Abstract
Stress-induced endogenous and ectopically expressed GADD34 proteins were present both in the cytoplasm and in membranes, with their membrane association showing similar biochemical properties. Deletion of N-terminal sequences in GADD34-GFP proteins highlighted an amphipathic helix, whose hydrophobic surface, specifically valine 25 and leucine 29, mediated endoplasmic reticulum (ER) localization. Substitution of leucines for three arginines on the polar surface indicated that the same helix also mediated the association of GADD34 with mitochondria. Fluorescence protease protection and chemical modification of cysteines substituted in the membrane-binding domain pointed to a monotopic insertion of GADD34 into the outer layer of the ER membrane. Fluorescence recovery after photobleaching showed that ER association retards the mobility of GADD34 in living cells. Both WT GADD34 and the mutant, V25R, effectively scaffolded the α-isoform of protein phosphatase-1 (PP1α) and enabled eIF2α dephosphorylation. However, the largely cytosolic V25R protein displayed a reduced rate of proteasomal degradation, and unlike WT GADD34, whose ectopic expression resulted in a dilated or distended ER, V25R did not modify ER morphology. These studies suggested that the association of with ER modulates intracellular trafficking and proteasomal degradation of GADD34, and in turn, its ability to modify ER morphology.
Keywords: Endoplasmic Reticulum(ER), Phosphatase, Proteasome, Translation Regulation, Ubiquitination
Introduction
GADD34, a growth arrest and DNA damage (gadd) gene, collaborates with other gadd genes to inhibit cell growth following genotoxic and other forms of stress (1, 2). A subset of gadd genes was also identified as Myd genes that promote myeloid differentiation (3, 4). The C-terminal domain of GADD34/Myd116 shares sequence homology with the HSV-1 viral protein, ICP34.5 (or γ134.5) (2, 5), and a common function for the mammalian and viral proteins was hinted at by the functional substitution of the mouse GADD34 C-terminal domain for ICP34.5, which prevents HSV-1-mediated inhibition of host protein synthesis (5). Subsequent studies showed that ICP34.5 (6) and GADD34 (7, 8) bound the α-isoform of protein phosphatase-1 (PP1α)3 to facilitate the dephosphorylation of the translational initiation factor, eIF2α. Although eIF2α phosphorylation represses general protein synthesis, translation of the GADD34 mRNA is unhindered (9). Thus, GADD34 creates a feedback loop that promotes cell recovery from translational repression (8). The disruption of mouse genes encoding GADD34 and its structural homologue, CReP (constitutive repressor of eIF2α phosphorylation), highlighted that eIF2α dephosphorylation is essential for mouse development (10).
Paradoxically, GADD34 expression induces apoptosis or programmed cell death. Thus, apoptotic stimuli also stimulate GADD34 expression in many cells (1), and GADD34 expression in turn sensitizes cells to apoptosis by ionizing radiation (11) and oxidative stress (12). In this regard, our prior work established that GADD34 is polyubiquitinated and rapidly degraded by 26 S proteasome (13) and highlighted the remarkable diversity in the cellular content of GADD34 protein in human cells exposed to different stresses. Finally, the mathematical modeling of the cell responses to mild/reversible and chronic/terminal stress suggested that the short half-life of the GADD34 protein plays an important role in the survival versus death decision in mammalian cells (14).
Novoa et al. (8) identified a C-terminal fragment of hamster GADD34/MYD116 (amino acids 292–590) that showed diffuse cytosolic localization when compared with the full-length or WT GADD34, which possessed a reticular localization pattern consistent with ER. These studies also noted that the C-terminal fragment enhanced protein translation to significantly a greater extent than WT GADD34, hinting that ER localization attenuates GADD34 function to assemble an eIF2α phosphatase, but the molecular mechanism remained unresolved. Yet other studies identified candidate GADD34-binding proteins in the plasma membrane (15) and nucleus (11, 16–19), suggesting a role for GADD34 in other subcellular compartments.
Current studies utilized biochemical and cellular analyses to investigate the subcellular distribution of GADD34. Our studies demonstrated the presence of GADD34 in cytoplasm and several membrane compartments including ER, mitochondria, and Golgi bodies. We identified an N-terminal helical domain that mediated the association of GADD34 with ER and mitochondria and established that mutations that impair ER binding attenuated the proteasomal degradation of GADD34 and GADD34-mediated ER distension. These data suggest that redistribution or trafficking of GADD34 may regulate the steady state levels of GADD34 protein and potentially control protein translation and ER morphology in mammalian cells.
EXPERIMENTAL PROCEDURES
Materials
HEK293T, HeLa, and COS7 cells were obtained from the American Type Culture Collection (ATCC). Okadaic acid was obtained from Enzo Life Sciences, MG262 and digitonin were from Calbiochem, and Triton X-100 was from Bio-Rad. Cycloheximide, Ezview Red anti-FLAG M2 affinity gel, MG132, sodium deoxycholate, sodium arsenite, and thapsigargin were obtained from Sigma-Aldrich. Complete protease inhibitor mixture was purchased from Roche Applied Science. Poly(ethylene glycol)methyl ester 5000-maleimide (mPEG-Mal 5000) was from Nanocs.
Polyclonal antibodies against GADD34 were from Serotec, and anti-phospho(S51)-eIF2α was from Invitrogen. Antibodies against SEC23, GRP94, FLAG peptide epitope, and tubulin were purchased from Sigma-Aldrich. Anti-GFP, anti-PP1, and anti-L3 antibodies as well as horseradish peroxidase-linked goat anti-rabbit IgG, goat anti-mouse IgG, and donkey anti-goat IgG were from Santa Cruz Biotechnology, antibodies against RFP and calnexin were from Thermo Scientific, and antibodies against calnexin, CReP, cytochrome c, GM130, and GRP75 (also known as mtHSP70) were from Abcam. Alexa Fluor 594 goat anti-rabbit IgG antibody was obtained from Invitrogen. pDsRed2-ER plasmid (Clontech) containing the ER targeting sequence from calreticulin was modified to substitute GFP for RFP and is referred to as “GFP-calreticulin.” Rabbit anti-translocon associated protein α antibody was obtained from Chris Nicchitta; plasmids expressing GFP-cytochrome b5 were from Sally Kornbluth, and plasmids expressing GFP-Bax were from Jeff Rathmell, all from Duke University.
Cell Transfection
Cells were maintained at 37 °C in Dulbecco's minimal essential medium (DMEM) containing 10% (v/v) fetal bovine serum. Transfection of plasmid DNAs was undertaken in 12- or 6-well plates using FuGENE 6 (Roche Applied Science). Plasmids encoding WT and mutant GADD34 were generated as described previously (13).
Analysis of GADD34 Protein Turnover
HEK293T or HeLa cells expressing WT and mutant GADD34-GFP proteins were incubated in medium containing cycloheximide (30 μg/ml) to inhibit protein synthesis (13). Cells were harvested at intervals of 1–2 h in SDS-sample buffer prior to subjecting them to SDS-PAGE and immunoblotting with anti-GADD34 or anti-GFP antibodies.
Immunocytochemistry
COS7 or HeLa cells, grown on coverslips, were transfected with plasmids encoding WT and mutant GADD34 proteins with an N- or C-terminal GFP epitope. Approximately 20 h after transfection, cells were fixed with 4% (w/v) paraformaldehyde in phosphate-buffered saline (PBS) and incubated with anti-TRAPα followed by Alexa Fluor 594-conjugated anti-rabbit IgG secondary antibody. Similarly, cells were stained with anti-GRP94, anti-cytochrome c, and anti-FLAG antibodies. All cells were visualized with a Zeiss LSM710 laser confocal microscope.
Immunoprecipitation
HEK293T cells expressing FLAG-tagged WT and mutant GADD34 proteins were homogenized in 50 mm Tris-HCl, pH 7.4, containing 150 mm NaCl, 10% (v/v) glycerol, 1% (v/v) Triton X-100, 1 mm EDTA, and proteinase inhibitors. Cell lysates were subjected to centrifugation at 16,000 × g for 10 min, and supernatants were incubated with EZview red anti-FLAG-M2 affinity gel beads for 3–5 h at 4 °C. The beads were washed 3–5 times with lysis buffer before boiling in SDS-sample buffer at 95 °C for 10 min, and the released proteins were analyzed by SDS-PAGE and immunoblotting.
Cell Fractionation
HeLa or HEK293T cells expressing endogenous GADD34 induced by 1 μm thapsigargin, 5 μm MG132, or 75 μm arsenite for 8 h or ectopic GADD34 were washed with PBS, scraped into isotonic buffer (10 mm HEPES, pH 7.4, containing 0.32 m sucrose, 2 mm EDTA, and proteinase inhibitors), and disrupted by 20 strokes in a Dounce homogenizer at 4 °C. Centrifugation at 1,000 × g for 15 min at 4 °C was used to sediment cell debris and nuclei, and the resulting supernatant was centrifuged at 100,000 × g for 1 h to sediment plasma membranes, microsomes, and mitochondria. The 100,000 × g pellet was washed and resuspended in 50 mm HEPES, pH 7.4, containing 2 mm EDTA and proteinase inhibitors and stored at −80 °C. The 100,000 × g supernatant, representing cytosol, was stored similarly. In selected experiments, intermediate 15,000 × g centrifugation removed mitochondria and nuclei and was followed by a 100,000 × g centrifugation to sediment membranes.
Membrane Protein Extraction
Extraction of membrane proteins was undertaken by incubating the 100,000 × g pellet in 50 mm Tris-HCl, pH 7.5 (control) or the same buffer containing 0.2 m NaCl, 0.6 m NaCl, 1.0% (v/v) Triton X-100, 1.0% (w/v) deoxycholate, or 1.0% (w/v) SDS. Aliquots of membranes were washed in 0.1 m Na2CO3, pH 10.0, or 0.1 m Na2CO3, pH 11.0, solutions. All samples were incubated for 20 min at room temperature by end-over-end shaking and centrifuged at 100,000 × g for 1 h at 4 °C to sediment residual membranes. Supernatants containing released proteins and pellets containing non-solubilized proteins were subjected SDS-PAGE and immunoblotting.
Fluorescence Protease Protection Assay
COS7 cells grown on Lab-Tek chambered coverslips (Nalge Nunc) to 50–75% confluence were cotransfected with 1 μg of plasmid encoding WT GADD34, with N- or C-terminal GFP epitope, and 0.2 μg of mRFP plasmid using FuGENE 6. After 24 h, the medium were replaced by 20 mm HEPES, pH 7.5, containing 110 mm potassium acetate and 2 mm MgCl2, and images were acquired using a Nikon Eclipse TE 2000-E inverted spinning disc confocal microscope before and after incubation with 50 μm digitonin and subsequent treatment with 0.02% (w/v) trypsin. All images were processed using MetaMorph 7.1 software.
Modification of Cysteines Substituted in Membrane-binding Domain
Plasmids encoding the N-terminal 60 amino acids of WT GADD34 fused at its C terminus to GFP and mutants substituting cysteines for alanine 6, methionine 26, serine 30, alanine 32, or alanine 58 were transfected in COS7 cells using FuGENE 6. After 24 h, cells were washed in 20 mm HEPES, pH 7.4, containing 110 mm sodium acetate, 2 mm magnesium acetate, 0.5 mm EGTA, and protease inhibitors. Parallel cell samples were incubated in the above buffer containing 100 nm digitonin or 100 nm digitonin and 1% (v/v) Triton X-100 for 20 min at 4 °C. Cells were sedimented at 6,000 × g for 10 min and subsequently incubated in buffer containing 100 nm digitonin and 1 mm mPEG-Mal 5000 or 100 nm digitonin, 1% (v/v) Triton X-100, and 2 mm mPEG-Mal 5000 for 30 min at 4 °C. The reaction was terminated by the addition of 20 mm DTT, and cells were lysed by incubating with 10% (w/v) deoxycholate. Cell lysates were subjected to centrifugation at 18,000 × g for 20 min, and the resulting supernatant was analyzed by SDS-PAGE and immunoblotting.
Fluorescence Recovery after Photobleaching
COS7 Cells expressing GFP and GFP-GADD34 proteins were analyzed with a Nikon A1R confocal microscope. A target area of 2.5-μm radius was bleached for 500 ms using 95% laser power, and time-dependent recovery of fluorescence was monitored. An adjacent unbleached target area of similar size was analyzed as a control and used to correct for background fluctuations in GFP fluorescence. Initial rates of fluorescence recovery from 3–6 replicate studies were calculated as described by McNally (20).
RESULTS
GADD34 is undetectable in most resting cells but significantly elevated following stress (13). In HeLa cells treated with arsenite (oxidative stress) or thapsigargin (ER stress), immunostaining with anti-GADD34 antibody showed that GADD34 was predominantly at the ER, which was costained with DiOC6, which also weakly stains mitochondria, and to an even lesser extent, Golgi bodies (supplemental Fig. S1).
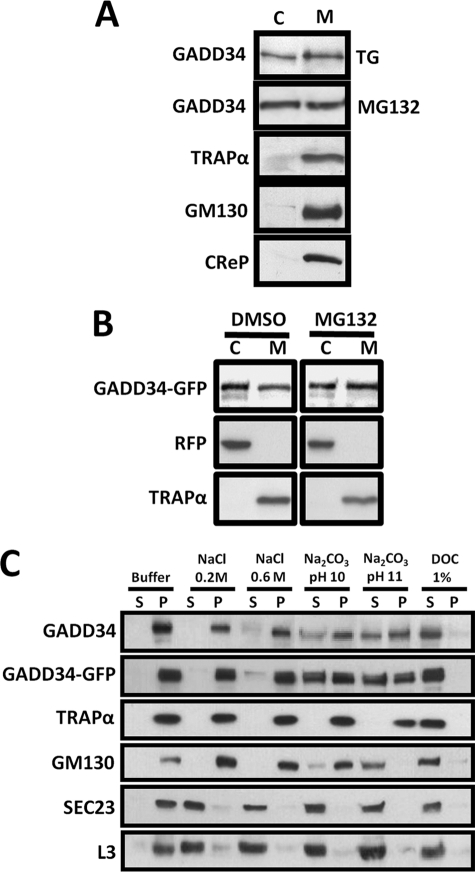
When lysates of HeLa cells treated with arsenite (data not shown), thapsigargin, or MG132 were subjected to high-speed centrifugation, approximately half the endogenous GADD34 sedimented with the 100,000 × g membrane fraction, whereas the remainder was present in the supernatant or cytoplasm (Fig. 1A). By comparison, TRAPα, an ER transmembrane protein, and GM130, a Golgi membrane protein, were both present exclusively in the membrane fraction. Interestingly, CReP, a structural and functional homologue of GADD34 (21), sedimented solely with the membranes, and no CReP was detected in the cytosol. In HeLa cells coexpressing GADD34-GFP and RFP, treated with either DMSO or MG132, similar distribution of GADD34-GFP was observed in membranes, containing TRAPα, and in cytoplasm, containing RFP (Fig. 1B).
FIGURE 1.
Subcellular distribution of human GADD34. A, HeLa cells subjected to ER stress by exposure to 1 μm thapsigargin (TG) or 5 μm MG132 for 5 h were homogenized as described under “Experimental Procedures.” Parallel samples of 100,000 × g supernatant or cytosol (C) and membrane pellet (M) were subjected to SDS-PAGE and immunoblotting with anti-GADD34, anti-TRAPα, anti-GM130, and anti-CReP antibodies. B, HeLa cells coexpressing GADD34-GFP and RFP were treated with either DMSO or 5 μm MG132 for 5 h and subjected to fractionation as described in panel A, and the cytosol and membrane fractions were immunoblotted with anti-GFP, anti-RFP, and anti-TRAPα antibodies. C, purified membranes from HeLa cells exposed to 1 μm thapsigargin for 5 h to induce endogenous GADD34 or expressing WT GADD34-GFP were incubated in a variety of extraction buffers as described under “Experimental Procedures.” Following centrifugation, supernatant (S) and pellet (P) were subjected to SDS-PAGE and immunoblotting with anti-GADD34, anti-GFP, anti-TRAPα, anti-GM130, anti-SEC23, and anti-ribosomal L3.
To investigate the biochemical characteristics of the membrane-bound GADD34, we purified membranes, utilizing an intermediate 15,000 × g centrifugation to remove mitochondria and nuclei accompanied by repeated centrifugation to remove all contaminating cytosol. In the control buffer, endogenous GADD34, GADD34-GFP, TRAPα, GM130, SEC23, a peripheral ER membrane protein, and the ribosomal protein, L3, all sedimented with the membrane fraction (Fig. 1C). Although SEC23 and L3 were largely released in Tris-HCl, pH 7.5, containing 0.2 or 0.6 m NaCl, endogenous GADD34, GADD34-GFP, TRAPα, and GM130 all remained membrane-bound. Na2CO3 solution (pH 10) effectively solubilized SEC23 and L3, but only a fraction of the endogenous GADD34, GADD34-GFP, and GM130 was released. By contrast, Na2CO3, pH 11.0, completely solubilized GM130, but GADD34 and GADD34-GFP remained partially membrane-bound. All membrane proteins, including TRAPα, were extracted in Tris-HCl, pH 7.5, containing 1% (w/v) sodium deoxycholate, consistent with detergent solubilization of the microsomal membranes. Together, these data emphasized the very similar biochemical properties of membrane-bound GADD34, whether endogenous or overexpressed, and suggested a tighter association of GADD34 with membranes when compared with some peripheral membrane proteins, e.g. SEC23 and GM130. On the other hand, GADD34 was more readily extracted from membranes than TRAPα, an ER transmembrane protein.
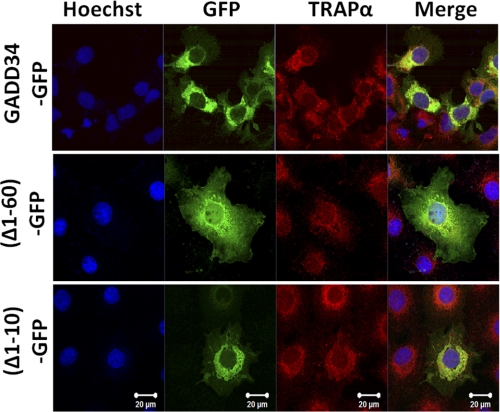
Earlier studies showed that deletion of the N-terminal one-third of GADD34 resulted in the diffuse cytosolic distribution of the protein (8, 22). Conversely, the fusion of the N-terminal 180 amino acids of GADD34 targeted GFP, an otherwise cytosolic protein, to the ER, displaying localization very similar to WT or endogenous GADD34 (22). To define the ER membrane-binding domain, we expressed WT GADD34-GFP, which colocalized with TRAPα, an ER transmembrane protein, in COS7 cells (Fig. 2). In contrast, (Δ1–60)-GADD34-GFP, lacking the N-terminal 60 amino acids, was distributed throughout the cytoplasm. However, (Δ1–10)-GADD34-GFP, lacking the N-terminal 10 amino acids, was still ER-associated, as shown by its colocalization with TRAPα. These data suggested that the ER-targeting domain in GADD34 resided between amino acids 10 and 60.
FIGURE 2.
An N-terminal domain mediates ER localization of GADD34. Localization of WT GADD34-GFP, GADD34 lacking N-terminal 60 residues, (Δ1–60)-GFP, and N-terminal 10 amino acids, (Δ1–10)-GFP from GADD34 fused to GFP in COS7 cells, is shown. Cells were fixed as described under “Experimental Procedures” and stained with Hoechst (blue) to show nuclei, visualized for GFP fluorescence (green), and immunostained with anti-TRAPα (red). The merged images (Merge) are also shown.
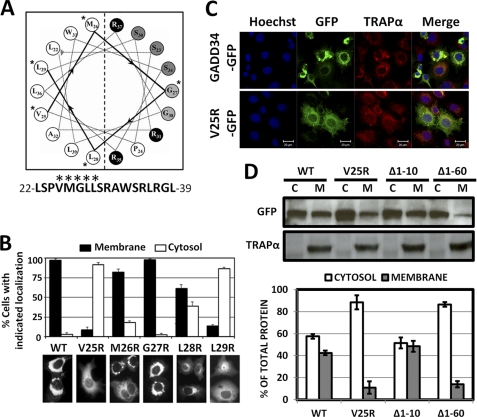
Hydropathy analysis of GADD34 highlighted a hydrophobic region, between amino acids 16 and 65 (supplemental Fig. S2), which was highly conserved in mammalian species and represented a potential membrane-binding domain. Analysis of this region by AmphipaSeeK (23) predicted an amphipathic helix between amino acids 22 and 39 that could function as an in-plane membrane anchor (Fig. 3A). Individual residues in the hydrophobic sequence, VMGLL, were substituted with arginines. Although WT GADD34-GFP localized with ER in 97% of cells, which also showed a dilated or distended ER morphology, 90% of cells expressing V25R showed a broad cytosolic distribution of this protein (Fig. 3B). Furthermore, M26R, G27R, and L28R remained predominantly ER-associated, but L29R was largely cytosolic. Location of Val25 and Leu29 in the center of the hydrophobic surface was consistent with their in-plane insertion into the outer lipid layer of the ER membrane.
FIGURE 3.
Amphipathic helix mediates membrane binding by GADD34. A, AmphipaSeeK predicts a potential amphipathic membrane anchoring helix between amino acids 22 and 39 in human GADD34, with residues (marked with asterisks) contributing to the hydrophobic surface. B, individual hydrophobic residues in the VMGLL sequence were substituted with arginines, and the mutant GADD34-GFP proteins were expressed in COS7 cells. Representative WT and mutant GADD34-GFP-expressing cells are shown below the bar graph, in which cells with GFP fluorescence localized at the ER (Membrane) or more broadly distributed (Cytosol) were counted and the results of three independent experiments, each counting ∼100 cells, shown with S.E. C, confocal microscopy was undertaken in HeLa cells expressing WT GADD34-GFP and V25R-GFP. Cells were stained with Hoechst (blue) to show nuclei and immunostained with anti-TRAPα (red). GFP fluorescence (green) for the GADD34 proteins and the merged images (Merge) are also shown. D, fractionation of HeLa cells expressing WT GADD34-GFP, V25R-GFP, (Δ1–10)-GFP, and (Δ1–60)-GFP was undertaken as described under “Experimental Procedures,” and the cytosol (C) and membrane (M) fractions were subjected to immunoblotting with anti-GFP and anti-TRAPα antibodies. The bar graph displays the fraction (in percentage) of the total GADD34 protein in cytosol (open bars) and membrane (filled bars) fractions derived from each cell population.
Confocal microscopy confirmed that WT GADD34-GFP colocalized with TRAPα, whereas V25R was widely distributed throughout the cytoplasm (Fig. 3C). Biochemical fractionation of HeLa cells expressing GADD34 proteins showed that WT GADD34-GFP and (Δ1–10)-GADD34-GFP were equally distributed in cytosol and membranes, whereas (V25R)-GADD34-GFP and (Δ1–60)-GADD34-GFP were predominantly cytosolic, although ∼10% was still membrane-bound (Fig. 3D).
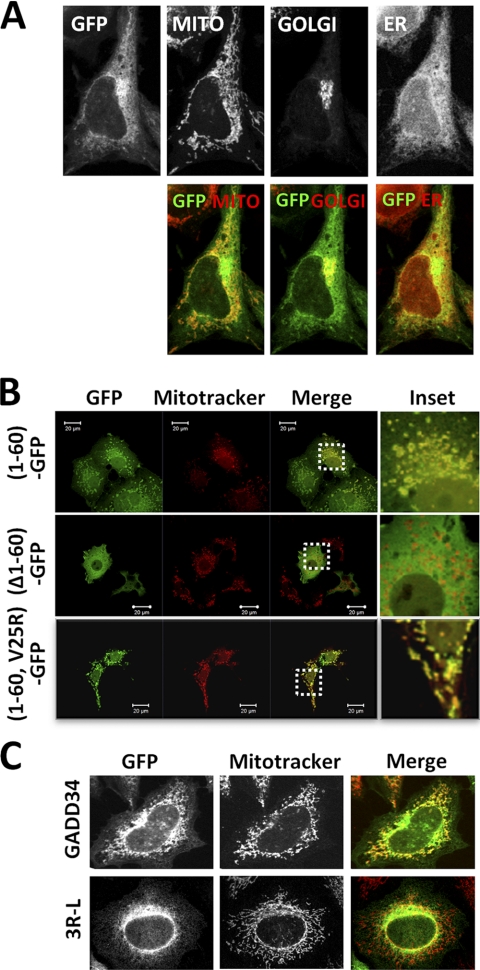
Analysis of WT GADD34-GFP-expressing cells stained with MitoTracker, a mitochondrial dye, and immunostained with antibodies against calnexin, an ER membrane protein, or GM130, a Golgi marker, suggested that although the majority of GADD34 was localized at the ER, a smaller fraction may also be present at the mitochondria with even smaller fraction showing overlap with the Golgi marker (Fig. 4A). Immunostaining with antibody against mtHSP70 or GRP75, a mitochondrial protein, confirmed this finding (data not shown).
FIGURE 4.
GADD34 localization in multiple membrane compartments. A, HeLa cells expressing GADD34-GFP were stained with MitoTracker red dye to show mitochondria (MITO). Cells were also immunostained with anti-GM130 (GOLGI) or anti-calnexin (ER) and counterstained with Alexa Fluor 647- or Alexa Fluor 350-conjugated secondary antibody respectively. Top row, individual fluorescent images (in black and white) showing GFP, mitochondria, Golgi bodies, and ER. Lower row, merged color images of GADD34-GFP (green) and MITO, Golgi bodies, or ER (red). B, (1–60)-GFP, (Δ1–60)-GFP, and (1–60,V25R)-GFP were expressed in COS7 cells, which were then stained with MitoTracker dye. GFP fluorescence (green), MitoTracker (red), and the merged images (Merge) are shown. Enlarged images of areas highlighted by boxes (Inset) are also shown. C, WT and mutant GFP-(3R-L)-GADD34 proteins (green) expressed in HeLa cells, in which the mitochondria were stained with MitoTracker dye (red). Merged images (Merge) are shown.
As expression of WT GADD34-GFP in mammalian cells often results in a dilated or condensed ER, which can mask the visualization of mitochondria, we also analyzed the (1–60)-GFP-expressing COS7 cells stained with MitoTracker (Fig. 4B). Significant overlap of GFP fluorescence was observed with the MitoTracker staining (see inset). By comparison, there was little overlap between the MitoTracker dye and (Δ1–60)-GADD34-GFP. This suggested that the N-terminal 60 amino acids that targeted GADD34 to the ER also mediate its targeting to mitochondria. By contrast, insertion of the V25R substitution, which abrogated ER binding, in (1–60,V25R)-GFP showed a surprising concentration with mitochondria stained by MitoTracker. These data showed that distinct elements of the same N-terminal domain targeted GADD34 to the ER and mitochondria and hinted at competition for GADD34 binding between these organelles.
MitoProt II predicted a mitochondrial targeting sequence in GADD34 (24) within the amphipathic helix shown to mediate ER association. Proteins that target mitochondria often rely on the presence of one or more arginines on the polar surface of an amphipathic helix (25). Hence, we substituted arginines 31, 35, and 37 with leucines. In cells expressing lower levels of WT GADD34-GFP, significant overlap of GFP fluorescence with the MitoTracker dye was observed. By contrast, GADD34-(3R-L)-GFP showed a much reduced or no overlap with MitoTracker (Fig. 4C).
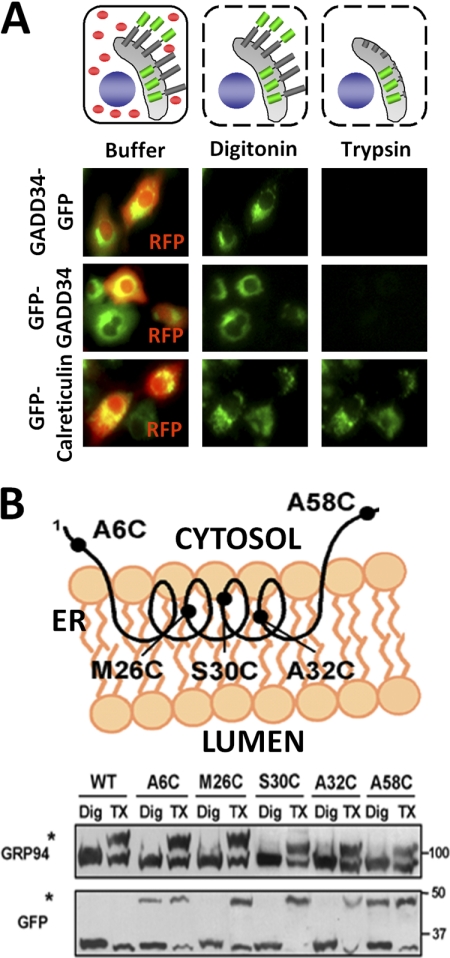
To establish the membrane topology of the ER-bound GADD34, we analyzed COS7 cells expressing GFP-GADD34 or GADD34-GFP with the fusion of either N-terminal or C-terminal GFP epitopes, along with GFP-calreticulin, which was targeted to the ER lumen. RFP was expressed as a cytosolic marker (Fig. 5A). Treatment of cells with 40 mm digitonin resulted in complete release of RFP, establishing the permeabilization of the plasma membrane and the release of cytosol. However, the ER-localized fluorescence associated with GFP-GADD34, GADD34-GFP, or GFP-calreticulin was unchanged by this treatment (Fig. 5A). Further treatment of the digitonin-permeabilized cells with trypsin resulted in the complete loss of GFP fluorescence accredited to either GFP-GADD34 or GADD34-GFP but not that for the ER luminal GFP-calreticulin. These data suggested that the N and C termini of GADD34 both resided in the cytoplasm and were accessible to trypsin in the digitonin-permeabilized cells. By contrast, the ER luminal GFP-calreticulin was completely protected.
FIGURE 5.
Topology of membrane-bound GADD34. A, the fluorescence proteinase protection assay is shown (schematic) in a COS7 cell that expressed GFP-GADD34 or GADD34-GFP with N- and C-terminal GFP (green), respectively, shown as potential ER transmembrane proteins with RFP as the cytosolic marker (red). Cells were subjected to digitonin permeabilization to release RFP or cytosol. Subsequent trypsin treatment digested fluorescent epitopes that were presented in cytosol, whereas those located in the ER lumen (shown in gray) were protected from proteolysis. GFP-calreticulin, an ER luminal protein, was used as control, and its fluorescent epitope is not released by either digitonin or trypsin treatment. Cells expressing GFP-GADD34 or GADD34-GFP (green) and RFP (red) incubated in buffer, digitonin, or digitonin and trypsin are shown below. B, an alternative approach in which cysteines were substituted in place of alanine 6, methionine 26, serine 30, alanine 32, and alanine 58. The schematic shows the putative orientation of a GADD34 polypeptide encompassing the N-terminal amphipathic helix that predicted A6C and A58C outside the membrane, whereas other substitutions, M26C, S30C, and A32C, are inserted in the lipid bilayer. COS7 cells expressing WT and mutant GFP-(1–60)-GADD34 proteins were permeabilized with digitonin (Dig) or Triton X-100 (TX) followed by incubation with mPEG-Mal 5000. Cysteine modification of GRP94, an ER luminal protein, was used as a control. The mPEG-Mal 5000-conjugated GRP94 and GFP-(1–60)-GADD34 proteins showed reduced electrophoretic mobility (marked by asterisks).
Further support for the membrane topology of GADD34 was obtained by substituting cysteines in place of selected residues, namely A6C, M26C, S30C, A32C, and A58C, in the N-terminal 60 amino acids of GADD34 fused to GFP (schematically shown in Fig. 5B), which localized at the ER. Cells expressing WT (1–60)-GFP were treated with digitonin to permeabilize plasma membrane or Triton X-100 to permeabilize both the ER and the plasma membranes. Following incubation with the cysteine-modifying compound, mPEG-Mal 5000, which does not traverse cell membranes, WT (1–60)-GFP, which contained no cysteines and was not modified by mPEG-Mal 5000 in cells treated with either digitonin or Triton X-100, displayed an unaltered electrophoretic mobility on SDS-PAGE (Fig. 5B). By contrast, GRP94, an ER luminal protein, was modified in cells permeabilized with Triton X-100 but not with digitonin (marked by asterisk). The (1–60)-GFP mutants, A6C and A58C, were modified by mPEG-Mal 5000 in cells treated with either digitonin or Triton X-100. By contrast, M26C, S30C and A32C, were only modified following the treatment of cells with Triton X-100. Together, these data suggested that methionine 26, serine 30, and alanine 32, but not alanine 6 and alanine 58, were inserted into the lipid bilayer and supported a monotopic ER membrane association of GADD34 with both its N terminus and its C terminus located in the cytoplasm.
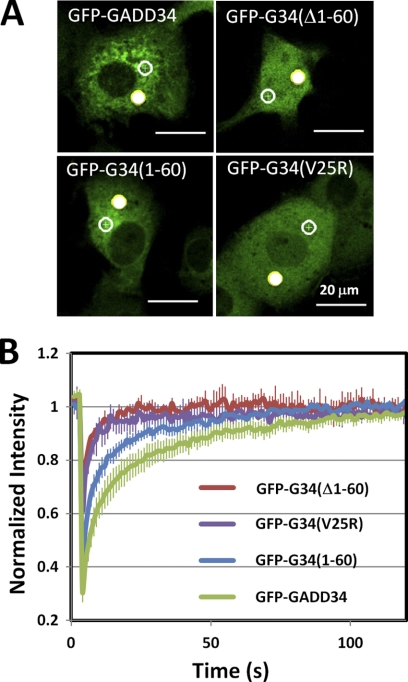
To explore the potential role of membrane binding in cellular dynamics of GADD34, we undertook fluorescence recovery after photobleaching in COS7 cells expressing GFP-GADD34, GFP-(Δ1–60)-GADD34, GFP-(1–60)-GADD34 and GFP-(V25R) GADD34 (Fig. 6A). The analysis of initial rates of fluorescence recovery in more than 3 independent replicates (Fig. 6B) showed a retarded recovery rate for GFP-GADD34 consistent with its ability to bind membranes. The deletion of the N-terminal 60 amino acids or the substitution, V25R, which resulted in the increased cytosolic distribution of GADD34, accelerated the recovery for fluorescence such that these proteins displayed rates of recovery approaching that of GFP (supplemental Table 1).
FIGURE 6.
Fluorescence recovery after photobleaching of GFP-GADD34-expressing cells. A, representative COS7 cells expressing WT GFP-GADD34, GFP-(Δ1–60)-GADD34 (GFP-G34((Δ1–60)), GFP-(1–60)-GADD34 (GFP-G34(1–60)), and GFP-(V25R)-GADD34 (GFP-G34(V25R)) are shown. Two target areas, one subjected to photobleaching (solid circle) and one adjacent to the unbleached area (open circle), were monitored over time. B, the results for fluorescence recovery for 3–6 independent replicate experiments with error bars showing the S.E.
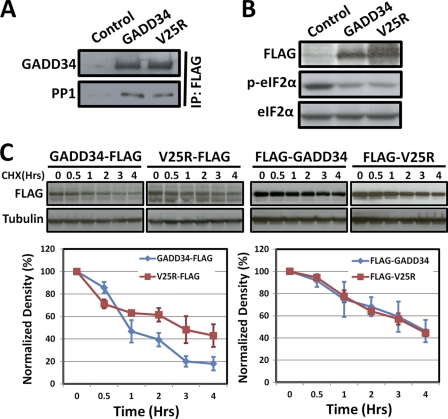
Coimmunoprecipitation of WT FLAG-GADD34 and FLAG-(V25R)-GADD34 with endogenous PP1 (Fig. 7A) correlated with their ability to promote eIF2α dephosphorylation (Fig. 7B). However, our prior studies showed a rapid proteasomal degradation of WT GADD34 with a half-life under 1 h (13). Comparison of the decay rates for WT GADD34-FLAG and V25R-GADD34-FLAG (epitope containing no lysines that can be ubiquitinated, and thus, influence GADD34 degradation) in cells following cycloheximide treatment showed a significantly slower rate for degradation of V25R-GADD34-FLAG when compared with that WT GADD34-FLAG (Fig. 7C, left panel). By contrast, the presence of an N-terminal FLAG epitope stabilized both proteins, essentially eliminating any difference in the turnover of WT FLAG-GADD34 and FLAG-V25R (Fig. 7C, right panel).
FIGURE 7.
ER-bound and cytosolic GADD34 assemble active eIF2α phosphatases. A, control untransfected and GADD34-expressing HEK293T cells. Immunoblotting with an anti-GADD34 and an anti-PP1 antibody was performed. All cells contained equal levels of PP1 catalytic subunit by immunoblotting with anti-PP1c antibody (data not shown). Immunoprecipitation (IP) with anti-FLAG M2 beads from lysates of control HEK293T cells and cells expressing WT GADD34-FLAG and V25R-GADD34-FLAG sedimented equal amounts of WT and mutant GADD34. Equal amounts of PP1 catalytic subunit were also coprecipitated. No PP1 was sedimented in control cells, which lacked GADD34-FLAG. B, WT GADD34-FLAG and V25R-GADD34-FLAG were expressed in HEK293T cells at similar levels by immunoblotting with anti-FLAG. Both proteins catalyzed similar eIF2α dephosphorylation, seen by immunoblotting with anti-phosphoserine 51 (eIF2α). All cells were treated with 100 nm okadaic acid for 10 min to elevate eIF2α phosphorylation (p-eIF2α). Immunoblotting with anti-eIF2α shows equivalent loading of all cell lysates. C, HEK293T cells expressing WT FLAG-GADD34 or mutant V25R-GADD34-FLAG were treated with cycloheximide (CHX) to inhibit protein synthesis, and lysates prepared at the indicated time intervals were subjected to immunoblotting with anti-GFP antibody. Tubulin was used as control for protein loading. A representative immunoblot and summed results of three independent experiments displayed as a graph (with S.D.) are shown below. Similar experiments were undertaken in HEK293T cells expressing WT FLAG-GADD34 and FLAG-V25R-GADD34, containing N-terminal FLAG epitopes. The quantification of three independent experiments is shown the graph below.
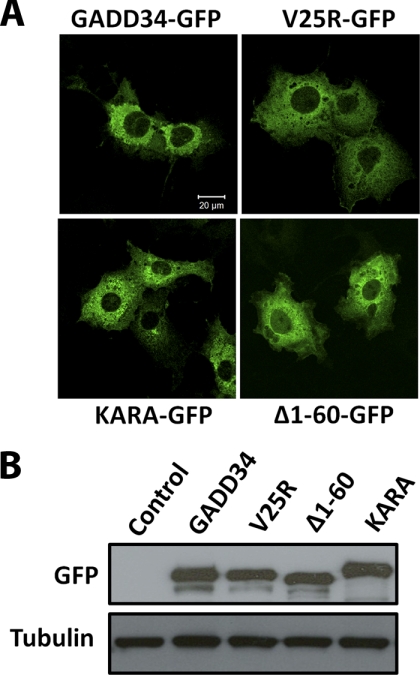
Finally, the ectopic expression of WT GADD34 in mammalian cells consistently resulted in a dilated or distended ER, as seen in the COS7 cells expressing WT GADD34-GFP (Fig. 8A). At similar levels of expression (Fig. 8B), (V25R)-GADD34-GFP and (Δ1–60)-GADD34-GFP did not alter ER morphology. KARA, a mutant GADD34 that localizes to the ER but does not bind PP1 (22), also did not change ER morphology.
FIGURE 8.
ER association and assembly of an active eIF2α phosphatase contribute to GADD34-mediated ER distension. A, COS7 cells expressing WT GADD34-GFP display a distended or dilated ER, which is not seen in cells expressing either V25R-GFP or (Δ1–60)-GFP, both of which are broadly distributed in the cytosol. By contrast, the mutant GADD34, KARA, which fails to bind PP1, is still localized at the ER and also did not alter ER morphology when compared with control untransfected cells (not shown). B, equivalent expression of the GADD34 proteins by immunoblotting with anti-GFP antibody. Anti-tubulin immunoblotting established equivalent protein loading.
DISCUSSION
Eukaryotic cells respond to adverse environmental and metabolic conditions by phosphorylating the translation initiation factor, eIF2α (at serine 51), and inhibiting general protein translation. This allows cells to focus their energy on resolving, and thereby, surviving the stress. GADD34 is a stress-induced protein whose translation is not attenuated by eIF2α phosphorylation. Thus, GADD34 is synthesized even in the presence of an ongoing stress and recruits PP1α (22) to facilitate eIF2α dephosphorylation (7) and restores general protein synthesis. Premature or excessive GADD34 expression can enhance protein translation and can aggrevate the already stressed ER protein quality control machinery in the ER, triggering cell death. In this regard, the short half-life of the GADD34 protein (and mRNA) is essential not only to restore effective eIF2α kinase signaling but also to ensure cell survival in the face of subsequent bouts of stress (7, 26).
In mammalian cells, proteins are translated on ribosomes associated with ER and those present in the cytoplasm. To evaluate the GADD34 contribution to protein translation in these compartments, we undertook cell biological studies in HeLa and COS7 cells that hinted at the presence of GADD34 primarily at the ER (22). However, current cell fractionation studies in HEK293T and HeLa cells provided the evidence for a broader subcellular distribution of GADD34 in mammalian cells that was independent of its expression levels, suggesting that its presence in membranes and cytosol was an intrinsic property of the protein. Both endogenous stress-induced and ectopically expressed GADD34 showed similar distributions in membranes and cytosol in several different cell types. The membrane-bound GADD34, whether endogenous or overexpressed, showed more avid membrane binding than peripheral proteins like SEC23 (ER) and GM130 (Golgi bodies) but was more readily solubilized than transmembrane proteins like TRAPα (ER) and CFTR (plasma membrane, data not shown). Moreover, the membrane-bound GADD34, whether endogenous or overexpressed, showed identical biochemical characteristics for extraction by various buffers. These data allowed GADD34 expression in cells to be used to gain insights into the molecular mechanism for membrane binding.
Mutagenesis studies highlighted an N-terminal amphipathic helix, between amino acids 10 and 60, that targeted GADD34 to ER. Specifically, the single amino acid substitution, V25R, dispersed GADD34 throughout the cell. In addition, two cell-based assays, fluorescence protease protection and mPEG-5000 modification of substituted cysteines, pointed to a monotopic insertion of GADD34 in the outer surface of the ER membrane with its N and C termini, critical for GADD34 regulation (13) and function (22), facing the cytoplasm.
ER-associated protein degradation or ER-associated protein degradation is activated by stress (27). Basal and sterol-induced ER-associated protein degradation of HMG-CoA reductase requires its binding to the ER membrane (28). ER binding also enhanced the proteasomal degradation of GADD34 as the cytosolic mutant, V25R, was degraded more slowly. GADD34 degradation, however, was not inhibited by the ER-associated protein degradation inhibitor, eeyarestatin 1 (data not shown). The presence of an N-terminal epitope that stabilizes GADD34 (13) eliminated the differences in protein turnover between WT GADD34 and V25R. Thus, we hypothesize that the N-terminal ubiquitination of GADD34 (13), thought to promote its turnover, may be facilitated by its association with ER.
ER morphology is often altered by elevated expression of ER-associated proteins, such as HMG-CoA reductase (28) or microsomal aldehyde dehydrogenase (29), which result in a crystalloid ER. By comparison, cells expressing WT GADD34 showed a dilated or distended ER that was clearly distinguishable using electron microscopy (data not shown). Insertion of monotopic helical membrane-binding domains often induce membrane curvature (30) and could account for the GADD34-induced changes in ER morphology. However, N-terminal fragments of GADD34, such as (1–60)-GADD34, that encompassed the amphipathic helix, were effectively targeted to the ER but did not modify ER morphology. ER dilation was reported in cells expressing eIF2α (S51A) (31) and the ER chaperone, HSP47 (32), and may reflect the increased protein synthesis and/or molecular crowding of proteins in the ER, awaiting processing by the protein quality control machinery (33). The changes in ER morphology induced by WT GADD34 were not observed when equivalent levels of the cytosolic V25R or the mutant, KARA, which does not assemble an eIF2α phosphatase (22), were expressed in mammalian cells. Thus, we speculate that increased protein translation and/or delayed protein processing accounts for the ER dilation or distension seen in GADD34-expressing cells (summarized in Table 1). This in turn may hint at the ER association of GADD34 preferentially enhancing the translation of mRNAs by the ER-bound polysomes (34). The current studies showed similar PP1 binding and eIF2α dephosphorylation by WT GADD34 and V25R. By contrast, a C-terminal fragment of mouse GADD34 (amino acids, 241–657), which was broadly distributed in cells, was reported to be more active than WT GADD34 in the suppression of the integrated stress response, as judged by the expression of a CHOP-GFP reporter protein, despite showing similar ability to promote eIF2α dephosphorylation. This suggests that eIF2α dephosphorylation may not accurately reflect the changes in protein translation, and analysis of specific mRNAs translated on ER-bound and/or cytosolic polysomes may provide greater insights into the role of GADD34 localization in the control of protein translation, the stress response, and ER morphology.
TABLE 1.
Function and regulation of GADD34 proteins
m, mouse; h, human. ND, not detected.
| Suppression of ISR | Protein stability | ER localization | ER distension | Ref. | |
|---|---|---|---|---|---|
| FLAG-mGADD34-(241–657) | ++ | NDa | − | − | 8 |
| FLAG-mGADD34-(1–657) | + | ND | +++ | +++b | 8 |
| FLAG-hGADD34-(1–674) | ND | +++ | +++ | +++ | Ref. 13, this work |
| hGADD34-(1–674)-FLAG | ND | + | +++ | +++ | Ref. 13, this work |
| hGADD34-(V25R)-FLAG | ND | +++ | − | − | This work |
| FLAG-hGADD34-(V25R) | ND | +++ | − | − | This work |
a Although analysis of N-terminally truncated GADD34 proteins (13) suggests that the C-terminal fragment of mouse GADD34 would be more stable than WT mGADD34, the presence of an N-terminal FLAG epitope would be predicted to abolish this difference in protein turnover.
b Although not specifically discussed (8), cells expressing this polypeptide showed a distended ER.
A novel finding of our studies was that GADD34 associates with mitochondria, as predicted by MitoProt II (24). Like proteins targeted to mitochondria by an amphipathic helix (25), multiple arginines in the N-terminal amphipathic helix appeared to target GADD34 to mitochondria. This places GADD34 among a unique class of proteins that bind ER and mitochondria by a single “chimeric” membrane-targeting domain. For example, cytochrome P450B1 utilizes a common N-terminal domain to target ER and mitochondria, with the unphosphorylated form of this protein displaying ER localization and the PKA phosphorylated protein being mitochondrial (35). In this regard, our studies showed that GADD34 localization in cells is dynamic. Moreover, GADD34 is highly phosphorylated (36), and future work will explore the role of covalent modification in determining the subcellular distribution of GADD34.
Growing evidence points to some ER-bound proteins being translocated to mitochondria to trigger apoptosis following severe stress (37–39). Although GADD34 expression was associated with mitochondrial fragmentation (supplemental Fig. S3), unlike the proapoptotic mitochondrial protein, Bax (40), GADD34 did not induce cytochrome c release (supplemental Fig. S3), and the physiological role of mitochondrial GADD34 is currently unknown.
In summary, our studies provided the first evidence for dynamic localization of GADD34 in multiple subcellular compartments, specifically ER, mitochondria, and cytosol. We also established that a bifunctional N-terminal domain targets GADD34 to both mitochondria and ER, with ER localization playing an important role in GADD34 turnover and changes in ER morphology. These data suggest that cellular mechanisms that can modulate GADD34 intracellular trafficking may also define the physiological function and regulation of GADD34 following stress.
Supplementary Material
Acknowledgment
We thank the Singapore Bio-imaging Consortium-Nikon Imaging Center for the use of the facility and advice in completing the fluorescence recovery after photobleaching studies.
The work was supported by research funds provided by Duke-NUS Graduate Medical School and an individual research grant (IRG09nov003) from the National Medical Research Council (to S. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3 and Table S1.
- PP1α
- protein phosphatase-1 α-isoform
- CReP
- constitutive repressor of eIF2α phosphorylation
- ER
- endoplasmic reticulum
- mPEG-Mal 5000
- poly(ethylene glycol)methyl ester 5000-maleimide
- DMSO
- dimethyl sulfoxide.
REFERENCES
- 1. Hollander M. C., Sheikh M. S., Yu K., Zhan Q., Iglesias M., Woodworth C., Fornace A. J., Jr. (2001) Int. J. Cancer 96, 22–31 [DOI] [PubMed] [Google Scholar]
- 2. Hollander M. C., Zhan Q., Bae I., Fornace A. J., Jr. (1997) J. Biol. Chem. 272, 13731–13737 [DOI] [PubMed] [Google Scholar]
- 3. Lord K. A., Abdollahi A., Hoffman-Liebermann B., Liebermann D. A. (1990) Cell Growth Differ. 1, 637–645 [PubMed] [Google Scholar]
- 4. Zhan Q., Lord K. A., Alamo I., Jr., Hollander M. C., Carrier F., Ron D., Kohn K. W., Hoffman B., Liebermann D. A., Fornace A. J., Jr. (1994) Mol. Cell. Biol. 14, 2361–2371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chou J., Roizman B. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 5247–5251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. He B., Gross M., Roizman B. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 843–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Connor J. H., Weiser D. C., Li S., Hallenbeck J. M., Shenolikar S. (2001) Mol. Cell. Biol. 21, 6841–6850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Novoa I., Zeng H., Harding H. P., Ron D. (2001) J. Cell Biol. 153, 1011–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lee Y. Y., Cevallos R. C., Jan E. (2009) J. Biol. Chem. 284, 6661–6673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Harding H. P., Zhang Y., Scheuner D., Chen J. J., Kaufman R. J., Ron D. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 1832–1837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Adler H. T., Chinery R., Wu D. Y., Kussick S. J., Payne J. M., Fornace A. J., Jr., Tkachuk D. C. (1999) Mol. Cell. Biol. 19, 7050–7060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lu P. D., Jousse C., Marciniak S. J., Zhang Y., Novoa I., Scheuner D., Kaufman R. J., Ron D., Harding H. P. (2004) EMBO. J. 23, 169–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Brush M. H., Shenolikar S. (2008) Mol. Cell. Biol. 28, 6989–7000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rutkowski D. T., Arnold S. M., Miller C. N., Wu J., Li J., Gunnison K. M., Mori K., Sadighi Akha A. A., Raden D., Kaufman R. J. (2006) PLoS Biol. 4, e374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shi W., Sun C., He B., Xiong W., Shi X., Yao D., Cao X. (2004) J. Cell Biol. 164, 291–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Garrido J. L., Maruo S., Takada K., Rosendorff A. (2009) Virol. J. 6, 231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Imai H., Harland J., McCulloch J., Graham D. I., Brown S. M., Macrae I. M. (2002) Eur. J. Neurosci. 15, 1929–1936 [DOI] [PubMed] [Google Scholar]
- 18. Stelzl U., Worm U., Lalowski M., Haenig C., Brembeck F. H., Goehler H., Stroedicke M., Zenkner M., Schoenherr A., Koeppen S., Timm J., Mintzlaff S., Abraham C., Bock N., Kietzmann S., Goedde A., Toksöz E., Droege A., Krobitsch S., Korn B., Birchmeier W., Lehrach H., Wanker E. E. (2005) Cell 122, 957–968 [DOI] [PubMed] [Google Scholar]
- 19. Wu D. Y., Tkachuck D. C., Roberson R. S., Schubach W. H. (2002) J. Biol. Chem. 277, 27706–27715 [DOI] [PubMed] [Google Scholar]
- 20. McNally J. G. (2008) Methods Cell Biol. 85, 329–351 [DOI] [PubMed] [Google Scholar]
- 21. Jousse C., Oyadomari S., Novoa I., Lu P., Zhang Y., Harding H. P., Ron D. (2003) J. Cell Biol. 163, 767–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Brush M. H., Weiser D. C., Shenolikar S. (2003) Mol. Cell. Biol. 23, 1292–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sapay N., Guermeur Y., Deléage G. (2006) BMC. Bioinformatics 7, 255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Claros M. G., Vincens P. (1996) Eur. J. Biochem. 241, 779–786 [DOI] [PubMed] [Google Scholar]
- 25. Lemire B. D., Fankhauser C., Baker A., Schatz G. (1989) J. Biol. Chem. 264, 20206–20215 [PubMed] [Google Scholar]
- 26. Harding H. P., Novoa I., Bertolotti A., Zeng H., Zhang Y., Urano F., Jousse C., Ron D. (2001) Cold Spring Harb. Symp. Quant. Biol. 66, 499–508 [DOI] [PubMed] [Google Scholar]
- 27. Jingami H., Brown M. S., Goldstein J. L., Anderson R. G., Luskey K. L. (1987) J. Cell Biol. 104, 1693–1704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chin D. J., Luskey K. L., Anderson R. G., Faust J. R., Goldstein J. L., Brown M. S. (1982) Proc. Natl. Acad. Sci. U.S.A. 79, 1185–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yamamoto A., Masaki R., Tashiro Y. (1996) J. Cell Sci. 109, 1727–1738 [DOI] [PubMed] [Google Scholar]
- 30. Drin G., Antonny B. (2010) FEBS. Lett. 584, 1840–1847 [DOI] [PubMed] [Google Scholar]
- 31. Scheuner D., Vander Mierde D., Song B., Flamez D., Creemers J. W., Tsukamoto K., Ribick M., Schuit F. C., Kaufman R. J. (2005) Nat. Med. 11, 757–764 [DOI] [PubMed] [Google Scholar]
- 32. Marutani T., Yamamoto A., Nagai N., Kubota H., Nagata K. (2004) J. Cell Sci. 117, 5913–5922 [DOI] [PubMed] [Google Scholar]
- 33. Shnyrova A., Frolov V. A., Zimmerberg J. (2008) Curr. Biol. 18, R474–R476 [DOI] [PubMed] [Google Scholar]
- 34. Stephens S. B., Dodd R. D., Brewer J. W., Lager P. J., Keene J. D., Nicchitta C. V. (2005) Mol. Biol. Cell 16, 5819–5831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Anandatheerthavarada H. K., Biswas G., Mullick J., Sepuri N. B., Otvos L., Pain D., Avadhani N. G. (1999) EMBO. J. 18, 5494–5504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Molina H., Horn D. M., Tang N., Mathivanan S., Pandey A. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 2199–2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Qi X., Mochly-Rosen D. (2008) J. Cell Sci. 121, 804–813 [DOI] [PubMed] [Google Scholar]
- 38. Morishima N., Nakanishi K., Tsuchiya K., Shibata T., Seiwa E. (2004) J. Biol. Chem. 279, 50375–50381 [DOI] [PubMed] [Google Scholar]
- 39. Gill M. B., Perez-Polo J. R. (2009) J. Neurosci. Res. 87, 2047–2065 [DOI] [PubMed] [Google Scholar]
- 40. Arnoult D. (2007) Trends Cell Biol. 17, 6–12 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.