning site-ADH1 terminator cassette). An ORF of LsCOS was amplified from pESC-Ura::CPR/LsCOS plasmid using primers 13a and 13b. The amplified PCR products were first cloned into pGEM-T Easy vector and then digested with SpeI. The digested products were ligated into the SpeI site of the pESC-Leu2d::GAS/LsGAO/CPR-Gal10_Cassette. This cloning created a quadruple expression plasmid, pESC-Leu2d::GAS/LsGAO/CPR/LsCOS.
Yeast Cultivation and Metabolite Extraction
For standard yeast culture, the transgenic yeast strain of interest was inoculated in 3 ml of synthetic complete medium omitting the appropriate amino acids with 2% Glc. The inocula were cultured overnight in 30 °C at 200 rpm. The start culture was diluted 25-fold in the synthetic complete medium omitting the appropriate amino acids with 1.8% Gal and 0.2% Glc. When using EPY300 for the in vivo production of sesquiterpenoids, methionine was added to the culture at a final concentration of 1 mm. To avoid acid-induced cyclization of germacrene A and germacrene A acid, a final concentration of 100 or 150 mm HEPES/NaOH (pH 7.5) was added to the culture medium to maintain a culture pH of >6.0. After yeast was cultured for 72 to 120 h in 30 °C at 200 rpm, the culture medium was adjusted to pH 6 with 5 n HCl, and the medium was extracted with ethyl acetate. The ethyl acetate fractions were evaporated in N2 gas or by a rotary evaporator, and the metabolites were dissolved in methanol.
Preparation of Germacrene A Acid
The EPY300 yeast strain transformed with pESC-Leu2d::GAS/LsGAO/CPR was cultured in neutralized medium with 100 mm HEPES/NaOH (pH 7.5) at 30 °C for 3 to 4 days. Crude GAA for the C12 in vitro enzyme assay was prepared by extracting the culture medium with ethyl acetate and replacing the extract solvent with methanol. For in vitro enzyme assays of LsCOS, GAA was purified through a HPLC system (Waters 2795 Separation Module; Waters SunFire C18 column, 3.5 μm, 4.6 × 150 mm; Waters 2996 Photodiode Array Detector with UV wavelength at 195 nm). The separation was achieved with a solvent gradient of 30:70 (A:B) to 28:72 (A:B) over 8 min at 1 ml min−1 and 40 °C column temperature (A, H2O with 0.1% acetic acid; B, 100% acetonitrile). To avoid acid-induced cyclization of GAA, the GAA fractions were collected into 250 mm ammonium acetate solution in which pH was kept at >6. Then, the ammonium acetate solution was adjusted to pH 6.0 with acetic acid, and GAA was recovered from ammonium acetate solution using the Sep-Pak Plus C18 cartridge (Waters). After the elution of GAA from the cartridge with 100% acetonitrile, the acetonitrile fraction was evaporated under the N2 stream. The purified GAA was dissolved in dimethyl sulfoxide.
Microsome Preparation and in Vitro Enzyme Assay
For in vitro enzyme assay, the protease-deficient Saccharomyces cerevisiae YPL154C:PEP4 KO strain was transformed with pESC-Ura::CPR, pESC-Ura::C12/CPR, or pESC-Ura::CPR/LsCOS. After cultivation in 2% Glc, the transgenic yeasts were transferred to the fresh medium with 2% Gal at a density of 0.4 at A600 and further cultivated for additional 17 h. Microsomes were prepared as described previously (21). The in vitro enzyme reactions were carried out in 1 ml of 50 mm HEPES/NaOH (pH 7.5) buffer containing microsomal protein, substrate, and 500 μm NADPH. C12 and its vector control reactions were conducted with 4 mg of microsomal proteins and crude GAA as a substrate at 28 °C for 2 h, whereas LsCOS and its vector control reactions were carried out with 1 mg of microsomal proteins and 100 μm of purified GAA at 30 °C for 1 h. The reaction products were extracted with ethyl acetate three times, and the ethyl acetate was replaced with methanol for LC-MS analysis.
LC-MS Analysis
LC-MS analysis was performed using an Agilent 1200 Rapid Resolution LC system coupled with Agilent 6410 MS. 10–40 μl of samples were injected onto a reverse phase C18 column (2.1 × 50 mm, 1.8 μm, Eclipse plus C18 Zorbax) with the flow rate of 0.4 ml min−1 at 40 °C column temperature. For detection of the in vitro enzyme assay product of C12 as well as the de novo synthesized costunolide, solvent gradient of 80:20 (A:B) to 20:80 (A:B) for 12 min (A: H2O with 1% acetic acid; B: 100% acetonitrile) was used. For detection of the in vitro enzyme assay product of LsCOS, solvent gradient of 80:20 (A:B) to 0:100 (A:B) for 10.5 min followed by a 4 min post-run was used. The initial 0.5 min of LC was operated at an isocratic mode with solvent composition 80:20 (A:B) in all analyses. For product ion scan of costunolide by LC-MS/MS, fragmenting and collision induced dissociation voltages were optimized to be 92 and 20 V, respectively.
Preparation of C6-Hydroxy-GAA
The lactone moiety of costunolide was opened by alkaline treatment. The reaction was carried out in 62.5% methanol solution containing 1 mm costunolide standard and 10 mm NaOH at 55 °C for 1 h. After the reaction, three products were mainly detected with the reversed-phase HPLC analysis. One of the products, which showed the earliest retention time, was C6-hydroxy-GAA, whereas the second compound remained unknown, and the last compound was costunolide. The purification was conducted by HPLC with a solvent gradient of 50:50 (A:B) to 40:60 (A:B) over 8 min at 1 ml min−1 and 40 °C column temperature (A, H2O with 0.1% acetic acid; B, 100% acetonitrile). The eluted C6-hydroxy-GAA was collected in 500 mm HEPES/NaOH (pH 7.5) to avoid acid-induced cyclization (final concentration of HEPES/NaOH after collection was ∼50 mm). The HPLC analysis confirmed that the purified C6-hydroxy-GAA was spontaneously converted to costunolide even in a neutral pH condition, and the conversion rate was facilitated in elevated temperatures.
Purification of C12 Enzymatic Reaction Product
To purify C12 enzymatic product, crude GAA was prepared as described above, and the solvent was replaced by dimethyl sulfoxide. The crude GAA in dimethyl sulfoxide was fed to the yeast culture of YPL154C:PEP4 KO strain containing pESC-Ura::C12/CPR. The culture was incubated for 4 days, and the pH of culture medium was adjusted to 6, followed by ethyl acetate extraction. The ethyl acetate solvent was replaced with methanol for purification through the HPLC. The extract was fractionated by a solvent gradient of 40:60 (A:B) to 38.4:61.6 (A:B) over 8 min at 1 ml min−1 and 40 °C column temperature (A, H2O with 0.1% acetic acid; B, 100% acetonitrile). The eluted C12 metabolic product was collected in 200 mm HEPES/NaOH (pH 7.5) to avoid acid-induced cyclization (final concentration of HEPES/NaOH after collection was ∼140 mm). The purified fraction was diluted 4 times with H2O, and its pH was adjusted to 6 by 1 n HCl. The C12 metabolic product was purified by the Sep-Pak Plus C18 cartridge or ethyl acetate extraction. When using ethyl acetate, repeated cycles of extraction and washing with H2O was conducted to avoid the contamination of HEPES/NaOH buffer into the sample.
Purification of LsCOS Enzymatic Reaction Product
The EPY300 strain transformed with pESC-Leu2d::GAS/LsGAO/CPR/LsCOS was incubated for 4 days in neutralized medium in 30 °C at 200 rpm. The culture medium was pH-adjusted and extracted with ethyl acetate, and the solvent was replaced with methanol for purification by HPLC. The separation condition was the same as the one used for C12 product purification except that acetic acid was removed from the solvent A. After collecting targeted peak, the solvent was evaporated using a rotary evaporator. After the evaporation of acetonitrile in the solution, the sample was subjected to the lyophilizer to get a powder of compound for NMR analysis.
NMR Analyses
For structural analysis of 8β-hydroxy-GAA, 1H and 13C NMR spectra were measured in 5-mm standard NMR tubes at 400.13 and 100.6 MHz on a Bruker AVANCE 400 spectrometer equipped with 5-mm inverse probe with triple axis gradients. Chemical shifts (δ) for both 13C and 1H were referenced to internal tetramethylsilane. Experiments performed were standard one- and two-dimensional NMR analyses, including COSY, NOESY, TOCSY, HSQC, and HMBC. The structure of the de novo-synthesized costunolide was confirmed by matching its NMR signals to the authentic standard. NMR spectra for standard and de novo-synthesized costunolide were obtained on a Varian 700 MHz spectrometer equipped with an inverse detection, cryo-cooled triple resonance, and Z-gradient probe. 1H-NMR chemical shifts are reported using the residual proton resonance of solvents as reference, CDCl3 δH; 7.24, and 13C-NMR chemical shifts are reported relative to CDCl3 δC; 77.0. For structural analysis of 8β-hydroxyilicic acid, NMR spectra were recorded in 3 mm standard NMR tubes on a Varian Unity Inova 500 MHz spectrometer equipped with a 3-mm ID-PFG probe. The 1H and 13C NMR chemical shifts were referenced to solvent signals at δH/C 7.14/127.68 (C6D6) relative to TMS. One- and two-dimensional homonuclear NMR spectra were measured with standard Varian pulse sequences, and the experiments performed included COSY, TOCSY, ROESY, HSQC, and HMBC.
Small Molecule Modeling
Energy minimized structures and heat of formation (ΔHf0) of 6α- and 8β-hydroxy-GAA and their lactones were obtained through molecular modeling by PCMODEL (version 8.0, Serena Software). The structures were optimized by sequential application of Minim, Dynam, and GMMX. To obtain minimum distances between the hydroxyl groups and C12, the dihedral driver routine was applied to simulate rotations around the C7–C11 bond. The same method was employed to model GAA, and the energy minimized GAA was used as a substrate in the homology modeling studies (supplemental Fig. 4).
Sequence Deposition
The sequences of cDNAs described in this work were deposited in the GenBank data library under the following accession numbers: C49, HQ439587; C63, HQ439588; C113, HQ439589; C12, HQ439590; C7, HQ439591; C100, HQ439592; C28, HQ439593; S1, HQ439594; S2, HQ439596; S3, HQ439598; and LsCOS, HQ439599.
Homology Modeling and Docking
Structural models for LsCOS and HaG8H were created by the program MODELLER via the ModBase server (32) using human cytochrome P450 CYP2E1 as a template. Molecular graphics images were produced and analyzed using the University of California, San Francisco Chimera package from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (33). Energy-minimized GAA was manually positioned in homology models to orient C6 or C8 toward the heme center, based on established regiochemstry of LsCOS and HaG8H.
RESULTS
Construction of cDNA Library from Sunflower Capitate Glandular Trichomes
Costunolide synthase, which catalyzes 6α-hydroxylation of GAA, was shown as a cytochrome P450 by using cell-free assay of chicory root (23). Because P450s are a diverse protein superfamily, and a few hundred P450s are encoded in the genomes of higher plants (34), a selection strategy is critical to narrow down the candidate P450 genes. Lettuce latex has STLs at millimolar levels and was initially considered as a source of transcripts for STL biosynthesis due to easy sample accessibility (17). However, qPCR analysis of lettuce GAS, the enzyme catalyzing the first committed step in STL biosynthesis, showed that this gene is expressed 150 times higher in stem than in latex. This result suggested that the latex may not be the correct cellular site where the transcription of STL biosynthetic genes occurs. On the other hand, almost exclusive expression of GAS in sunflower trichomes has been observed (20). Accordingly, H. annuus cv. HA300 was chosen for STL transcript cataloguing. This particular sunflower cultivar has abundant trichomes on its florets (∼200 per floret), where a diverse array of STLs including a costunolide derivative (haageanolide, Fig. 2B) were isolated and structurally characterized (16). Importantly, the floret trichomes can be visualized by a dissecting microscope, allowing physical separation of pure trichomes by mechanical methods. Total RNA isolated from pure trichomes was used to generate a plasmid cDNA library. A total of 1,130 clones were sequenced by single pass 5′-end Sanger sequencing, and the resulting ESTs were assembled into 116 contigs and 651 singletons, yielding 767 unigenes. Using an E-value cut-off of 1e-3, 539 unigenes (70.2%) were annotated by the UniProt database (for a full list of annotated genes, see supplemental Table 2). Previously reported GAS and GAO transcripts were present in three and nine copies, respectively. Intriguingly, GAO was the fifth most abundant transcript (0.8% of total transcripts) in the trichome EST database, whereas only four copies of GAO were found from 86,398 ESTs generated from various tissues of sunflower (0.004%) (CGP2 database, University of California, Davis). This result suggests that the trichome cDNA library is highly enriched for the transcripts of STL biosynthesis.
Functional in Vivo Screening of Candidate P450s
Our in silico results encouraged us to pursue the functional identification of costunolide synthase from sunflower cDNAs. Previously, we reported the metabolically engineered yeast strain (EPY300-GAA), which can de novo synthesize GAA, a substrate for costunolide synthase, from simple sugar (21). By also transforming individual P450 genes in this yeast background, the P450s for oxidative modification of GAA could be easily screened in vivo. This approach can circumvent the lengthy microsome preparation and the cumbersome substrate purification from EPY300-GAA. Considering that (i) GAO is present in high copies in the cDNA library and that (ii) sesquiterpene-modifying P450s mostly belong to the CYP71 subfamily, eight P450 genes constituting contigs (C49, C113, C63, C12, C7, C51, C100, and C28) and three singletons (S1, S2, and S3) belonging to the CYP71 subfamily (i.e. a total of 11 clones) were subjected to further characterizations (for detailed information, see supplemental Table 3). Unfortunately, all targeted P450 clones were partial clones in the EST database, and therefore 5′-RACE were performed to retrieve their respective full-length clones. Among 11 targeted P450s, all full-length clones were isolated, except for C51, and their corresponding ORFs were cloned in a plasmid with Ura selection marker (pYES-DEST52 or pESC-Ura). For in vivo biochemical evaluations of these candidate P450s, the EPY300-GAA strain was transformed with the second plasmid expressing individual P450 and cultivated for 3 days. Organic extracts of each culture were then analyzed by (+)LC-ESI-MS for the presence of costunolide. Costunolide [M+H]+ ion could be detected at m/z 233 under the instrument conditions used.
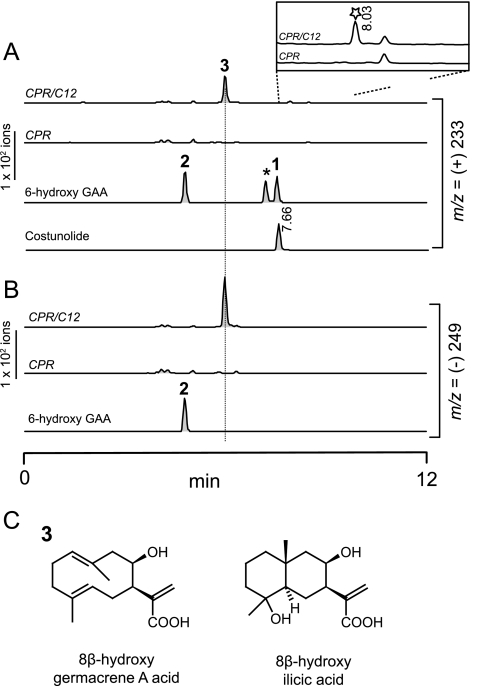
Initial screening identified a peak at m/z 233 in the medium extract of the EPY300-GAA yeast expressing C12 clone (supplemental Fig. 1). However, its retention time did not coincide with the costunolide standard, and (−)LC-MS analysis of this compound showed m/z 249. Because a common dehydration of the compound could result in [M-H2O+H]+ ion at m/z 233 in (+)LC-MS, these results suggested that molecular mass of this compound is 250 (GAA mass plus 16). Along the same line, total (+) ion scan of this compound identified [M+H]+ and [M+Na]+ ion at m/z 251 and 273, respectively, in addition to the ion at m/z 233; also, further fragmentation of m/z 251 by tandem LC-MS confirmed that m/z 233 was derived from a parental [M+H]+ ion at m/z 251 (supplemental Fig. 1). It was obvious that the compound synthesized by C12 was hydroxylated GAA based on the data from mass spectrometry. Other than C12, none of other nine P450 clones showed a unique m/z signal different from the control. Immunoblot analyses using V5 or FLAG-antibodies confirmed that C100 and S2 were not expressed in yeast, but the eight other clones were expressed (supplemental Fig. 2).
To verify the C12 enzymatic activity in vitro, CPR was added to pESC-Ura::C12 to generate the pESC-Ura::C12/CPR. Microsomes isolated from the yeast expressing C12 and CPR were incubated with substrate, GAA. In these assays, 6-hydroxy-GAA (the lactone ring open form of costunolide) was chemically prepared from costunolide and included as a standard. The [M-H]− ion of 6-hydroxy-GAA (molecular weight = 250) was detected at m/z 249 in (−)LC-MS, and its [M-H2O+H]+ ion was detected at m/z 233 in (+)LC-MS. The major new product produced from the microsomes containing recombinant C12 and CPR showed positive and negative ions at m/z 233 and m/z 249, respectively. This compound was essentially the identical compound identified from the in vivo screening (Fig. 3, A and B). However, the retention time of this new compound did not match to that from 6-hydroxy-GAA. In addition, a closer observation of the chromatogram at m/z 233 showed a very minor peak unique to the C12/CPR-expressing microsomes (Fig. 3A, inset). Although this peak could be a lactone compound, it also displayed a different retention time from that of costunolide. Further chemical characterizations of this minor compound in in vitro and in vivo (see below) conditions could not be achieved due to its low abundance. These results from in vitro enzyme assays showed that the major compound enzymatically synthesized by C12 recombinant enzyme is likely to be hydroxy-GAA. However, it was not 6-hydroxy-GAA, and thus, the hydroxyl group appears to be attached on another carbon of GAA.
FIGURE 3.
Biochemical and chemical characterizations of germacrene A acid 8β-hydroxylase. A and B, (±)LC-MS analyses of C12 (HaG8H) enzymatic products are shown. Microsomes from the yeast expressing C12 and CPR catalyzes the synthesis of a new compound (labeled as 3) with [M-H2O+H]+ ion at m/z 233 in A and with [M-H]− ion at m/z 249 in B. The 6-hydroxy-GAA (compound 2) was prepared by alkaline hydrolysis of authentic costunolide standard (1). The identity of 2 was confirmed by reverting it to costunolide. The chemical identity of the peak marked by asterisk is unknown. The compound marked by the star in A (inset) is a minor compound displaying m/z 233, but it showed different retention time from the costunolide (8.03 min versus 7.66 min). C, structures of the new compound (3) purified from the in vivo feeding assay (8β-hydroxygermacrene A acid) and its rearranged product in an acidic condition (8β-hydroxyilicic acid). In the 8β-hydroxyilicic acid, the stereochemistry of a C15 methyl and a hydroxyl group attached to C4 could not be determined due to NMR signal overlapping.
Structure Analyses of C12 Enzymatic Product
To elucidate the structure of the hydroxy-GAA synthesized by C12, the C12 substrate GAA was extracted from EPY300-GAA and fed to the medium of yeast cultures expressing C12 and CPR. Throughout the cultivation, the culture medium was maintained above pH 6 to prevent the reported acid-induced rearrangement of GAA (21). The HPLC-purified hydroxy-GAA (i.e. C12 enzymatic reaction product) was subjected to the standard one- and two-dimensional NMR spectroscopy. Four conformers were detected for the hydroxy-germacrene A, as reported previously for compounds having the germacrene A backbone, complicating the NMR interpretations. Nonetheless, the 13C-and 1H-NMR signals of the major conformer could be clearly assigned, and the structure of the C12 enzymatic reaction product was determined to be 8β-hydroxy-GAA (Fig. 3C; supplemental Table 4). While we were developing the HPLC-purification methods, it was noticed that 8β-hydroxy-GAA could be rapidly rearranged to another unknown compound in an acidic condition (pH 2.5). This compound did not form conformers, and its purification by HPLC was easy due to its distinctively early elution pattern. This new compound induced by acid was purified and analyzed by 13C-, 1H-NMR, and LC-MS, revealing its structure as 8β-hydroxyilicic acid with its [M-H]− at m/z 267 (Fig. 3C and supplemental Table 4). The ilicic acid was a major acid-induced rearranged product of GAA in our previous study (21). Therefore, 8β-hydroxy-GAA appears to be unstable in an acidic conditions and is rearranged to 8β-hydroxyilicic acid. The structural elucidations of the C12 enzymatic reaction product (8β-hydroxy-GAA) and its major acid-rearranged product (8β-hydroxyilicic acid) clearly demonstrated that C12 catalyzes the 8β-hydroxylation of GAA. Therefore, C12 was functionally named as sunflower (H. annuus) GAA 8β-hydroxylase (HaG8H), and also an official P450 name, CYP71BL1, was assigned to this gene by the P450 nomenclature committee.
Isolation of Costunolide Synthase Gene from Lettuce
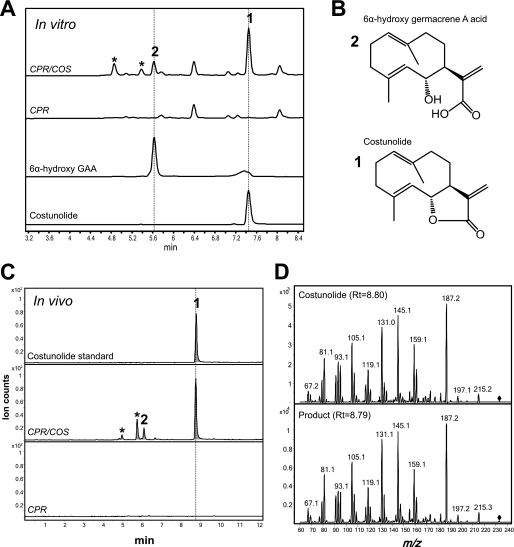
Although HaG8H could not synthesize costunolide from GAA, the substrate-binding pocket of HaG8H could accommodate GAA. This result suggested that natural HaG8H variants in other Asteraceae plants might catalyze related reactions from GAA. Lettuce and closely-related chicory showed evidence of costunolide biosynthesis (23, 26). Hence, we examined the HaG8H homologs from lettuce (Lactuca sativa). The BLAST search of 76,043 lettuce ESTs showed that the closest HaG8H homolog in lettuce shares 65% amino acid identity. This in silico analysis was intriguing because the diverged amino acid sequences (65% identity) suggested that this lettuce homolog might catalyze a related but distinct reaction. Its full-length gene was recovered from lettuce leaf cDNA by using RACE, and an official name CYP71BL2 was assigned to this P450 gene. Microsomes were isolated from the transgenic yeast expressing CYP71BL2 and CPR and incubated with GAA to examine GAA-modifying activities. (+)LC-MS monitoring at m/z 233 of the organic extract identified one dominant and three early eluting minor compounds, which were not present in the control reaction (Fig. 4A). The minor compound 2 showed an identical retention time with [M-H2O+H]+ ion for 6-hydroxy-GAA standard, and the dominant compound 1 coincided with [M+H]+ ion for costunolide standard. In the (−)LC-MS analyses, compound 2 and 6-hydroxy-GAA standard showed [M-H]− ion at m/z 249. These results suggested that 6α-hydroxy-GAA and costunolide are produced in microsome containing CYP71BL2 and CPR (Fig. 4B), and therefore, these results showed that CYP71BL2 is the lettuce costunolide synthase (LsCOS).
FIGURE 4.
Biochemical and chemical characterization of costunolide synthase. A, (+)LC-MS scan at m/z 233 demonstrated that compounds 1 and 2 showed identical retention times with costunolide and 6-hydroxygermacrene A acid, respectively. Asterisks indicate unknown compounds, which are likely due to the unspecific activities of LsCOS. B, the structures of the standards are depicted. C, metabolite profile of the culture extraction from the EPY300 strain expressing GAS, LsGAO, CPR, and with or without LsCOS by (+)LC-MS scan at m/z 233. D, product ion scans of the costunolide standard and compound 1 by (+)LC-MS-MS showed identical fragmenting patterns. Diamonds indicate the parental ion at m/z 233.
A question arose as to why 6α-hydroxy-GAA forms a lactone, whereas 8β-hydroxy-GAA does not. Because heat and acidity can promote a lactonization reaction, stability of the two types of hydroxy-GAA was evaluated in low pH and elevated temperature. The 8β-hydroxy-GAA was stable even in pH 4.5 at 65 °C. However, the purified 6α-hydroxy-GAA from the alkaline hydrolysis of costunolide was easily converted to costunolide in a neutral condition at room temperature, and its lactonization was promoted by heat.
De Novo Synthesis of Costunolide in Yeast
To perform more reliable chemical analysis of the costunolide produced by LsCOS, a larger quantity of reaction product was required. Accordingly, a high copy plasmid coding four plant genes (GAS, LsGAO, LsCOS, and CPR) was constructed in pESC-Leu2d. This quadruple expression plasmid was transformed to EPY300 for the microbial de novo synthesis of costunolide. After cultivating the transgenic yeast for 4 days, whole culture was extracted with ethyl acetate, and the metabolites were analyzed by (+)LC-MS. The [M+H]+ ion for costunolide and [M-H2O+H]+ ion for 6α-hydroxy-GAA were clearly detected at m/z 233 (Fig. 4C), and the (+)LC-MS/MS analysis by collision-induced dissociation demonstrated the identical fragmenting patterns for the in vivo reaction product with the costunolide standard (Fig. 4D). In two independent experiments, transgenic yeast synthesized costunolide at the levels of 6.2 ± 0.8 mg liter−1 and 9.3 ± 0.9 mg liter−1 (n = 6 of independent flasks each). The microbially produced costunolide was purified from the culture, and its structure was determined by NMR analysis. The 13C-and 1H-NMR signals of the de novo-synthesized costunolide and standard were identical (supplemental Table 4), further confirming the correct structure of the microbially synthesized costunolide.
Comparative Genomics Analysis in Asteraceae
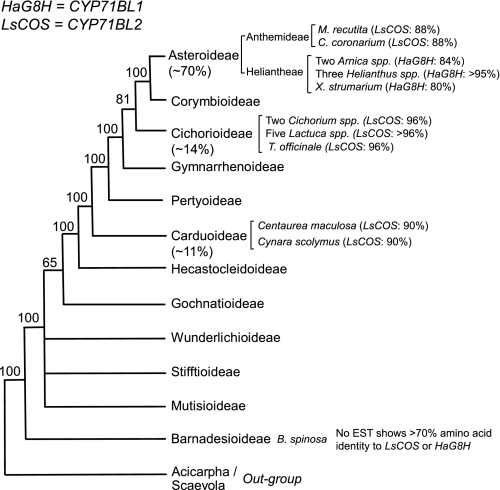
The sequence information of sunflower HaG8H (CYP71BL1) and lettuce LsCOS (CYP71BL2) allowed us to infer the biochemical evolution of these enzymes in a wider range of Asteraceae plants. Three major subfamilies of Asteraceae are Carduoideae, Cichorioideae, and Asteroideae, which constitute 95% of the 23,000 plant species of Asteraceae family (Fig. 5). The bioinformatics analyses revealed that the ESTs highly homologous to the LsCOS (>88% identity at amino acid sequence level) were found in the EST databases of all three major subfamilies of Asteraceae family except for the Heliantheae tribe of the Asteroideae (Fig. 5). There are also highly homologous genes to GAS and GAO in each EST database of these species. These results suggest that costunolide biosynthetic pathway is conserved in the majority of genera in Asteraceae. On the other hand, none of the species examined in the Heliantheae tribe (genera of Arnica, Helianthus, and Xanthium) has ESTs highly homologous to the costunolide synthase, LsCOS. This informatics result was derived from ∼1.2 million ESTs for Arnica and ∼0.5 million ESTs for Xanthium by 454 pyrosequencing3 and ∼280,000 ESTs for Helianthus by the Sanger sequencing (The Compositae Genome Project). Thus, insufficient sequence data is an unlikely cause for the complete absence of LsCOS homolog in Heliantheae tribe. Instead, they have acquired ESTs highly homologous to the HaG8H (CYP71BL1), with their amino acid sequence identity ranging from 80 to 99%. These HaG8H homologs could not be found in any of the species where the LsCOS ortholog was identified, and therefore the occurrence of HaG8H appears to be restricted to the Heliantheae and possibly some other tribes in the Asteroideae subfamily. The ESTs displaying high homology (> 70%) to the HaG8H and LsCOS were not identified in the EST database (28,483 ESTs) from a phylogenetically basal species, Barnadesia spinosa. The most homologous EST from B. spinosa showed 58 and 68% amino acid sequence identity to HaG8H and LsCOS, respectively.
FIGURE 5.
Identifications of the homologous genes for HaG8H (CYP71BL1) and LsCOS (CYP71BL2) in Asteraceae. The Asteraceae phylogeny was adapted from Panero and Funk (15). Estimated percentages of species constituting Asteraceae plants in three major subfamilies are given in parentheses. Shown are: Matricaria recutita (German Chamomile); Chrysanthemum coronarium (Garland chrysanthemum); two Arnica species are A. montana and A. chamissonis; three Helianthus species are H. annuus, H. argophyllus, and H. ciliaris; Xanthium strumarium; two Cichorium species are Cichorium intybus and C. endivia; T. officinale is Taraxacum officinale; and five Lactuca species are L. sativa, L. serriola, L. saligna, L. virosa, and L. perennis; Barnadesia spinosa.
DISCUSSION
Although thousands of STLs from plants in the family of Asteraceae (or Compositae) have been reported, their biosynthesis has been poorly understood at the molecular level. We propose that the studies of STL metabolism in Asteraceae provide an insight into how enzymes evolve to influence the appearance of new chemical phenotypes. In turn, naturally occurring enzyme variants will offer new insights into the evolutionary emergence of catalytic function. Asteraceae is particularly suited for this purpose as this family has a well defined phylogeny over 50 million years (13, 15) with a wealth of natural product structural data in the literature.
Together with the C15 sesquiterpene hydrocarbon skeletons created by sesquiterpene synthases, the regio- and stereoselective formation of the γ-lactone ring is central to the structural diversity of STLs in nature. Obviously, the coordinated reactions of sesquiterpene synthases and P450s elaborate the diverse structures of STLs. The co-occurrence of these enzymes at specific evolutionary time points should in principle create unique chemical profiles in different lineages of Asteraceae plants. However, until now only a limited number of the molecular clones responsible for the synthesis of STLs have been identified and characterized. The difficulties of STL research has been in part due to the instability and unavailability of pathway intermediates and low abundance of the enzymes for STL biosynthesis. In this report, we overcame these problems by using a blend of techniques (single cell genomics, microbial metabolic engineering, and standard chemistry) and identified two unique enzymes involved in the oxidative modifications of GAA from lettuce (L. sativa) and sunflower (H. annuus). These two novel P450 enzymes help us better understand the biogenesis and evolution of STL metabolisms in Asteraceae.
One interesting observation was that 6α-hydroxy-GAA is unstable and undergoes spontaneous lactonization to form costunolide, but the 8β-hydroxy-GAA does not. Although the cryptic lactonization activity of LsCOS in addition to its standard P450-mediated hydroxylation was suspected, this possibility was ruled out because the HPLC-purified 6α-hydroxy-GAA prepared by the alkaline hydrolysis of costunolide was easily reverted to costunolide. This non-enzymatic, spontaneous conversion was very efficient and did not allow us to assess the possible lactonization activity in LsCOS. We believe that the lactonization from 6α-hydroxy-GAA to costunolide is entirely non-enzymatic or, if any, the LsCOS enzymatic role of lactonization is negligible.
On the contrary, the 8β-hydroxy-GAA is stable and does not spontaneously undergo lactonization. To address this discrepancy, we conducted computational modeling to simulate the three-dimensional structures of 6α-hydroxy-GAA, 8β-hydroxy-GAA, and their corresponding lactones. The energy-minimized conformations of these compounds were simulated, and the heat of formation (ΔHf0) and the distance between the hydroxyl oxygen atom and C-12 carbonyl carbon were estimated (supplemental Fig. 3). The ΔHf0 values for both of the hydroxy-GAAs were not significantly different; 6α-hydroxy-GAA was 1.0 kcal mol−1 lower than 8β-hydroxy-GAA. However, comparison of the ΔHf0 values between the corresponding lactones indicates that costunolide has a lower value by 4.3 kcal mol−1. This suggests that the lactone formation from 6α-hydroxy-GAA is significantly more favorable than that from 8β-hydroxy-GAA.
In line with the aforementioned thermodynamic consideration, the atomic distance between hydroxyl oxygen and carbonyl carbon that forms C-O bond in lactonization suggests kinetically favorable formation of costunolide. In the case of the 8β-hydroxy-GAA, the minimum distance between O-8 and C-12 that can be brought about by rotation of the C7-C11 bond was estimated to be 3.41 Å, whereas the distance between O-6 and C-12 of the 6α-hydroxy-GAA was significantly shorter by 0.87 Å (supplemental Fig. 3). Therefore, this closer atomic distance of 6α-hydroxy-GAA would allow more facile collision of O-6 with C-12 than O-8 with C-12, resulting in spontaneous formation of the lactone in costunolide. Ultimately, the x-ray diffraction data of the 8β-hydroxy-GAA crystal can provide an unambiguous answer to this question.
The bioinformatics analyses of HaG8H and LsCOS in Asteraceae provided an evolutionary track of STL biochemistry (Fig. 5). One key discovery is that LsCOS is not restricted to lettuce and its related species, but it is widely conserved in many lineages of Asteraceae plants, encompassing all three major subfamilies. LsCOS appears to have emerged in a relatively early phase of Asteraceae evolution. Since then, the costunolide synthesized from GAS, GAO, and LsCOS has been an integral part of the isoprene secondary metabolism in the majority of Asteraceae plants. On the other hand, occurrence of HaG8H is restricted to the Heliantheae and possibly other related tribes in Asteroideae subfamily. It is noteworthy that among the plants where HaG8H homologs were identified, LsCOS homologs could not be found, and vice versa. In all species examined, LsCOS and HaG8H are still the closest homologs to each other (∼65% amino acid sequence identity) in the cross-species BLAST analyses. Based on these observations, we postulate that HaG8H has evolved from LsCOS in certain Asteraceae lineages (e.g. Heliantheae) perhaps after the loss of LsCOS function. In other words, HaG8H could be an evolutionary descendant of LsCOS in some later evolved genera of Asteraceae.
At least three distinct regio- and stereoselective STLs were correlated with the ESTs highly homologous to the HaG8H from the plants belonging to the Heliantheae tribe, where LsCOS disappeared. These STLs form C6-C7 trans- (niveusin B from sunflower), C7-C8 trans- (xanthatin from Xanthium strumarium), and C7-C8 cis-(helenalin from Arnica montana) configurations (Fig. 2). Although sunflower retains the costunolide-type C6-C7 trans-lactone, new distinct type of lactones (C7-C8 cis- and trans-lactones) appeared in two genera, Xanthium and Arnica. It would be of great interest to examine whether the HaG8H homologs from A. montana and X. strumarium have acquired unique activities for C7-C8 cis-lactonization (helenalin) and C7-C8 trans-lactonization (xanthatin), respectively.
The STL biosynthesis of sunflower requires further studies. Sunflower has GAS and GAO (20, 21), and its capitate glandular trichomes produce abundant STLs of costunolide-type C6-C7 trans-lactone (16); however, its trichome ESTs and the ESTs from several related Helianthus spp. do not contain LsCOS. Some possibilities to explain this are as follows: (i) sunflower has costunolide synthase gene that is not homologous to LsCOS, and (ii) 8β-hydroxy-GAA or its derivatives, instead of costunolide, is a pathway intermediate for the C6-C7 STL biosynthesis in sunflower. In any case, the biochemical and genomics data here support the converged evolution of STL biogenesis in Asteraceae. Costunolide and its derivatives have bioactivities, and hence as long as a set of the three genes (GAS, GAO, LsCOS) is functional, Asteraceae plants are not under pressure to develop new routes for STL biosynthesis. However, disappearance of LsCOS in some lineages of Asteraceae might evoke new metabolic strategies for STL synthesis by recruiting different enzymes or by developing different metabolic routes.
To deduce the mechanisms for LsCOS and HaG8H, energy-minimized homology models of these two P450s were generated against the human CYP2E1 (35). To analyze these homology models, substrate GAA was positioned in the active site by orienting the C6 and C8 of GAA perpendicular to the center of the heme group for LsCOS and HaG8H, respectively (supplemental Fig. 4). In these models, eight residue variants including one deletion variation in HaG8H, were located within a 5-Å radius from the substrate. All of the residues identified belong to the six substrate recognition sites (SRSs) defined by Gotoh (36). In particular, SRS5 (immediately outside of K-helix, see supplemental Fig. 4) has a deletion variation and Val residue in HaG8H, whereas the equivalent positions were occupied by Pro and Thr residues in LsCOS. A threonine residue in LsCOS could potentially stabilize GAA binding through a hydrogen bond. On the other hand, flipping the GAA to pose the C8 of GAA perpendicular to the heme might allow a different interactions through the two Ser residues in SRS4. In these models, residue variants in other SRS domains might help orient GAA properly by interacting the C10 ring moiety of GAA. This proposed model and the roles of the residues identified form the basis of future systematic site-directed mutagenesis studies to ascertain their potential contributions to the evolution of alternative regiospecificities seen in the modern enzymes.
In this work, expression of some sunflower P450s was low or absent, making functional characterization and annotation difficult (supplemental Fig. 2). Advances in gene synthesis technology and its affordability will enable full codon-optimized synthetic genes to be made, allowing more thorough biochemical studies with different substrates (e.g. 8β-hydroxy-GAA) to be conducted to clarify the metabolic pathways of STLs in sunflower. In addition, current intensive deep sequencing of hundreds of plant species will soon provide ample resources to trace the occurrence of LsCOS and HaG8H in the evolution and diversifications of STLs in Asteraceae.
Supplementary Material
Acknowledgments
We thank Dr. Deane McIntyre (the Bio-NMR Center at the University of Calgary), Drs. John Vederas and Jesse Li (Department of Chemistry, University of Alberta), and Dr. Jürgen Conrad (Bioorganic Chemistry, University of Hohenheim) for assistance in NMR spectra analysis. We thank Dustin Cram and Jacek Nowak (National Research Council-Plant Biotechnology Institute, Saskatoon, Canada) for the transcript assembly by the FIESTA system. We also thank Dr. Guillermo Martínez Massanet (Departamento de Química Orgánica, University of Cádiz) for advice on 8β-hydroxy-GAA and Gillian MacNevin for critical reading of this article. The Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco was supported by National Institutes of Health Grant P41 RR001081.
This work was supported by Natural Sciences and Engineering Research Council of Canada Discovery Grant, Canada Research Chair program Alberta Innovates Bio Solutions, and Canada Foundation for Innovation (to D.-K. R.) and by the Japan Society for the Promotion of Science Postdoctoral Fellowships for Research Abroad (to N. I.). This work was also supported by German Research Foundation Grant (Deutsche Forschungsgemeinschaft) SP292/20-2 (to O. S. and J. C. G.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1–4 and Figs. 1–4.
The nucleotide sequence(s) reported in this paper has been submitted to the Gen-BankTM/EBI Data Bank with accession number(s) HQ439587-HQ439594, HQ439596, HQ439598, and HQ439599.
D.-K. Ro, unpublished data.
- STL
- sesquiterpene lactone
- P450
- cytochrome P450 monooxygenase
- CPR
- cytochrome P450 reductase
- GAA
- germacrene A acid
- GAS
- germacrene A synthase
- GAO
- germacrene A oxidase(s)
- LsCOS
- L. sativa costunolide synthase
- HaG8H
- H. annuus germacrene A acid 8β hydroxylase
- EST
- expressed sequence tag
- contig
- group of overlapping clones
- RACE
- rapid amplification of cDNA ends.
REFERENCES
- 1. Eckstein-Ludwig U., Webb R. J., Van Goethem I. D., East J. M., Lee A. G., Kimura M., O'Neill P. M., Bray P. G., Ward S. A., Krishna S. (2003) Nature 424, 957–961 [DOI] [PubMed] [Google Scholar]
- 2. Kwok B. H., Koh B., Ndubuisi M. I., Elofsson M., Crews C. M. (2001) Chem. Biol. 8, 759–766 [DOI] [PubMed] [Google Scholar]
- 3. Satou T., Koga M., Matsuhashi R., Koike K., Tada I., Nikaido T. (2002) Vet. Parasitol. 104, 131–138 [DOI] [PubMed] [Google Scholar]
- 4. Christensen S. B., Andersen A., Kromann H., Treiman M., Tombal B., Denmeade S., Isaacs J. T. (1999) Bioorg. Med. Chem. 7, 1273–1280 [DOI] [PubMed] [Google Scholar]
- 5. Picman A. K. (1986) Biochem. Syst. Ecol. 14, 255–281 [Google Scholar]
- 6. Rodriguez E. R., Towers G. H., Mitchell J. C. (1976) Phytochemistry 15, 1573–1580 [Google Scholar]
- 7. Fischer N. H. (1990) in Biochemistry of the Mevalonic Acid Pathway to Terpenoids (Towers G. H., Stafford H. A. eds) pp. 161–202, Plenum Press, New York [Google Scholar]
- 8. Park S. H., Choi S. U., Lee C. O., Yoo S. E., Yoon S. K., Kim Y. K., Ryu S. Y. (2001) Planta Med. 67, 358–359 [DOI] [PubMed] [Google Scholar]
- 9. Dall'Acqua S., Viola G., Giorgetti M., Loi M. C., Innocenti G. (2006) Chem. Pharm. Bull. 54, 1187–1189 [DOI] [PubMed] [Google Scholar]
- 10. Brecknell D. J., Carman R. M. (1979) Aust. J. Chem. 32, 2455–2471 [Google Scholar]
- 11. Holub M., Toman J., Herout V. (1987) Biochem. Syst. Ecol. 15, 321–326 [Google Scholar]
- 12. Seaman F. C. (1982) Bot. Rev. 48, 121–594 [Google Scholar]
- 13. Barreda V. D., Palazzesi L., Tellería M. C., Katinas L., Crisci J. V., Bremer K., Passalia M. G., Corsolini R., Rodríguez Brizuela R., Bechis F. (2010) Science 329, 1621 [DOI] [PubMed] [Google Scholar]
- 14. Jansen R. K., Palmer J. D. (1987) Proc. Natl. Acad. Sci. U.S.A. 84, 5818–5822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Panero J. L., Funk V. A. (2008) Mol. Phylogenet. Evol. 47, 757–782 [DOI] [PubMed] [Google Scholar]
- 16. Göpfert J. C., Heil N., Conrad J., Spring O. (2005) Plant Biol. 7, 148–155 [DOI] [PubMed] [Google Scholar]
- 17. Sessa R. A., Bennett M. H., Lewis M. J., Mansfield J. W., Beale M. H. (2000) J. Biol. Chem. 275, 26877–26884 [DOI] [PubMed] [Google Scholar]
- 18. Bennett M. H., Mansfield J. W., Lewis M. J., Beale M. H. (2002) Phytochemistry 60, 255–261 [DOI] [PubMed] [Google Scholar]
- 19. Bouwmeester H. J., Kodde J., Verstappen F. W., Altug I. G., de Kraker J. W., Wallaart T. E. (2002) Plant Physiol. 129, 134–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Göpfert J. C., Macnevin G., Ro D. K., Spring O. (2009) BMC Plant Biol. 9, 86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nguyen D. T., Göpfert J. C., Ikezawa N., Macnevin G., Kathiresan M., Conrad J., Spring O., Ro D. K. (2010) J. Biol. Chem. 285, 16588–16598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ro D. K., Paradise E. M., Ouellet M., Fisher K. J., Newman K. L., Ndungu J. M., Ho K. A., Eachus R. A., Ham T. S., Kirby J., Chang M. C., Withers S. T., Shiba Y., Sarpong R., Keasling J. D. (2006) Nature 440, 940–943 [DOI] [PubMed] [Google Scholar]
- 23. de Kraker J. W., Franssen M. C., Joerink M., de Groot A., Bouwmeester H. J. (2002) Plant Physiol. 129, 257–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rao A. S., Kelkar G. R., Bhattacharyya S. C. (1960) Tetrahedron 9, 275–283 [Google Scholar]
- 25. Kassuya C. A., Cremoneze A., Barros L. F., Simas A. S., Lapa Fda R., Mello-Silva R., Stefanello M. E., Zampronio A. R. (2009) J. Ethnopharmacol. 124, 369–376 [DOI] [PubMed] [Google Scholar]
- 26. Takasugi M., Okinaka S., Katsui N., Masmune T., Shirata A., Ohuchi M. (1985) J. Chem. Soc. Chem. Commun. 621–622 [Google Scholar]
- 27. Robinson A., Kumar T. V., Sreedhar E., Naidu V. G., Krishna S. R., Babu K. S., Srinivas P. V., Rao J. M. (2008) Bioorgan. Med. Chem. Lett. 18, 4015–4017 [DOI] [PubMed] [Google Scholar]
- 28. Eliza J., Daisy P., Ignacimuthu S., Duraipandiyan V. (2009) Chem. Biol. Interact. 179, 329–334 [DOI] [PubMed] [Google Scholar]
- 29. Wedge D. E., Galindo J. C., Macías F. A. (2000) Phytochemistry 53, 747–757 [DOI] [PubMed] [Google Scholar]
- 30. Chen H. C., Chou C. K., Lee S. D., Wang J. C., Yeh S. F. (1995) Antivir. Res. 27, 99–109 [DOI] [PubMed] [Google Scholar]
- 31. Ro D. K., Ouellet M., Paradise E. M., Burd H., Eng D., Paddon C. J., Newman J. D., Keasling J. D. (2008) BMC Biotechnol. 8, 83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pieper U., Eswar N., Webb B. M., Eramian D., Kelly L., Barkan D. T., Carter H., Mankoo P., Karchin R., Marti-Renom M. A., Davis F. P., Sali A. (2009) Nucleic Acids Res. 37, D347–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 34. Nelson D. R., Schuler M. A., Paquette S. M., Werck-Reichhart D., Bak S. (2004) Plant Physiol. 135, 756–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Porubsky P. R., Meneely K. M., Scott E. E. (2008) J. Biol. Chem. 283, 33698–33707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gotoh O. (1992) J. Biol. Chem. 267, 83–90 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.