Abstract
The RNA virus that causes the Crimean Congo Hemorrhagic Fever (CCHF) is a tick-borne pathogen of the Nairovirus genus, family Bunyaviridae. Unlike many zoonotic viruses that are only passed between animals and humans, the CCHF virus can also be transmitted from human to human with an overall mortality rate approaching 30%. Currently, there are no atomic structures for any CCHF virus proteins or for any Nairovirus proteins. A critical component of the virus is the envelope Gn glycoprotein, which contains a C-terminal cytoplasmic tail. In other Bunyaviridae viruses, the Gn tail has been implicated in host-pathogen interaction and viral assembly. Here we report the NMR structure of the CCHF virus Gn cytoplasmic tail, residues 729–805. The structure contains a pair of tightly arranged dual ββα zinc fingers similar to those found in the Hantavirus genus, with which it shares about 12% sequence identity. Unlike Hantavirus zinc fingers, however, the CCHF virus zinc fingers bind viral RNA and contain contiguous clusters of conserved surface electrostatics. Our results provide insight into a likely role of the CCHF virus Gn zinc fingers in Nairovirus assembly.
Keywords: Negative-strand RNA Viruses, NMR, Protein Structure, RNA-binding Protein, Zinc Finger
Introduction
Recent outbreaks of the Crimean Congo Hemorrhagic Fever (CCHF)2 virus along with the reported ability of the virus to transfer between humans have raised concerns of a widespread pandemic (1). The virus is transmitted to humans by tick bite or by direct handling of infected animal meat or blood (1, 2). Infection causes a hemorrhagic fever and myalgia resulting in mortality rates approaching 30% (1–3). The virus contains an antisense RNA genome divided into three segments, and named according to lengths as the S, M, and L (for Small, Medium, and Large) segments (4). The viral proteins are the nucleocapsid protein, two membrane glycoproteins Gn and Gc (also referred to as G1 and G2 in other Bunyaviridae) (5, 6), a nonstructural protein (NSm) (7), and an RNA polymerase (4). In the mature virion, the Gn glycoprotein contains a 176 residue ectodomain followed by a 24 residue transmembrane region and terminates in a long cytoplasmic tail consisting of ∼100 residues (5, 7).
Recent results from other related Bunyaviridae viruses suggest the role of the Gn tail in viral assembly. For example, alanine mutagenesis of the cytoplasmic tails of Uukuniemi virus (genus Phlebovirus) (8) and Bunyamwera virus (genus Orthobunyavirus) (9) affect the ability of virus-like particles (VLPs) to effectively incorporate ribonucleoproteins, thus intimating a role for Gn tails in genome packaging. More recently, the Gn tail of Puumala virus (genus Hantavirus) was shown to co-immunoprecipitate with the Puumala nucleocapsid protein (10). These results suggest that the CCHF virus Gn tail plays an equally important role in viral assembly of genus Nairovirus.
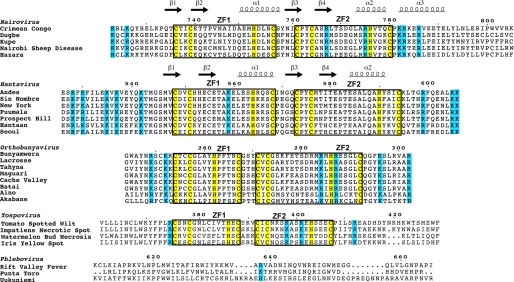
The sequence of the CCHF virus cytoplasmic tail is somewhat variable in Nairoviruses (∼24% identity) and even more so when compared with other Bunyaviruses (12% identity with Hantavirus Gn tails). However, one characteristic feature present in four of the five genera of Bunyaviridae is a conserved dual C-X-C-X-H-X-C motifs of cysteine and histidine residues with X representing any amino acid (Fig. 1). Others have suggested that the high cysteine content of the CCHF virus Gn tail could be due to extensive disulfide bonding (5). Recently, we reported that the cysteines in the Andes hantavirus Gn tail fold into a novel arrangement of back-to-back classical ββα zinc fingers (11). Despite low sequence identity between the Gn tail of Nairoviruses and Hantaviruses, the spacing of the dual CCHC motif in the CCHF virus most closely resembles that of Hantaviruses, suggesting the presence of a similar dual zinc finger structure. To test this hypothesis, we determined the NMR structure of the CCHF virus Gn cytoplasmic tail from residues 729–805. We report here the first known atomic structure of any protein component of the CCHF virus and demonstrate that the high cysteine content of the Gn cytoplasmic tail is partly due to the presence of dual, back to back ββα-type zinc fingers similar to those found in Hantaviruses. Unlike Hantaviral zinc fingers, however, the electrostatic surface of the CCHF virus zinc finger reveals a clear distribution of conserved electrostatic charges. Moreover, we demonstrate using electrophoretic mobility shift assays (EMSA) that these conserved electrostatics may play a role in forming a surface for binding viral RNA. Together, these data provide insight into the role of the Gn tail in Nairovirus assembly.
FIGURE 1.
Sequence alignment of the Gn tails of representative members of family Bunyaviridae. Bunyaviridae is comprised of five genera: Nairovirus, Hantavirus, Orthobunyavirus, Tospovirus, and Phlebovirus. The conserved CCHC-zinc finger motifs (boxed) are present in four of the five genera, with Phlebovirus the lone exception. Another recurring feature is the clustering of conserved basic residues (in blue) in the vicinity of the CCHC-motifs. Notably, these basic residues overlap with ZF2 in Nairovirus, Orthobunyavirus, and Tospovirus, but are located outside ZF2 in Hantavirus.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
Various constructs of the CCHF virus (strain SPU103/87) Gn cytoplasmic region (spanning residues 719–819) were subcloned from a synthetic gene (GenScript) into the expression vectors pDZ1 and pDZ3 (12), which expressed His6-tagged GB1 fusion proteins with TEV protease cleavage sites. For NMR structure determination, the soluble Gn construct spanning residues 729–805 (Gn729–805) was expressed and purified under native conditions following the method reported previously for the Andes hantavirus zinc finger domain (11). Briefly, 15N- and 15N/13C-labeled proteins were expressed in Escherichia coli BL21(DE3) grown in 1 liter M9 minimal media supplemented with 0.1 mm ZnSO4 before and after induction. Cells were grown at 37 °C, induced with 1 mm isopropyl-β-d-thiogalactopyranoside at A600 ∼0.8, and cell growth was continued in a 15 °C shaker incubator overnight (to a final A600 ∼2.0). Cells were harvested by centrifugation, resuspended in buffer A (20 mm Tris-HCl pH 8.0, 20 mm NaCl, 1 mm DTT, 0.1 mm ZnSO4), and lysed by sonication. Cellular debris was removed by centrifugation, and to the supernatant was added one-tenth volume of 1% polyethyleneimine (pH 8) to precipitate the nucleic acids. Following centrifugation, the supernatant was bound to a 40 ml of Q column (GE Healthcare) and eluted with a 280 ml linear gradient of buffer B (20 mm Tris-HCl, pH 8.0, 0.5 m NaCl, 1 mm DTT, 1 mm ZnSO4). For TEV protease digestion, fractions containing the fusion protein were pooled and dialyzed at 25 °C overnight in buffer (50 mm Tris-HCl pH 8.0, 20 mm NaCl, 1 mm DTT, 1 mm ZnSO4) with 0.16 mg recombinant TEV protease (13) per 10 ml of fusion protein. The TEV digestion mixture was dialyzed back into buffer A and passed again through a 40 ml Q column (GE Healthcare). The GB1 tag (theoretical pI of 5.6) was retained on the column while Gn729–805 (theoretical pI of 8.6) was present in the flow-through. The 50 ml flow-through fraction was concentrated using Ultra-15 centrifugal filters (Amicon) and dialyzed in NMR buffer (10 mm NaPO4 pH 7.0, 10 mm NaCl, 1 mm DTT, 0.1 mm ZnSO4). The Gn729–805 construct retained three residues (Gly-His-Met) cloning artifacts at the N terminus.
NMR Spectroscopy
NMR data were acquired at 25 °C using a Bruker Avance 800 MHz spectrometer equipped with a cryoprobe, processed with NMRPipe (14), and analyzed with NMRView (15). Backbone assignments were obtained from two-dimensional 1H-15N HSQC (16) and three-dimensional HNCA (17), CBCA(CO)NH (17), and HNCACB (18). Secondary structures were identified from the Cα, Cβ, and Hα chemical shifts (19). Side chain assignments were obtained from two-dimensional 1H-13C HMQC (20), three-dimensional HBHA(CO)NH (21), and three-dimensional 13C-edited HMQC-NOESY (22) (tmix = 120 ms). The histidine ring nitrogen atoms coordinated to Zn2+ ions were identified from two-dimensional 15N HMQC (23) using a nitrogen sweep width of 160–230 ppm. NOE (nuclear Overhauser effect) crosspeaks were identified from three-dimensional 15N-edited NOESY-HSQC (24) (tmix = 120 ms) and three-dimensional 13C-edited HMQC-NOESY (22) (tmix = 120 ms).
Backbone 15N relaxation parameters were acquired on a 0.5 mm 15N-labeled sample in NMR buffer. The steady-state heteronuclear {1H}-15N NOE was acquired as a pair of two-dimensional datasets in an interleaved manner (where portions of each two-dimensional spectrum were acquired sequentially until both datasets were completed) (25). The first two-dimensional dataset contained a 3-s proton saturation (achieved with a series of 120° pulses) whereas the second two-dimensional dataset contained a 3-s delay. The heteronuclear {1H}-15N NOE was calculated as the ratio of the intensities for each peak in the two datasets. Each two-dimensional dataset was acquired with 2048 (1H) × 128 (15N) complex points, 32 scans per point, and a 5 s recycle delay. Error bars were estimated using the standard deviation of the background signal of each spectrum. The 15N backbone relaxation rates R1 and R2 were acquired as described (26). The time delays used to determine R1 were 10, 60, 120, 240*, 400, 900, and 1100 ms, and the time delays used to determine R2 were 20, 40*, 50, 60, 70, 90, 100, 120, and 150 ms (asterisk denotes spectra acquired in duplicate to estimate reproducibility). Peak intensities were obtained from NMRView (15) and fitted using GNUPLOT (27). Deviations from fitting were reported as error bars. Because of peak overlap, residues 749, 787, 791, and 796 were not used in the analysis.
Structure Calculation
The protocol used for NMR structure calculation has been described previously (11). Briefly, unique NOE distance restraints were classified into upper bounds of 2.7, 3.5, 4.5, and 5.5 Å and lower bound of 1.8 Å based on peak volumes. Backbone dihedral angles in the α-helical regions identified by the secondary Cα, Cβ, and Hα chemical shifts (19) were restrained to ϕ (−60 ± 20°) and ψ (−40 ± 20°). Initial structures were generated using CYANA (28), followed by molecular dynamics and simulated annealing in AMBER7 (29); first in vacuo, then with the generalized Born (GB) potential. Initial structural calculations were performed in CYANA without the Zn2+ restraints to confirm that the zinc finger domain will fold from NOE-derived restraints only. Once the topology of the Zn2+-coordinated residues were confirmed, subsequent CYANA structure calculations used distance restraints that imposed tetrahedral Zn2+-coordination to Cys and His residues (22). Iterative cycles of AMBER calculations followed by refinement of NMR-derived restraints were performed until the structures converged with low restraint violations and good statistics in the Ramachandran plot. A family of twenty lowest energy structures were analyzed using PROCHECK (30) and molecular graphics were generated using PYMOL (31). The surface electrostatic potentials were calculated using APBS (32) and visualized in PYMOL (31).
In Vitro Transcription
A DNA oligonucleotide representing the M genomic segment panhandle was assembled by PCR primer extension and used for in vitro transcription. In vitro transcription was carried out following manufacturer's protocol (MAXIscript Kit, Ambion). Briefly, a 20-μl reaction was carried out for 1.5 h (37°) and terminated by adding 2 μl 0.25 m EDTA and heating to 90° followed by rapid cooling on ice. Reaction mixtures were then treated with DNase I and subjected to ethanol precipitation. The RNA transcripts were resuspended in RNase-free ddH2O and analyzed for purity on a native 12% acrylamide gel stained with SYBR Green II dye (Invitrogen).
RNA Binding Assays
To assess protein-RNA binding by gel electrophoresis, RNA transcripts were incubated on ice for 15 min with increasing amounts of either CCHF virus Gn729–805 or Andes hantavirus G1543–599 in binding buffer (30 mm NaPO4, 30 mm NaCl, pH 7.4). Samples were mixed with one-half volume 50% glycerol and loaded onto a native 12% acrylamide Tris borate gel. The gel was run in a cooling water bath at 90 V for 1 h in Tris borate buffer, pH 8.3 and visualized by staining with SYBR Green II dye. For nucleic acid size determination, each gel included a 100 bp DNA ladder (NEB, NO467S).
CD Spectroscopy
CD spectra were collected in triplicate at 25° on a JASCO J-815 Spectro-polarimeter using a scanning speed of 50 nm/min. Protein concentrations were kept at 1 μm in buffer (10 μm NaPO4, 10 μm NaCl, 0.1 mm ZnSO4). EDTA and ZnSO4 titrations were applied to the same sample.
RESULTS
Protein Expression and Purification
Our previous work with Hantavirus glycoprotein cytoplasmic tails indicates expression of the tail is toxic to E. coli (11). Therefore, all constructs of the CCHF virus Gn cytoplasmic tail were expressed as GB1 fusion proteins. The GB1 tag contained His6 for nickel affinity purification and a TEV protease cleavage site to recover the native Gn zinc finger domain. The fusion protein was expressed in soluble form in E. coli, purified under native conditions, and digested with TEV protease to obtain the Gn zinc finger domain. Longer constructs comprising the entire predicted cytoplasmic tail (Gn719–819) expressed as insoluble inclusion bodies. Gn729–819, which was missing the first ten residues following the transmembrane region, expressed as soluble protein but with low yield. Gn729–805 represented the longest construct containing the conserved C-X2-C-X11–12-H-X3-C (where X is any amino acid) that also expressed in high enough yield to give high resolution NMR data.
Zn2+ Is Required for Proper Folding
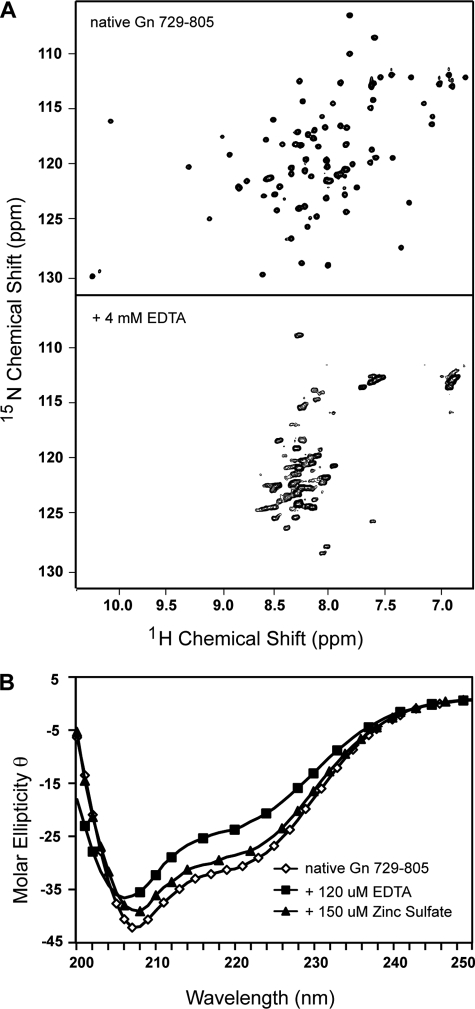
To examine the reliance of Zn2+-coordination on the proper folding of the CCHF virus Gn tail, we recorded the two-dimensional 15N HSQC of the Gn729–805 in the presence of 4 mm EDTA (Fig. 2A). The spectrum in the presence of EDTA is collapsed between ppm values of 6.5 and 8.6, whereas the folded spectrum in the absence of EDTA is well dispersed between 6.5 and 9.3 ppm. Narrowing of the spectrum suggests a loss of tertiary structure upon removal of Zn2+, indicating the requirement for Zn2+ binding in folding of the domain. A similar titration using circular dichroism (CD) spectroscopy demonstrates that the presence of EDTA causes a downward spectral shift, indicating a transition toward an unfolded protein (Fig. 2B). Here we also demonstrate that the addition of Zn2+ ion back into the sample recovers the trace of the original native spectrum. Therefore, Zn2+ is required for proper folding of the CCHF virus Gn tail.
FIGURE 2.
CCHF virus Gn zinc finger domain (residues 729–805) relies on Zn2+ for proper folding. Addition of 4 mm EDTA to a sample of 15N-labeled Gn729–805 effectively narrows the HSQC spectrum into a characteristic of an unfolded protein (A). Likewise, addition of a metal chelator causes a downward shift at 208 nm in the CD spectra toward random coil (Y axis: molar ellipticity θ per residue, deg·cm2 dmol−1residue−1 × 104) (B). Titration of zinc sulfate back into the sample recovers the original CD trace (B).
NMR Structure Determination
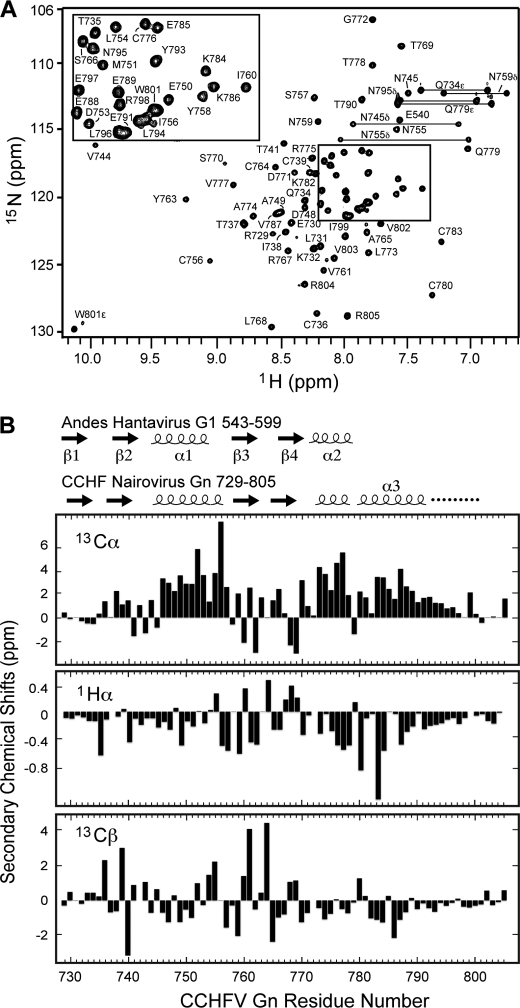
CCHF virus Gn729–805 showed a well dispersed two-dimensional 1H-15N HSQC (Fig. 3A). Complete backbone assignments were obtained from three-dimensional HNCA, CBCA(CO)NH, HNCACB, and 15N-edited NOESY-HSQC. The Cα, Hα, and Cβ secondary chemical shifts (Fig. 3B) showed the presence of three short α-helices with an intervening random coil region between the second and third helix. Two more regions in random coil orientations flanked the central sequence of the domain as indicated by the heteronuclear {1H}-15N NOE (Fig. 4C). Side chain assignments were completed using two-dimensional 1H-13C HMQC, three-dimensional HBHA(CO)NH, and three-dimensional 13C edited HMQC-NOESY. There were six invariant cysteine and two histidine residues (His-752 and His-776) in Gn729–805 (Fig. 1), all of which were involved in Zn2+ coordination. Long distance NOE's confirmed that His-752 and His-776 were involved in Zn2+ coordination. Notably, His-752 Hϵ1 shares an NOE with Cys736 Hβ′s. Likewise, His-776 Hϵ1 and Hδ2 share NOEs with Cys-761 and Cys-780 Hβ′s. A two-dimensional 15N HMQC (23) spectrum showed that His-752 and His-776 coordinated Zn2+ through the Nδ1 and Nϵ2 atoms (supplemental Fig. S1), respectively. Manual analysis of three-dimensional 15N- and 13C-edited NOESY spectra identified 1193 unambiguous interproton NOE distance restraints. The NOE restraints together with 26 ϕ and 26 ψ dihedral angle restraints and zinc coordination restrains (Table 1) were used in structure calculation and refinement in CYANA and AMBER. The 20 low energy NMR structures of Gn729–805 converged into a family of structures (Fig. 5) with low restraint violations and good Ramachandran plot statistics (Table 1).
FIGURE 3.
The CCHF virus Gn zinc finger yielded a well dispersed two-dimensional 1H-15N HSQC spectrum (A). The smaller peak in the tryptophan (W801) side-chain suggested a minor conformation of the tryptophan ring possibly due to ring flip-flop. Secondary chemical shifts for 13Cα, 1Hα, and 13Cβ suggest the presence of three short α helices interspersed with two short β hairpins (B).
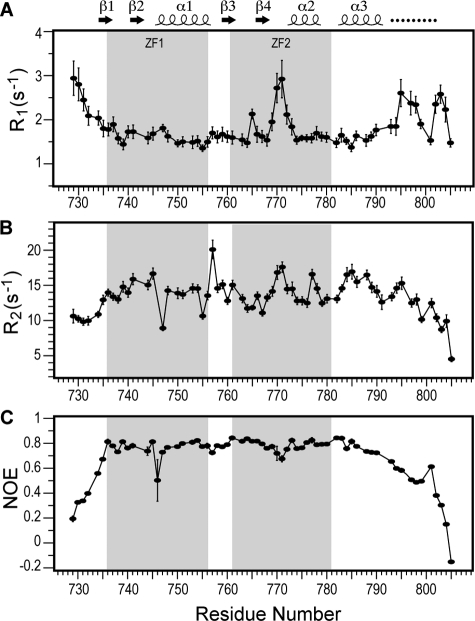
FIGURE 4.
Amide backbone relaxation rates R1 (A), R2 (B), and heteronuclear {1H}-15N NOE (C) of the CCHF virus Gn zinc finger.
TABLE 1.
NMR restraints and structural statistics for the 20 refined NMR structures
| Total distance restraints | 1193 |
| Intraresidue (i, i) | 246 |
| Sequential (i, i +1) | 407 |
| Long Range (i-j) > 4) | 307 |
| Total dihedral angle restraints | 52 |
| Phi | 26 |
| Psi | 26 |
| RMS deviation from mean structure | |
| Backbone atoms (N, Cα, Ć) (Å) | 0.25 |
| All heavy atoms (C, N, O) (Å) | 0.76 |
| NOE violations | |
| Max distance violation (Å) | 0.47 |
| Max dihedral angle violation (°) | 5.3 |
| Energies (kcal/mol) | |
| Mean GBa-AMBER energy | −3359 |
| Mean restraint energy | 79 |
| Ramachandran plot | |
| Most favorable region (%) | 79.2 |
| Additionally allowed regions (%) | 20.1 |
| Generously allowed regions (%) | 0.6 |
| Disallowed regions (%) | 0.2 |
a Generalized Born potential.
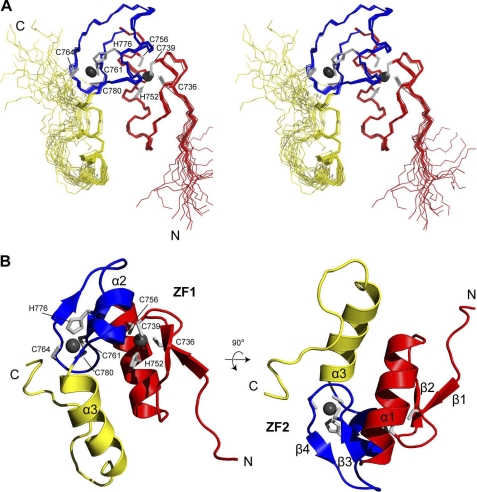
FIGURE 5.
NMR structure of CCHF virus Gn tail zinc finger. Stereoview of the superposition of 20 lowest energy NMR structures of CCHF virus Gn zinc finger (A). CCHF virus Gn zinc finger domain folds into a compact three-helix structure consisting of two back-to-back ββα zinc fingers with helix α3 pinned underneath the core zinc finger structure (B).
Structure of CCHF Virus Zinc Finger
The NMR structure of Gn729–805 reveals a rigid, compact three-helix structure with four short β-strands (Fig. 5). The structure contains a pair of tightly associated, back to back ββα zinc fingers connected by a short four residue linker (Ser757-Ile760) (Fig. 5). The first CCHC-zinc finger array (ZF1) consists of a Zn2+ ion coordinated to residues Cys-736, Cys-739, His-752, and Cys-756 and forms the classical ββα zinc finger fold. Cys-736 and Cys-739 form part of a short β-hairpin. Thr-737 and Ile-738 form a loop with Cys-736 and Cys-739 on either side of the hairpin. The structure contains helix α1 formed by Ile-747 to Ser-757 that folds back toward the β-hairpin, forming the ββα zinc finger fold. Cys-756 forms the fourth Zn2+-coordinating residue and is located on the same surface of helix α3 with His-752.
Likewise, the second CCHC-zinc finger array (ZF2) consists of a second Zn2+ ion coordinated to residues Cys-761, Cys-764, His-776, and Cys-780 into a classical ββα zinc finger fold. Cys-761 and Cys-764 are positioned on either side of a short β-hairpin. Pro-762 and Tyr-763 form a loop between Cys-761 and Cys-764. The structure is followed by a short helix α2 formed by Leu773-Cys-780 and folded back toward the β-hairpin, forming the ββα zinc finger fold. His-776 is located toward the middle of helix α3, and the final coordinating cysteine (Cys-780) is located at the end of helix α3. Although ZF2 also resembles the classical ββα fold, a minor difference exists when compared with ZF1. The helix α2 of ZF2 is shorter than helix α1 by three residues. This is due to the presence of helix breakers Gly-772 and Pro-781 located at either end of helix α2.
Unlike many classical ββα zinc fingers which form independently folded domains like “beads-on-a-string,” the two CCHF virus zinc fingers were tightly stuck together, with over 65 NOEs observed between ZF1 and ZF2. These NOEs fix the relative orientation of ZF1 with respect to ZF2. Among these NOEs, the strongest were observed between Met-751 (ZF1) and Tyr-763 (ZF2), Cys-739 (ZF1) and Ala-774 (ZF2), His-752 (ZF1) and Val-777 (ZF2), and Thr-741 (ZF1) and Val-777 (ZF2).
In addition to the two classical ββα fold zinc fingers, the structure contains an additional helix, helix α3, formed by Lys-782 to Glu-791 that packs against the dual zinc finger fold. A hydrophobic interaction between Val-744 of ZF1 and Val-787 keeps helix α3 pinned to the core structure. The orientation of helix α3 to ZF2 is partially determined by the helix breaker Pro-781, the residue immediately following ZF2. Pro-781 is 100% identical among Nairoviruses (Fig. 1) and serves as a kink between helix α2 and α3. Strong Cys Cα to Pro-718 Cδ NOEs indicated a trans proline isomer. The C-terminal 13 residues (Leu-792 to Lys-805) are primarily unstructured. NOEs between Ile-799 Cγ2 and Met-751 Cγ indicate the unstructured tail is pinned to the rest of the structure.
The 15N backbone relaxation rates (R1 and R2) as well as the heteronuclear {1H}-15N NOE (Fig. 4) showed that the ZF1, linker (residue 757–760), and ZF2 regions behave with nearly similar amide backbone dynamics. The average R1 values for ZF1, ZF2 and linker regions were well within each other, with values of 1.60 (+ 0.15), 1.81 (+ 0.40), and 1.65 (+ 0.04) s−1, respectively (Fig. 4A). ZF1 and the linker had essentially similar R1 values, however, within ZF2, the R1 values increased for residues 770 and 771 of the loop connecting β4 and α2, indicating increased mobility of this region. Likewise, the average R2 values for ZF1, ZF2, and linker regions were similar to each other, with values of 13.8 (+ 1.8), 13.8 (+ 1.8), and 15.6 (+ 3.1) s−1, respectively. Interestingly, the first linker residue, Ser-756, showed increased R2 without a corresponding increase in R1, which suggested chemical exchange on the μs-ms timescale for Ser-756. The average heteronuclear {1H}-15N NOE for the ZF1, linker and ZF2 regions was 0.8 (Fig. 4C), indicating reduced flexibility for the dual zinc finger domain including the linker region. In brief, the NMR amide backbone relaxation parameters (Fig. 4) confirmed that the two zinc fingers essentially tumble as one entity, that the linker between the two zinc fingers was rigid and tumble at the same rate as the zinc fingers, and that the loops and tails were flexible.
CCHF Virus Zinc Finger Contains Conserved Electrostatic Surfaces
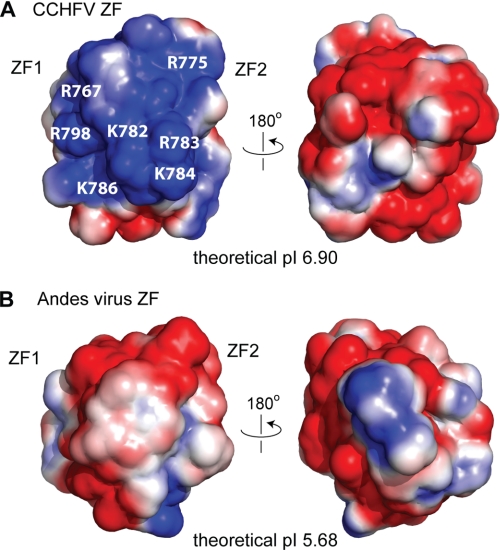
Analysis of the surface electrostatics of the CCHF virus Gn729–805 reveals clustering of positive and negatively charged surfaces on opposite faces of the structure (Fig. 6). Surface residues Glu-740, Glu-750, and Asp-753 of ZF1 converge with surface residues Glu-789 and Glu-791 of helix α3 and Glu-797 of the C-terminal unstructured region to form a large, nearly contiguous negatively charged surface. Similarly, Arg-767 and Arg-775 of ZF2 converge with Lys-782, Lys-784, and Lys-786 of helix α3 and Arg-798 of the C-terminal unstructured region to form a large contiguous positively charged surface. Of the charged surface residues, only Glu-750, Glu-797, Lys-784, and Arg-798 are not conserved in Nairoviruses. Most of the surface electrostatics, therefore, is a conserved feature of the Nairoviruses zinc finger domain.
FIGURE 6.
Comparison of the surface electrostatics of the CCHF virus (A) and Andes hantavirus (B) zinc fingers. Analysis of the surface electrostatics of the CCHF virus Gn zinc finger reveals a contiguous cluster of basic charges (colored blue) that cover the entire half of the structure (A). Rotating the structure 180° reveals an equally large cluster of acidic charges (colored red) on the opposite face (A). Surface electrostatics of the Andes hantavirus zinc fingers (PDB 2K9H) in the same orientation as the CCHF virus structure (B). Notably, the clustering of conserved basic surface in Nairovirus (A) is absent in Hantavirus (B).
CCHF Virus Gn Tail Binds RNA
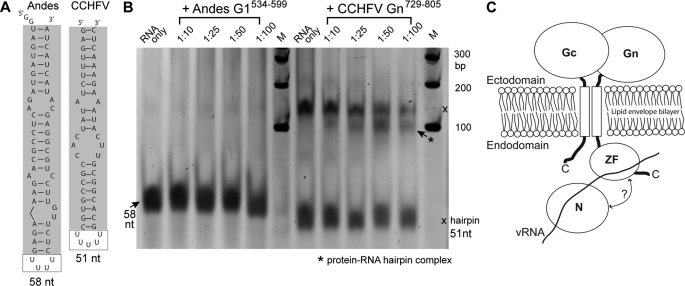
RNA electrophoretic mobility shift binding assays (EMSA) were carried out using two different proteins: the zinc finger domain of CCHF virus consisting of Gn729–805, and the zinc finger domain G1534–599 of the related Andes hantavirus. The RNA sequences used in the EMSA were a 58-mer RNA of the Andes hantavirus and a 51-mer RNA of the CCHF virus (Fig. 7A). Both RNA sequences contain 23–26 nucleotides at the 5′ and 3′ termini of the M genomic segments of the Andes and CCHF viruses, and these 5′ and 3′ strands are complementary to each other and are expected to form into hairpin-like panhandle structures (Fig. 7A). On a 12% native acrylamide gel, the Andes hantavirus RNA traveled as a single band consistent with a 58-mer hairpin (Fig. 7B), however, the CCHF virus 51-mer RNA migrated as two bands, a higher molecular weight form and a lower band migrating at a size consistent with a 51-mer hairpin (Fig. 7B). Incubation of the Andes hantavirus protein with the 58-mer RNA failed to affect the migration of RNA (Fig. 7B). The RNA bands in the presence of the Andes hantavirus protein were similar to the free RNA band (Fig. 7B). However, incubation of the CCHF virus zinc finger protein notably affected the migration of the CCHF virus RNA, as demonstrated by the appearance of an additional band (marked with asterisk, Fig. 7B) migrating between the hairpin and the higher MW form of the CCHF virus RNA. The protein-RNA complex band was most likely formed between the zinc finger protein and the RNA hairpin. Because the 51-mer CCHFV RNA existed in two forms (the hairpin and the higher MW form), the relative amounts of each form must be in equilibrium and binding of the CCHFV zinc finger protein to the hairpin RNA will also affect the amount of the RNA B form as seen in Fig. 7B. We are designing new RNA sequences that will only form hairpins for future studies of the protein-RNA interaction of the CCHF virus zinc finger. Nevertheless, the main conclusion from the EMSA results demonstrated that the CCHF Nairovirus Gn tail (residues 729–805) interacted with RNA whereas the Andes hantavirus G1 tail (residues 534–599) did not.
FIGURE 7.
RNA electrophoretic mobility shift assay comparing the Andes Hantavirus and the CCHF virus zinc fingers. RNA sequences used in the EMSA: Andes Hantavirus RNA (58 nt) and CCHF virus RNA (51 nt) (A). CCHF virus RNA traveled in two forms (marked with ×), lower band consistent with a 51-nt hairpin form, and a higher molecular weight band above 100 bp (B). While the Andes hantavirus zinc finger fails to alter the mobility of Andes hantavirus RNA, increasing the amount of CCHF virus zinc finger causes the appearance of an additional band (marked with *) for the CCHF virus RNA, thus suggesting a protein-RNA complex (B). Each lane contained 0.24 μmol RNA, proteins came from 0.4 mm stock diluted accordingly (B). Surface electrostatics combined with EMSA results of the Gn tail provide mechanistic insight into RNP packaging (C). We propose a model in which packaging consists of a zinc finger-RNA complex. Studies in related viruses suggest a nucleocapsid, Gn tail interaction, presented here as a possible additional packaging site (C).
To test our hypothesis that conserved basic residues in the CCHF virus zinc finger protein may be involved in RNA binding, point mutations were introduced in conserved lysine and arginine residues into aspartic acid and the mutant proteins were used in EMSA. The point mutants K783D, K786D, and R767D retained the ability to bind RNA suggesting these basic residues may not be critical in RNA binding (supplemental Fig. S2A). However, the K782D mutation disrupted the protein-RNA interaction, as its shift pattern resembles that of RNA alone. Coincidentally, Lys-782 is located at the kink between helix α2 and α3, and is pointed away from the zinc finger domain (supplemental Fig. S2B), suggesting that the kink between helix α2 and α3 may be important for RNA binding interaction of the CCHF virus Gn zinc finger.
DISCUSSION
We report here that an envelope glycoprotein of a Nairovirus contains a dual CCHC-type zinc finger (Figs. 2–5) domain that binds RNA. Although zinc fingers have been known in viruses, in particular, the HIV-1 nucleocapsid protein zinc fingers (33) are critical in RNA packaging and viral assembly, zinc fingers are rarely found in viral envelope glycoproteins. Including the zinc finger in this report, there are currently only three known structures of viral envelope glycoprotein zinc fingers. First is the zinc finger of the Hantavirus G1 envelope glycoprotein (11), and second is the zinc finger of the Junin virus envelope glycoprotein (34) (also an RNA virus, of family Arenaviridae). In all three cases, the cytoplasmic tails of the envelope glycoproteins contain the zinc finger domains. The Junin virus zinc fingers (34) form a unique fold that do not show any structural nor sequence similarity with the Nairovirus and Hantavirus zinc fingers. Although the Nairovirus and the Hantavirus zinc fingers (11), which both belong to family Bunyaviridae (Fig. 1), show an overall similar global fold (supplemental Fig. S3), they also have major structural differences (Fig. 6) and properties (Fig. 7).
Key Differences between Nairovirus and Hantavirus Gn Zinc Fingers
Examination of the surface electrostatics of the CCHF virus Gn tail reveals key differences when compared with the Andes hantavirus structure (Fig. 6). Whereas the CCHF Gn tail displays sharp clustering of conserved charges that form a large contiguous swath on the protein surface, the Andes hantavirus zinc fingers display charges that are predominately negative and apparently randomly dispersed (Fig. 6). The variation in charge conservation is also evident in the sequence analysis (supplemental Fig. S3). Whereas the spacing of CCHC motif is mostly conserved, the spacing between conserved charges is highly variable. The CCHF virus displays conserved negative charges on ZF1 and conserved positive charges on ZF2. The Andes hantavirus sequence, however, displays clustering of conserved positive charges on the sequences flanking the negatively charged core zinc finger structure.
Moreover, the CCHF virus Gn tail contains a structural motif that is absent in the Andes hantavirus structure. Helices α2 and α3 (Fig. 3B), residues Leu-773 to Leu-792, form a helix-kink-helix motif due the positioning of the conserved helix breaker Pro-781. While not an uncommon motif, this structural aspect in the CCHF Gn tail forms the core scaffold for a large positively charged surface partly composed of the conserved charges at Arg-775, Lys-782, and Lys-786 (Fig. 6A). By contrast, the Andes hantavirus structure contains neither the corresponding proline nor the charges to support a similar motif (Fig. 1). Instead, it contains the non-conserved helix breaker Gly-598 followed by conserved charges exclusively on the C-terminal end of ZF2 (Fig. 1). In this respect, the surface electrostatics of the CCHF virus Gn tail may more closely resemble that of Orthobunyaviruses, with conserved helix breakers flanked by conserved basic charges (Fig. 1). Perhaps not coincidentally, Nairoviruses and Orthobunyaviruses are both arthropod-borne, whereas Hantaviruses are rodent-borne (3). Overall, the general preservation of the fold indicates that the dual zinc finger motif plays a general but important role in the life cycle of both Nairoviruses and Hantaviruses. However, the major differences in the surface electrostatics of the CCHF virus and Hantavirus cytoplasmic tail structures (Fig. 6) also suggests that while general, the function of the tail may be species specific.
Proposed Role in Viral Assembly
Given the data available regarding the Bunyaviridae Gn role in viral assembly (8–10), it is likely that the surface electrostatics play an important role in assembly of the CCHF virus, presumably via direct interaction with some component of the ribonucleoprotein. The large positively charged surface of the Gn tail would suggest RNA binding, as is the traditional role for zinc fingers in retroviruses (33, 35). Our EMSA results (Fig. 7) suggest that this may in fact be the case. Using increasing amounts of Gn729–805 revealed the migration of an additional RNA band (Fig. 7B), which likely represents a protein-RNA complex consisting of Gn729–805 and the hairpin-like M segment panhandle. While the observed complex is weakly bound and therefore likely to be nonspecific, these results suggest RNA interaction mediated by some of the conserved residues in the Gn tail. Additionally, these results suggest the possibility of an interaction between the Gn tail and the RNA component of the ribonucleoproteins (Fig. 7C). A reverse genetics system for studying the CCHF virus has only recently been developed (36), thus allowing testing of this model in the future.
In summary, we present the NMR structure of a zinc finger domain in the Gn tail of the CCHF virus. Currently, this is the only available atomic structure for a protein component of the Nairovirus genus. The global fold of this zinc finger is similar to that of the Hantavirus zinc finger, which represents a unique fold of two classical-type ββα-zinc fingers that are stuck together (in contrast, individual classical ββα-zinc fingers behave as independent domains, like beads-on-a-string). We also demonstrated that the CCHF virus Gn tail binds RNA in vitro, thus suggesting the possibility of an interaction between the Gn tail with the viral RNA. Taken together, our results contribute novel mechanistic insight toward understanding the CCHF virus life cycle.
Supplementary Material
Acknowledgments
We thank Lou Altimura (United States Army Medical Research Institute of Infectious Diseases), Gaya Amarasinghe (Iowa State University), Sunhwan Jo (University of Kansas), Gerard Kroon (Scripps Research Institute), Dan McElheny (University of Illinois at Chicago), and Asokan Anbanandam (University of Kansas) for helpful discussions.
This work was supported in part by AHA 0755724Z, K-INBRE, and NIH P20 RR016475 (to R. N. D.) and the Madison and Lila Self Graduate Fellowship (to D. F. E.).
The atomic coordinates and structure factors (code 2L7X) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
NMR assignments were deposited at the Biological Magnetic Resonance Bank (BMRB ID 17383).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- CCHF
- Crimean Congo Hemorrhagic Fever
- EMSA
- electrophoretic mobility shift assay
- GB1
- the B1 domain of Streptococcus protein G
- HSQC
- heteronuclear single-quantum coherence spectroscopy
- NOE
- nuclear Overhauser effect
- R1
- longitudinal or spin-lattice relaxation rate
- R2
- transverse or spin-spin relaxation rate
- ZF1
- first CCHC zinc binding array, residues 736–756
- ZF2
- second CCHC zinc binding array, residues 761–780.
REFERENCES
- 1. Ergönül O. (2006) Lancet Infect. Dis. 6, 203–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Whitehouse C. A. (2004) Antiviral. Res. 64, 145–160 [DOI] [PubMed] [Google Scholar]
- 3. Elliott R. M., Bouloy M., Calisher C. H., Goldbach R., Moyer J. T., Nichol S. T., Pettersson R., Plyusnin A., Schmaljohn C. (2000) in Bunyaviridae. in Virus Taxonomy: The classification and Nomenclature of Viruses. The Seventh Report of the International Committee on Taxonomy of Viruses (Van Regenmortel M. H. V., Fauquet C. M., Bishop D. H. L., Carsten E. B., Estes M. K., Lemon S. M., Maniloff J., Mayo M. A., McGeoch D. J., Pringle C. R., Wickner R. B. eds.), Academic Press, San Diego [Google Scholar]
- 4. Schmaljohn C. S., Hooper J. W. (2001) in Bunyaviridae: the Viruses and Their Replication, 4th Ed. Fields Virology (Knipe D. M., Howley P. M. eds.), Lippincott, Williams & Wilkins, Philadelphia [Google Scholar]
- 5. Sanchez A. J., Vincent M. J., Nichol S. T. (2002) J. Virol. 76, 7263–7275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sanchez A. J., Vincent M. J., Erickson B. R., Nichol S. T. (2006) J. Virol. 80, 514–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Altamura L. A., Bertolotti-Ciarlet A., Teigler J., Paragas J., Schmaljohn C. S., Doms R. W. (2007) J. Virol. 81, 6632–6642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Overby A. K., Pettersson R. F., Neve E. P. (2007) J. Virol. 81, 3198–3205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shi X., Kohl A., Li P., Elliott R. M. (2007) J. Virol. 81, 10151–10160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hepojoki J. M., Strandin T., Wang H., Vapalahti O., Vaheri A., Lankinen H. (2010) J. Gen. Virol. 91, 2341–2350 [DOI] [PubMed] [Google Scholar]
- 11. Estrada D. F., Boudreaux D. M., Zhong D., St Jeor S. C., De Guzman R. N. (2009) J. Biol. Chem. 284, 8654–8660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chatterjee S., Zhong D., Nordhues B. A., Battaile K. P., Lovell S., De Guzman R. N. (2011) Protein Sci. 20, 75–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Geisbrecht B. V., Bouyain S., Pop M. (2006) Protein Expr. Purif. 46, 23–32 [DOI] [PubMed] [Google Scholar]
- 14. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 15. Johnson B. A. (2004) Methods Mol. Biol. 278, 313–352 [DOI] [PubMed] [Google Scholar]
- 16. Grzesiek S., Bax A. (1993) J. Am. Chem. Soc. 115, 12593–12594 [Google Scholar]
- 17. Grzesiek S., Döbeli H., Gentz R., Garotta G., Labhardt A. M., Bax A. (1992) Biochemistry 31, 8180–8190 [DOI] [PubMed] [Google Scholar]
- 18. Wittekind M., Mueller L. (1993) J. Magn. Reson. 101, 201–205 [Google Scholar]
- 19. Wishart D. S., Nip A. M. (1998) Biochem. Cell Biol. 76, 153–163 [DOI] [PubMed] [Google Scholar]
- 20. Tolman J. R., Chung J., Prestegard J. H. (1992) J. Magn. Reson. 98, 462–467 [Google Scholar]
- 21. Grzesiek S., Bax A. (1993) J. Biomol. NMR 3, 185–204 [DOI] [PubMed] [Google Scholar]
- 22. Hoffman R. C., Xu R. X., Klevit R. E., Herriott J. R. (1993) J. Magn. Reson. Ser. B 102, 61–72 [Google Scholar]
- 23. Pelton J. G., Torchia D. A., Meadow N. D., Roseman S. (1993) Protein Sci. 2, 543–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Marion D., Driscoll P. C., Kay L. E., Wingfield P. T., Bax A., Gronenborn A. M., Clore G. M. (1989) Biochemistry 28, 6150–6156 [DOI] [PubMed] [Google Scholar]
- 25. Stone M. J., Fairbrother W. J., Palmer A. G., 3rd, Reizer J., Saier M. H., Jr., Wright P. E. (1992) Biochemistry 31, 4394–4406 [DOI] [PubMed] [Google Scholar]
- 26. Farrow N. A., Muhandiram R., Singer A. U., Pascal S. M., Kay C. M., Gish G., Shoelson S. E., Pawson T., Forman-Kay J. D., Kay L. E. (1994) Biochemistry 33, 5984–6003 [DOI] [PubMed] [Google Scholar]
- 27. Janert P. K. (2009) Gnuplot in Action: Understanding Data with Graphs, Manning Publication Co., Greenwich, CT [Google Scholar]
- 28. Güntert P. (2004) Methods Mol. Biol. 278, 353–378 [DOI] [PubMed] [Google Scholar]
- 29. Case D. A., Pearlman D. A., Caldwell J. W., Cheatham T. E., Iii, Wang J., Ross W. S., Simmerling C. L., Darden T. A., Merz K. M., Stanton R. V., Cheng A. L., Vincent J. J., Crowley M., Tsui V., Gohlke H., Radmer R. J., Duan Y., Pitera J., Massova I., Seibel G. L., Singh U. C., Weiner P. K., Kollman P. A. (2002) AMBER7, University of California, San Francisco [Google Scholar]
- 30. Laskowski R. A., Rullmannn J. A., MacArthur M. W., Kaptein R., Thornton J. M. (1996) J. Biomol. NMR 8, 477–486 [DOI] [PubMed] [Google Scholar]
- 31. DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific, San Carlos, CA [Google Scholar]
- 32. Baker N. A., Sept D., Joseph S., Holst M. J., McCammon J. A. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. De Guzman R. N., Wu Z. R., Stalling C. C., Pappalardo L., Borer P. N., Summers M. F. (1998) Science 279, 384–388 [DOI] [PubMed] [Google Scholar]
- 34. Briknarová K., Thomas C. J., York J., Nunberg J. H. (2011) J. Biol. Chem. 286, 1528–1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. D'Souza V., Summers M. F. (2005) Nat. Rev. Microbiol. 3, 643–655 [DOI] [PubMed] [Google Scholar]
- 36. Flick R., Flick K., Feldmann H., Elgh F. (2003) J. Virol. 77, 5997–6006 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.