Abstract
The antibiotic fusidic acid potently inhibits bacterial translation (and cellular growth) by lodging between domains I and III of elongation factor G (EF-G) and preventing release of EF-G from the ribosome. We examined the functions of key amino acid residues near the active site of EF-G that interact with fusidic acid and regulate hydrolysis of GTP. Alanine mutants of these residues spontaneously hydrolyzed GTP in solution, bypassing the normal activating role of the ribosome. A conserved phenylalanine in the switch II element of EF-G was important for suppressing GTP hydrolysis in solution and critical for catalyzing translocation of the ribosome along mRNA. These experimental results reveal the multipurpose roles of an interdomain joint in the heart of an essential translation factor that can both promote and inhibit bacterial translation.
Keywords: Antibiotics, Bacterial Signal Transduction, G Proteins, Ribosomes, Translation Elongation Factors
Introduction
Many antibiotics are known to inhibit translation in bacteria directly by binding to ribosomes. A few antibiotics can achieve the same end indirectly by targeting essential protein factors that regulate bacterial translation through hydrolysis of GTP. Examples of antibiotics in the latter category include fusidic acid and kirromycin, which target elongation factor (EF)2 G and EF-Tu, respectively. These antibiotics do not inhibit EF-G and EF-Tu from binding to ribosomes or hydrolyzing GTP. Rather, they prevent dissociation of the factors from ribosomes. Their potency is enhanced because multiple ribosomes often engage simultaneously in translation of a single mRNA molecule, thereby halting one ribosome and all others behind it on an mRNA.
Despite their specificity and potency, the widespread use of these (and other) antibiotics by humans has spurred the spread of resistant strains of pathogenic bacteria (1). Fusidic acid is a steroid antibiotic clinically introduced in 1962 and still used to treat staphylococcal infections (2). Fusidic acid-resistant (FusR) bacteria often harbor mutations in fusA, the principal chromosomal gene that encodes EF-G, as well as other horizontally acquired genes (3–7). The fusA mutations cluster around a central pocket formed by three domains of EF-G (3, 8, 9). Because fusidic acid binds only to the ribosome·EF-G complex after GTP hydrolysis (10), its target was not precisely determined until recently, with the report of the first crystal structure of the ribosome containing EF-G, GDP, and fusidic acid (11). This structure showed that fusidic acid indeed lodges into the interdomain pocket of EF-G, as suggested by the earlier genetic studies.
Antibiotic-resistant bacteria often pay a price of reduced biological fitness, which selects against their survival in the absence of antibiotic (12). FusR bacteria have served as good experimental models. FusR mutations involve highly conserved residues of EF-G that not only interact with fusidic acid but also probably serve important roles in the normal cellular functions of EF-G. FusR bacteria display multifarious phenotypes, including reduced rates of protein synthesis, impaired cellular growth, and abnormal levels of the alarmone ppGpp (4, 13). Intragenic secondary mutations in fusA can ameliorate the fitness of FusR bacteria to close to wild-type levels in liquid culture or animal models (6, 14, 15). The latter may account for persistent infections by FusR bacteria that harbor multiple fusA mutations in clinical isolates (5). However, FusR EF-G proteins remain mostly uncharacterized at the biochemical level, except in a few cases (13, 16).
To explore the biochemical nature of FusR EF-G proteins, in this study, we examined the functions of highly conserved residues in the interdomain pocket of EF-G that are mutated in bacteria most resistant to fusidic acid. Our results led us not only to a better understanding of the inhibitory mechanism of fusidic acid but also to unexpected insights into the regulation of GTP hydrolysis and translocation of the ribosome during normal protein synthesis in bacteria. Our findings are reminiscent of the effects of kirromycin on EF-Tu and suggest avenues for finding novel antibiotics that target GTPases of bacteria.
EXPERIMENTAL PROCEDURES
Materials
Ribosomes were purified from Escherichia coli MRE600 cells, and other materials were obtained as described (17, 18). Mutations of desired EF-G codons were introduced genetically into a plasmid encoding E. coli EF-G with a C-terminal hexahistidine tag (17). EF-G proteins were purified by nickel-nitrilotriacetic acid affinity chromatography, followed by gel filtration chromatography (17, 19).
All assays were carried out in polymix buffer (pH 7.5) containing 5 mm potassium phosphate, 5 mm magnesium acetate, 0.5 mm calcium chloride, 95 mm potassium chloride, 5 mm ammonium chloride, 1 mm dithiothreitol, 8 mm putrescine, and 1 mm spermidine (20).
EF-G Binding and Hydrolysis of 2′,3′-O,N′-Methylanthraniloyl (mant)-GTP
EF-G interactions with mant-GTP were monitored by fluorescence spectroscopy (19, 21). Binding affinities were derived from experiments titrating EF-G with increasing concentration of mant-GTP. mant-GTP bound to EF-G was measured by Förster resonance energy transfer methods, i.e. exciting tryptophan residues in EF-G with photons and measuring the emission of photons from the bound mant-GTP (19, 21). The rates of mant-GTP hydrolysis were derived from stopped-flow fluorescence experiments measuring the initial rate of fluorescence increase associated with mant-GTP binding and hydrolysis (see supplemental “Experimental Procedures”).
EF-G Binding to the Ribosome
The degree of EF-G binding to the ribosome was assessed by the specific protection by EF-G of nucleotide A2660 in 23 S rRNA of the ribosome from methylation by dimethyl sulfate (DMS) (22). We quantified the binding of FusR EF-G mutants relative to control samples that contained or lacked wild-type EF-G. The relative intensities of the DMS-modified A2660 bands in polyacrylamide gels were measured by phosphorimaging (17).
GTP Hydrolysis
The hydrolysis of radioactive [γ-32P]GTP was analyzed by TLC (18). EF-G and ribosomes were heat-activated just before experiments (∼300 s, 37 °C). EF-G, GTP (containing [γ-32P]GTP), and ribosomes (when present) were mixed together. Aliquots were removed at four time points during the linear phase of the reaction kinetics and quenched with 15% formic acid. Reactant and product (γ-32P-labeled GTP and inorganic phosphate) were separated by TLC and quantified to calculate the values indicated on the y axis. These values were normalized for hydrolyzed GTP in the stock (between 3 and 4%).
Fusidic Acid Inhibition of EF-G
GTP hydrolysis assays were performed under multiple turnover conditions to measure the ability of fusidic acid to trap EF-G on the ribosome. Reactions contained EF-G (0.4 μm), ribosomes (2 μm), GTP (200 μm, containing [γ-32P]GTP), and fusidic acid (0–5 mm). Samples were quenched and analyzed by TLC as described above.
Ribosome Translocation
Translocation of the ribosome by one codon along an mRNA was monitored by assays of toeprinting and fluorescent mRNA (18). In both assays, pretranslocational ribosome complexes were formed with the E. coli ribosome containing T4 gene 32 mRNA, tRNAfMet bound in the ribosome P site, and tRNAPhe in the ribosome A site. Then, EF-G and GTP were added to the pretranslocational ribosomes. In toeprinting assays, a DNA primer (radioactively 5′-end-labeled) was annealed to the mRNA 3′-end; ribosome translocation was detected by primer extension with reverse transcriptase. In fluorescent mRNA assays, pyrene was attached to the mRNA 3′-end; EF-G·GTP and pretranslocational ribosomes were rapidly mixed together via a stopped-flow device; and ribosome translocation was detected by quenching of the fluorescence intensity as pyrene entered the ribosome.
RESULTS
FusR Mutants in E. coli EF-G
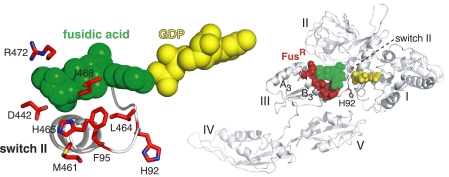
We selected seven residues of EF-G that are mutated in bacteria of clinical and laboratory origins that display the highest levels of resistance to fusidic acid (4–8). We studied these strong FusR mutants in the context of the well studied E. coli translation system (17, 18). Five of these residues make direct (van der Waals) contact with fusidic acid in the recent crystal structure of EF-G bound to the Thermus thermophilus ribosome (Fig. 1) (11). The selected residues of E. coli EF-G were Phe-95, Asp-442, Met-461, Leu-464, His-465, Ile-468, and Arg-472. A sequence alignment of EF-G proteins (relevant to this study and from other bacterial phyla) that highlights the seven FusR residues is shown in supplemental Fig. 1.
FIGURE 1.
FusR residues of EF-G selected for in vitro analysis. This structural schematic, based on ribosome-bound EF-G from T. thermophilus (11), shows the locations of fusidic acid and GDP on EF-G. The enlargement (left) highlights our seven selected FusR residues with the corresponding numbers of E. coli EF-G. It shows the side chain of Phe-95 (stick representation) in switch II of domain I (ribbon backbone) and the side chains of other FusR residues (sticks) in helices A3 and B3 (backbones are not shown on the left). Also shown is His-92, the catalytic residue that activates the reacting water molecule for nucleophilic attack of GTP. The same residues are highlighted in the overall EF-G structure (right) bound to the ribosome (not shown).
Several observations led us to suspect that these FusR residues not only bind fusidic acid but also may be important for normal functions of EF-G. Phe-95 of EF-G corresponds to a previously isolated mutation (F95L) that confers the strongest fusidic acid resistance in Staphylococcus aureus (8). Phe-95 is located prominently at the tip of the switch II element of domain I of EF-G. Switches I and II are conformationally mobile elements in the conserved G domains of GTPases (23). In EF-G, these switches are thought to convert free energy from GTP hydrolysis on its G domain (I) into mechanical movements of its other four domains (II–V), which ultimately drive the unidirectional translocation of the ribosome along an mRNA and the rapid cycling of EF-G on and off the ribosome during protein synthesis (21, 24–26). In crystal structures of EF-G proteins in different functional states, Phe-95 is oriented differently, at a loop-helix junction in switch II. The side chain of Phe-95 can be tucked into the loop (Fig. 1) (8, 11) or can project away from domain I and toward the neighboring domain III or V (27). Rearrangements in switch II (including Phe-95) appear to drive rotational movements of domains I, III, and V (11, 25). The other six FusR residues are located in domain III, where their side chains orient toward Phe-95 and fusidic acid (Fig. 1), suggesting their functional interactions. Finally, the identities of all seven residues are conserved across bacterial and mitochondrial phyla, with the one exception of Val-468 (supplemental Fig. 1), suggesting their cellular essentiality.
To study the roles of the seven FusR residues in normal EF-G functions, we introduced the FusR mutations into our recombinant hexahistidine-tagged E. coli EF-G (which we refer to as “wild-type EF-G”) (17, 18). We introduced the FusR mutants that were previously isolated and characterized in vivo. In addition, we replaced the same FusR residues individually with alanine. The resulting EF-G proteins were purified and characterized in a number of assays measuring specific EF-G activities in vitro.
EF-G Mutant F95A Spontaneously Hydrolyzes mant-GTP in Solution
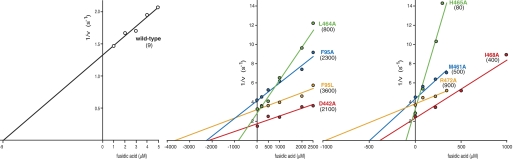
Our first experiments focused on the functional consequences of mutating Phe-95 of EF-G. We measured the ability of fusidic acid to inhibit the GTPase activity of EF-G in the presence of the ribosome and excess GTP (i.e. multiple substrate turnover conditions). The effects of titrating fusidic acid on the various FusR mutants are graphed in Fig. 2. The inhibitory constant (Ki) for fusidic acid using wild-type EF-G was 9 μm. Mutant F95L of EF-G, previously characterized in vivo (8), was strongly resistant to fusidic acid in vitro (Ki = 3600 μm). Mutant F95A was also strongly FusR (Ki = 2300 μm). These results suggested that Phe-95 is critical for the inhibitory effects of fusidic acid on EF-G.
FIGURE 2.
Alanine mutants of previously identified FusR residues in E. coli EF-G are also strongly resistant to fusidic acid in vitro. We measured the ability of fusidic acid to suppress GTP hydrolysis catalyzed by EF-G proteins in the presence of the ribosome, excess GTP, and variable concentrations of fusidic acid (as indicated on the x axis). Rates (v) of reactions were determined as described under “Experimental Procedures.” Data of 1/v as a function of fusidic acid concentration are plotted. The inhibitory constant (Ki) for fusidic acid binding to EF-G corresponds to the x axis intercept, i.e. the fitted line through the data points and extrapolated to the x axis. Ki values (in μm) for wild-type and mutant EF-G proteins are given in parentheses.
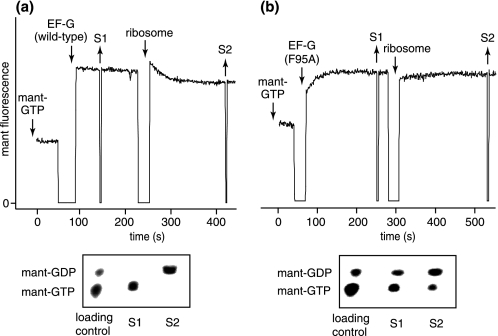
To examine whether Phe-95 might also play a role in normal functions of EF-G, apart from its interactions with fusidic acid, we compared the binding of F95A and wild-type EF-G to GTP in solution in the absence of the ribosome (supplemental Fig. 2). For these experiments, we used the fluorescent substrate mant-GTP. In accord with previous studies (18, 19, 21), when we titrated wild-type EF-G with mant-GTP, the fluorescence rapidly jumped in intensity and remained stable after each addition of mant-GTP, indicative of binding. The dissociation constant (Kd) was 6.6 ± 0.9 μm. However, when we titrated F95A with mant-GTP, the fluorescence gradually increased after each mant-GTP addition, eventually reaching a stable level. The response in the fluorescence became progressively faster as the concentration of mant-GTP increased. Thus, F95A appeared to bind mant-GTP very slowly, but with a similar Kd relative to wild-type EF-G.
We then compared the ability of the ribosome to activate GTP hydrolysis catalyzed by F95A and wild-type EF-G (Fig. 3, a and b, upper panels). Again, we took advantage of mant-GTP, the fluorescence of which is also sensitive to the hydrolysis reaction (18). When wild-type EF-G was added to mant-GTP, the fluorescence rapidly jumped and stabilized. When ribosomes were added, the fluorescence slowly decayed to an intermediate level, which is associated with the hydrolysis reaction triggered by ribosomes (18). However, when we repeated the same experiments using F95A instead, we obtained completely different results. When F95A was added to mant-GTP, the fluorescence gradually increased, eventually reaching a stable level after ∼100 s. When ribosomes were added, the fluorescence did not change significantly.
FIGURE 3.
F95A spontaneously hydrolyzes mant-GTP in solution. a and b, upper panels, fluorescent traces of experiments that monitored mant-GTP interactions with wild-type EF-G and mutant F95A, respectively. Downward arrows indicate the addition of mant-GTP (120 μm), EF-G (25 μm), and E. coli vacant ribosomes (1.4 μm). Upward arrows indicate the removal of samples S1 and S2, which were precipitated with 0.2 m HCl and analyzed by TLC (lower panels below fluorescent traces) (18).
To assess hydrolysis of mant-GTP directly, we removed samples after adding EF-G or ribosomes and resolved mant-GTP from mant-GDP by TLC (Fig. 3, a and b, lower panels). As expected (18), mant-GTP remained intact after wild-type EF-G was added and was completely hydrolyzed after ribosomes were added. Surprisingly, mant-GTP was partially hydrolyzed ∼200 s after F95A was added. Moreover, the addition of ribosomes only slightly stimulated the further hydrolysis of mant-GTP by F95A.
These behaviors of F95A are unusual for EF-G and other translational GTPases because GTP hydrolysis is normally coupled to their functional cycles on and off the ribosome. GTP hydrolysis by EF-G is suppressed in solution until EF-G binds to active ribosomes and catalyzes their translocation. GTP hydrolysis by EF-Tu is coupled to delivery of tRNA substrates to the ribosome. It is activated only upon proper pairing of the tRNA anticodon with its complementary mRNA anticodon in the ribosome. Unlike EF-Tu, GTP hydrolysis by EF-G can be efficiently activated by vacant ribosomes (lacking mRNA and tRNAs) (28), thereby partially decoupling wild-type EF-G (as in the above experiments). Unlike wild-type EF-G, the F95A mutant represents a completely decoupled protein.
FusR EF-G Mutants Are Intrinsically Active in GTP Hydrolysis
These behaviors of F95A stimulated our thoughts about how GTP hydrolysis is activated and coupled to the specific functions of translational GTPases on the ribosome. In EF-G, Phe-95 directly contacts the catalytic histidine residue (His-92), which is required for GTP hydrolysis (29) and conserved in translational GTPases. The imidazole side chain of this histidine is thought to activate a water molecule that attacks the γ-phosphate of GTP (30, 31). In EF-Tu, this histidine interacts with a conserved tyrosine (Tyr-88). However, the precise interactions between the aromatic side chains (of Phe-95 and Tyr-88) and the catalytic imidazole depend on the functional state of each GTPase (8, 11, 27, 30–33). Furthermore, Phe-95 interacts also with domain III of EF-G, which is important for both GTP hydrolysis and ribosome translocation (34), whereas Tyr-88 interacts with tRNA substrates (32).
Thus, we hypothesized that the conserved aromatic residues are key regulators of the functional cycles of translational GTPases. In EF-G, Phe-95 makes differential interactions with other FusR residues clustered in helices A3 and B3 of domain III (8, 11, 27), which suggests functionally important interactions among these residues. To test these ideas, we examined the activities of single alanine mutants of all seven of our selected FusR residues in EF-G. We compared their activities in quantitative assays for GTP hydrolysis and ribosome translocation.
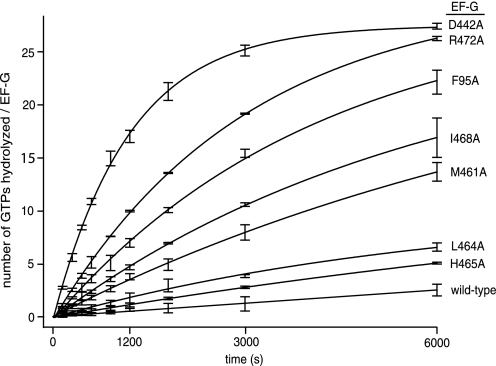
To assess intrinsic GTPase activity, we measured the reaction rates of the FusR mutants in catalyzing the hydrolysis of radioactive [γ-32P]GTP substrate, without the ribosome, under saturating multiple substrate turnover conditions (Fig. 4 and Table 1). F95A hydrolyzed GTP at a rate that was 11-fold faster compared with wild-type EF-G. The other FusR mutants also displayed elevated intrinsic GTPase activities, up to 32-fold faster than wild-type EF-G.
FIGURE 4.
GTP hydrolysis by FusR EF-G mutants in the absence of the ribosome. Reactions were conducted and samples were analyzed as described under “Experimental Procedures.” Error bars are centered on the average y axis values (±S.D.) measured from three separate reactions. The values in Table 1 were obtained from the linear slopes in the graph in the initial phase of the reactions.
TABLE 1.
Biochemical activities of FusR EF-G proteins (derived from wild-type E. coli EF-G)
Values represent the means ± S.D. from three or more separate reactions. ND, not detectable.
| EF-G protein | GTP binding affinity without ribosome (Kd or Km) | GTP hydrolysis rate (k)a |
Ribosome translocation rate (k)b | |
|---|---|---|---|---|
| Without ribosome | With ribosome | |||
| μm | s−1 | s−1 | s−1 | |
| Wild-type | 6.6 ± 0.9c | (5 ± 1) × 10−4 | 4.6 ± 0.4 | 1.8 ± 0.4 |
| F95A | 3.9 ± 0.3d | (56 ± 9) × 10−4 | ND | 0.002 ± 0.001 |
| D442A | 2.4 ± 0.2d | (162 ± 7) × 10−4 | 0.56 ± 0.02 | 0.044 ± 0.002 |
| M461A | 12 ± 1d | (41 ± 4) × 10−4 | 1.18 ± 0.07 | 0.8 ± 0.1 |
| L464A | 14 ± 2c | (15 ± 5) × 10−4 | ND | 0.033 ± 0.007 |
| H465A | 19 ± 2c | (11 ± 3) × 10−4 | 0.36 ± 0.09 | 0.9 ± 0.4 |
| I468A | 13 ± 1d | (29 ± 3) × 10−4 | ND | 0.046 ± 0.003 |
| R472A | 7.3 ± 0.9d | (86 ± 2) × 10−4 | ND | 0.072 ± 0.007 |
a Reactions at 20 °C contained EF-G (0.4 μm), GTP (100 μm), and (when present) vacant E. coli ribosome (0.4 μm). Values represent kcat (GTPs hydrolyzed per EF-G/s) under steady-state conditions at saturating GTP concentration.
b Values are the rate of translocation of fluorescent pyrene-mRNA bound to the ribosome (Fig. 7b and supplemental Fig. 3). EF-G (2.5 μm) and GTP (1 mm) were rapidly mixed with pretranslocational ribosomes (0.25 μm) at 20 °C. Values represent the first-order reaction rate constants under pre-steady-state conditions.
c Kd for mant-GTP (without hydrolysis).
d Km for hydrolysis of mant-GTP.
The most active mutants were D442A, R472A, F95A, M461A, and I468A. Asp-442 and Arg-472 form a salt bridge linking the C termini of helices A3 and B3 (8, 25). Phe-95 interacts alternatively either with Met-461 at the N terminus of helix B3 (27) or with Leu-464 and Ile-468 in helix B3 (8, 25). Thus, the alanine mutations would disrupt these bridges, thereby increasing the internal flexibility of domain III and its dynamic interactions with switch II.
To distinguish between potential effects on GTP binding or hydrolysis, we extended the fluorescence-based mant-GTP hydrolysis experiments to the FusR mutants in domain III. All mutants, except for L464A and H465A, exhibited gradual increases in mant-GTP fluorescence (data not shown), as we had observed for F95A. The rates of fluorescence increase depended on the mant-GTP concentration. They also correlated with the rates of [γ-32P]GTP hydrolysis. From these experiments, we derived binding affinities of the EF-G proteins for mant-GTP and mant-GDP (Table 1). The Michaelis constants (Km) associated with mant-GTP hydrolysis for the FusR mutants were within 3-fold of the wild-type Kd for mant-GTP. Likewise, the Kd values for mant-GDP were within 3-fold of each other (data not shown). Thus, the FusR mutations had relatively minor effects on guanine nucleotide binding to EF-G.
The Ribosome Fails to Stimulate the GTPase Activities of FusR EF-G Mutants
The experiments above suggested that residues of switch II and domain III interact to suppress spontaneous GTP hydrolysis by EF-G in solution. Although the FusR mutants were up to 32-fold more active in intrinsic GTP hydrolysis, the ribosome exerts a much stronger activation of GTP hydrolysis in both EF-G and EF-Tu by several orders of magnitude (35).
Thus, we were curious to examine the effect of the ribosome on GTP hydrolysis by the FusR mutants of EF-G (Table 1). Using stoichiometric amounts of EF-G and vacant ribosomes, we measured the rates of [γ-32P]GTP hydrolysis under the same conditions as described above. Despite its intrinsic GTPase activity, F95A was not detectably stimulated by the ribosome (above the negative control reaction containing the ribosome and lacking EF-G). By contrast, in the positive control reaction, the ribosome stimulated the GTPase activity of wild-type EF-G by a factor of 9000. The ribosome also did not affect the GTPase activities of other FusR mutants in domain III of EF-G. Only one mutant, M461A, was stimulated moderately by the ribosome.
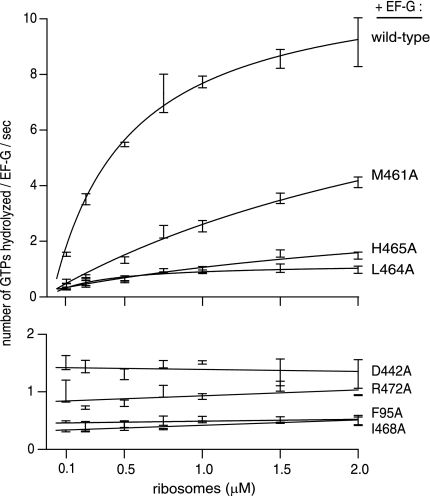
To examine ribosome effects on GTP hydrolysis quantitatively, we also titrated each FusR mutant with increasing concentrations of the ribosome (Fig. 5). The GTPase activity of wild-type EF-G as a function of ribosome concentration displayed a Km of 0.5 μm, in accord with previous studies (18, 19, 28). However, the FusR mutants largely failed to respond to increasing ribosome concentrations, except for M461A. Thus, it was unclear whether the FusR mutants were even binding to ribosomes.
FIGURE 5.
Effect of the ribosome on GTP hydrolysis by FusR mutants of EF-G. EF-G proteins were titrated with ribosomes, and the rates of GTP hydrolysis (v) were measured. Reactions conditions were as follows: [EF-G] = 0.4 μm, [GTP] = 100 μm (+trace [γ-32P]GTP), and [ribosome] = 0.06–2 μm at 20 °C. Smooth curves (upper four EF-G proteins) are fits of the data to the equation v = [EF-G] × [ribosome] × kcat/([ribosome] + Km). Error bars are the means ± S.D., each derived from three or more independent reactions.
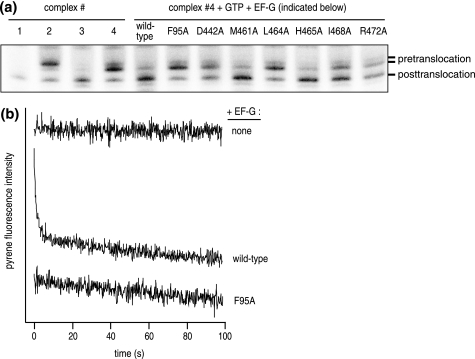
To examine ribosome binding more directly, we monitored the reaction of DMS with ribosomal nucleotide A2660, located at the tip of the sarcin-ricin loop (SRL; helix 95) of 23 S rRNA. When bound to the ribosome, EF-G specifically blocks methylation of A2660 by DMS, which can be quantitatively monitored by primer extension analysis of rRNA (22). We bound the FusR mutants of EF-G to the ribosome in the presence of a non-hydrolyzable substrate analog GDPNP. We chose this particular complex for several reasons: the FusR mutants hydrolyzed GTP to variable extents; we could not obviously use fusidic acid to stabilize the complex; EF-G shields A2660 both before and after GTP hydrolysis on the ribosome (22); and A2660 does not interact with the GTPase active site (11, 31).
The results are shown in Fig. 6. At stoichiometric (1 μm) amounts of EF-G and ribosomes, the FusR mutants all protected A2660 from DMS. The degree of protection was more or less similar to that obtained with the wild-type EF-G control, with the exception of two mutants, F95A and D442A (0.66 and 0.70, respectively, relative to wild-type EF-G). These results indicated that all FusR mutants could specifically and stably bind to the ribosome, although F95A and D442A were bound only partially to the ribosome under these conditions. Thus, the failure of the ribosome to further activate GTP hydrolysis of many of the FusR mutants was most likely not caused by their failure to form a proper pretranslocational ribosome complex, poised for GTP hydrolysis and ribosome translocation.
FIGURE 6.
Binding of FusR EF-G mutants to the ribosome. Binding was assessed by protection of nucleotide A2660 of 23 S rRNA from chemical modification by DMS. EF-G (1 μm) was bound to pretranslocational ribosomes (1 μm) with GDPNP (1 mm). The resulting complexes were probed with DMS (33 mm). Ribosomal RNAs were extracted from the ribosome and analyzed by primer extension (22). The gel band intensity of A2660 indicates the degree of DMS modification. A and G, sequencing lanes.
Phe-95 in Switch II of EF-G Is Essential for Catalyzing Ribosome Translocation along mRNA
For GTP hydrolysis to be used productively, it must be coupled to ribosome translocation, the principal function of EF-G during protein synthesis. The lower fitness of FusR bacteria might arise from excessive GTP hydrolysis by EF-G off the ribosome or from poor coupling of GTP hydrolysis and translocation on the ribosome.
We evaluated the activities of the FusR mutants to catalyze ribosome translocation using two complementary assays. First, toeprinting experiments allowed us to measure the extent and accuracy of a single round of ribosome translocation by tracking the movement along a model mRNA by one codon (Fig. 7a). In these experiments, F95A had no detected translocation activity compared with the pretranslocational ribosome control, whereas wild-type EF-G catalyzed efficient ribosome translocation. The other FusR mutants catalyzed partial ribosome translocation, less efficiently than wild-type EF-G but with similar accuracy.
FIGURE 7.
Ribosome translocation catalyzed by FusR EF-G mutants. a, toeprinting experiment. Complexes are as follows: 1, mRNA only (control); 2, ribosome + mRNA + tRNAfMet (P site); 3, ribosome + mRNA + tRNAPhe (P site); 4 pretranslocational ribosome), ribosome + mRNA + tRNAfMet (P site) + tRNAPhe (A site). EF-G proteins (1.2 μm) + GTP (2 mm) were added to pretranslocational ribosomes (1.0 μm). The ribosome position on the mRNA was assessed by primer extension (toeprinting) and analyzed by polyacrylamide gel electrophoresis (17). Gel bands corresponding to the ribosome before and after translocation are marked on the right. b, fluorescent mRNA experiment (18). Complex 4 (0.5 μm) was prepared with 3′-pyrene-mRNA and rapidly mixed with an equal volume of EF-G (5 μm) + GTP (2 mm) in a stopped-flow device at 20 °C. The pyrene fluorescence was monitored over time at 20 °C. The time-dependent decay in pyrene fluorescence corresponds to ribosome translocation (18). Shown here are representative traces for reactions containing wild-type EF-G or F95A. For clarity, traces are offset along the y axis. Traces for other FusR mutants are provided in supplemental Fig. 3.
Second, fluorescent pyrene-mRNA allowed us to measure the kinetics of a single round of ribosome translocation (Fig. 7b and Table 1). Consistent with the toeprinting results, M461A and H465A catalyzed rapid ribosome translocation (2-fold slower than wild-type EF-G). However, the translocation reactions catalyzed by the other FusR mutants were much slower than the wild-type reaction (from 24- to 51-fold) (supplemental Fig. 3). Most notably, translocation by F95A occurred extremely slowly, if at all (∼450-fold slower than wild-type EF-G and virtually the same as the negative control after 100 s) (Fig. 7b).
Thus, these results revealed that the major defect of the FusR mutants involves the productive coupling of GTP hydrolysis to translocation on the ribosome. This defect is particularly severe in F95A.
DISCUSSION
This study provides the first biochemical characterizations of conserved residues near the GTP hydrolytic site of EF-G that earlier genetic studies had identified as commonly mutated in bacteria that acquire strong resistance to fusidic acid (3–8). Although the mutability of these FusR residues suggests their dispensability, their phylogenetic conservation, their strategic locations in EF-G, and the reduced biological fitness of FusR bacteria all suggest otherwise.
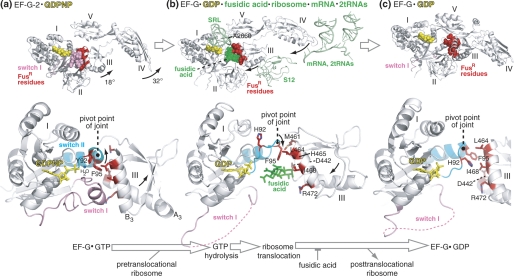
To illustrate their functional significance and to further interpret our results, we highlighted the corresponding FusR residues in crystal structures of EF-G proteins in three functional states (Fig. 8). These structures are EF-G-2·GDPNP (25), ribosome-bound EF-G·GDP·fusidic acid (11), and various versions of EF-G·GDP (8, 27). The FusR residues cluster at the interface of domains I and III (8), where they contact fusidic acid, whereas EF-G is on the ribosome (11). The same residues form a hydrophobic core that stabilizes three neighboring elements (switches I and II of domain I and helix B3 of domain III) in EF-G-2 off the ribosome (25).
FIGURE 8.
Mechanism for how conserved FusR residues at the interface between domains I and III of EF-G control GTP hydrolysis, ribosome translocation, and EF-G release from the ribosome. This figure highlights the FusR residues in the context of three previously published crystal structures of EF-G proteins in different functional states. a, EF-G-2·GDPNP (25); b, ribosome-bound EF-G·GDP·fusidic acid (11); c, FusR EF-G(T84A)·GDP (27). Shown in the upper row are the overviews of the three EF-G structures, along with selected ribosomal components and fusidic acid (highlighted in green in b). The middle row shows a close-up view of domains I and III of EF-G of the three structures, with the side chains of the FusR residues highlighted in red. The lower row shows the reaction scheme of the EF-G catalytic cycle represented by the three EF-G structures separated by large arrows. The small arrows in the EF-G structures indicate the directions of EF-G domain movements during GTP hydrolysis and ribosome translocation. The pivot point of the joint between switch II and domain III is also indicated in each of the three structures. The structural schematics are based on Protein Data Bank codes 1WDT, 2WRI, 2WRJ, and 2BM0. All three EF-G structures are from T. thermophilus. Residue numbers refer to E. coli.
We have found that the same FusR residues also regulate fundamental activities of EF-G as it cycles on and off the ribosome during normal protein synthesis. These residues regulate GTP hydrolysis by maintaining EF-G·GTP in an inactive state off the ribosome. They mediate the ability of the ribosome to strongly activate the latent GTPase activity of EF-G. They couple GTP hydrolysis to translocation of the ribosome on mRNA.
The data pertaining to Phe-95 are the most striking. The FusR mutant F95A is 10-fold more active in intrinsic GTP hydrolysis and 450-fold less active in ribosome translocation relative to wild-type E. coli EF-G. These functional data complement earlier structural work that compared EF-G mutants that are FusR or fusidic acid-hypersensitive (27). Positioned at the apex of switch II, the side chain of Phe-95 rotates widely in the cavity formed by domains I, III, and V in FusR and fusidic acid-hypersensitive mutant EF-G structures. Through its movements, Phe-95 interacts dynamically with other FusR residues in helix B3 and with residues in switch I (25).
We can unify the structural and functional data on the FusR residues into a mechanism of the catalytic cycle of EF-G (Fig. 8). At the crux of this mechanism is an interdomain joint between domains I and III. Rotational motions centered on this joint regulate GTP hydrolysis, ribosome translocation, and EF-G release from the ribosome. Below, we present the arguments for this mechanism, with new insights into normal activities of EF-G and its inhibition by fusidic acid.
Regulation of GTP Hydrolysis
Phe-95 is located three residues away from His-92, the key catalytic residue for GTP hydrolysis. In T. thermophilus EF-G-2, a tyrosine residue (Tyr-92) replaces the highly conserved His-92 in translational GTPases. Tyr-92 is oriented toward the nucleophilic water in the active site. EF-G-2 exhibits unusually high intrinsic GTPase activity in the absence of the ribosome (25). In T. thermophilus EF-G·GDP structures, His-92 is oriented away from the active site. Simultaneously, Phe-95 rotates around the apex of switch II and changes its multiple contacts with switch I, other FusR residues in helix B3 of domain III, and His-92 of switch II.
An earlier study excised the entire domain III from T. thermophilus EF-G and found that the resulting protein was defective in ribosome-activated GTP hydrolysis and ribosome translocation (34). Despite these major defects, the protein still maintained interactions with guanine nucleotides and the ribosome. This study provided the first indications of the functional importance of domain III of EF-G.
On the basis of our results and recent crystal structures, we suggest that activation of GTP hydrolysis requires rotational motion at the apex of switch II, the pivot point of the interdomain joint in EF-G. Hydrophobic interactions between Phe-95 and other FusR residues in domain III preclude GTP hydrolysis in EF-G in solution. Alanine mutations disrupt these interactions, thereby subluxating the joint and making EF-G too flexible.
Clearly, the ribosome strongly activates GTP hydrolysis and may contribute additional catalytic residues. The ribosomal element that comes closest to the GTPase active site is the SRL of 23 S rRNA in the large ribosomal subunit. In crystal structures of free EF-G proteins, the density of the side chain of His-92 is usually weak, suggesting its mobility. On the ribosome, the tip of the SRL wedges between switch II and domain III, suggesting that the SRL may promote the rotational rearrangements at the interdomain joint. In addition, His-92 becomes more ordered. Finally, a recent structure shows that the backbone of the SRL orients His-92 toward GTP bound to EF-Tu, which may also hold true for EF-G (31).
Regulation of Ribosome Translocation
GTP hydrolysis accelerates the subsequent steps of ribosome translocation (24). This process entails several large-scale movements in EF-G and the ribosome (Fig. 8). In EF-G, switch I becomes disordered and flips out from the pocket between domains I and III of EF-G (19, 26). Domains III–V of EF-G undergo large rotational motions, which become progressively amplified from switch II to the tip of domain IV (11, 25, 36). In the ribosome, the 30 S and 50 S subunits rotate relative to each other, and the head of the 30 S subunit also rotates, which ultimately moves the mRNA and two tRNAs in the ribosome (37).
Once again, the FusR residues of EF-G play a major role in regulating ribosome translocation. The rotation of Phe-95 around the apex of switch II drives rotation of domain III, which in turn drives rotation of domains IV and V. Other FusR residues in helix B3 interact with Phe-95 and switch I.
Our results show that Phe-95 is critical for ribosome translocation coupled to GTP hydrolysis. The F95A mutant was unresponsive to activation by the ribosome of GTP hydrolysis and was severely defective in catalyzing ribosome translocation. In conjunction with crystal structures, these results suggest a mechanical coupling between the rotations of Phe-95 and helix B3, which become amplified progressively from the pivot point of the interdomain joint.
Other FusR mutants had corresponding defects in ribosome-activated GTP hydrolysis and translocation (D442A, L464A, I468A, and R472A). Phe-95 makes initial contacts with Leu-464 and Ile-468 before GTP hydrolysis and then rotates around switch II to make contact with Met-461. Arg-472 interacts with switch I before GTP hydrolysis and then forms a salt bridge with Asp-442. Thus, the coupling of GTP hydrolysis to translocation appears to involve all three elements (switches I and II and helix B3) working in dynamic harmony.
Fusidic Acid Sensitivity and Resistance
After switch I flips out from the interdomain pocket, fusidic acid takes its place (Fig. 8). Phe-95 directly interacts with fusidic acid, which effectively traps switch II between its GTP and GDP conformations (11). Fusidic acid also acts like a wedge that blocks the retraction of domains III–V and thereby traps EF-G·GDP on the ribosome (26).
Because FusR residues were identified based on their resistance to fusidic acid, not surprisingly, the strongest FusR mutants involve residues of EF-G that make direct contact with fusidic acid (11). Because FusR residues are also critical for normal functions of EF-G in the absence of fusidic acid (this work), they are usually mutated to residues other than alanine in FusR bacteria. The most prevalent FusR bacterial mutants are F95L, D442N, M461I, L464F, H465Y/Q, and R472L/C/S/H (3–8). These relatively conservative mutations imply that they achieve a compromise, allowing bacterial growth by disfavoring the binding of fusidic acid while retaining sufficient EF-G functions to maintain protein synthesis for bacterial growth.
Toward New Antibiotics Targeting GTPases
The unusual intrinsic GTPase activities of FusR mutants of EF-G are reminiscent of the effects of kirromycin-like antibiotics on EF-Tu. Like fusidic acid, kirromycin traps EF-Tu·GDP on the ribosome. Unlike fusidic acid, kirromycin can bind to free EF-Tu and can induce its hydrolysis of GTP in solution, without the ribosome (38). In a crystal structure of EF-Tu·GDP·kirromycin, its switch I is disordered, and the catalytic histidine in switch II is oriented toward the nucleophilic water (39). Both the ribosome and kirromycin change the conformation of switch II, which may activate GTP hydrolysis (33).
These parallels suggest that the FusR mutants of EF-G may mimic the effects of fusidic acid on ribosome-bound EF-G by freezing an activated transition state conformation of EF-G. In one such structure of FusR EF-G(T84A)·GDPNP, the catalytic histidine is indeed oriented toward the γ-phosphate of GDPNP (40). Moreover, this is a common theoretical principle of action of many potent enzyme inhibitors, including antibiotics (1).
If so, by screening small molecule libraries, we might find other activators of the intrinsic GTPase activities of EF-G and EF-Tu. Positive hits might prove useful as probes of the physiological roles of these (and other) GTPases and possibly also as new antibiotic inhibitors of bacterial translation and cellular growth.
Supplementary Material
Acknowledgments
We thank Stephen Rader and Diane Taylor for comments on the manuscript.
This work was supported by the Canadian Institutes of Health Research.

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Experimental Procedures,” Figs. 1–4, and additional references.
- EF
- elongation factor
- FusR
- fusidic acid-resistant
- mant
- 2′,3′-O,N′-methylanthraniloyl
- DMS
- dimethyl sulfate
- SRL
- sarcin-ricin loop
- GDPNP
- guanosine 5′-(β,γ-imido)triphosphate.
REFERENCES
- 1. Fischbach M. A., Walsh C. T. (2009) Science 325, 1089–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Turnidge J. (1999) Int. J. Antimicrob. Agents 12, S23–S34 [DOI] [PubMed] [Google Scholar]
- 3. Johanson U., Hughes D. (1994) Gene 143, 55–59 [DOI] [PubMed] [Google Scholar]
- 4. Nagaev I., Björkman J., Andersson D. I., Hughes D. (2001) Mol. Microbiol. 40, 433–439 [DOI] [PubMed] [Google Scholar]
- 5. Besier S., Ludwig A., Brade V., Wichelhaus T. A. (2003) Mol. Microbiol. 47, 463–469 [DOI] [PubMed] [Google Scholar]
- 6. Lannergård J., Norström T., Hughes D. (2009) Antimicrob. Agents Chemother. 53, 2059–2065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Castanheira M., Watters A. A., Bell J. M., Turnidge J. D., Jones R. N. (2010) Antimicrob. Agents Chemother. 54, 3614–3617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Laurberg M., Kristensen O., Martemyanov K., Gudkov A. T., Nagaev I., Hughes D., Liljas A. (2000) J. Mol. Biol. 303, 593–603 [DOI] [PubMed] [Google Scholar]
- 9. Chen Y., Koripella R. K., Sanyal S., Selmer M. (2010) FEBS J. 277, 3789–3803 [DOI] [PubMed] [Google Scholar]
- 10. Willie G. R., Richman N., Godtfredsen W. P., Bodley J. W. (1975) Biochemistry 14, 1713–1718 [DOI] [PubMed] [Google Scholar]
- 11. Gao Y. G., Selmer M., Dunham C. M., Weixlbaumer A., Kelley A. C., Ramakrishnan V. (2009) Science 326, 694–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Andersson D. I., Hughes D. (2010) Nat. Rev. Microbiol. 8, 260–271 [DOI] [PubMed] [Google Scholar]
- 13. Macvanin M., Johanson U., Ehrenberg M., Hughes D. (2000) Mol. Microbiol. 37, 98–107 [DOI] [PubMed] [Google Scholar]
- 14. Johanson U., Aevarsson A., Liljas A., Hughes D. (1996) J. Mol. Biol. 258, 420–432 [DOI] [PubMed] [Google Scholar]
- 15. Björkman J., Nagaev I., Berg O. G., Hughes D., Andersson D. I. (2000) Science 287, 1479–1482 [DOI] [PubMed] [Google Scholar]
- 16. Martemyanov K. A., Liljas A., Yarunin A. S., Gudkov A. T. (2001) J. Biol. Chem. 276, 28774–28778 [DOI] [PubMed] [Google Scholar]
- 17. Wilson K. S., Nechifor R. (2004) J. Mol. Biol. 337, 15–30 [DOI] [PubMed] [Google Scholar]
- 18. Nechifor R., Murataliev M., Wilson K. S. (2007) J. Biol. Chem. 282, 36998–37005 [DOI] [PubMed] [Google Scholar]
- 19. Nguyen B., Ticu C., Wilson K. S. (2010) J. Mol. Biol. 397, 1245–1260 [DOI] [PubMed] [Google Scholar]
- 20. Jelenc P. C., Kurland C. G. (1979) Proc. Natl. Acad. Sci. U.S.A. 76, 3174–3178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wilden B., Savelsbergh A., Rodnina M. V., Wintermeyer W. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 13670–13675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Moazed D., Robertson J. M., Noller H. F. (1988) Nature 334, 362–364 [DOI] [PubMed] [Google Scholar]
- 23. Vetter I. R., Wittinghofer A. (2001) Science 294, 1299–1304 [DOI] [PubMed] [Google Scholar]
- 24. Rodnina M. V., Savelsbergh A., Katunin V. I., Wintermeyer W. (1997) Nature 385, 37–41 [DOI] [PubMed] [Google Scholar]
- 25. Connell S. R., Takemoto C., Wilson D. N., Wang H., Murayama K., Terada T., Shirouzu M., Rost M., Schüler M., Giesebrecht J., Dabrowski M., Mielke T., Fucini P., Yokoyama S., Spahn C. M. (2007) Mol. Cell 25, 751–764 [DOI] [PubMed] [Google Scholar]
- 26. Ticu C., Nechifor R., Nguyen B., Desrosiers M., Wilson K. S. (2009) EMBO J. 28, 2053–2065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hansson S., Singh R., Gudkov A. T., Liljas A., Logan D. T. (2005) J. Mol. Biol. 348, 939–949 [DOI] [PubMed] [Google Scholar]
- 28. Rohrback M. S., Bodley J. W. (1976) Biochemistry 15, 4565–4569 [DOI] [PubMed] [Google Scholar]
- 29. Daviter T., Wieden H. J., Rodnina M. V. (2003) J. Mol. Biol. 332, 689–699 [DOI] [PubMed] [Google Scholar]
- 30. Berchtold H., Reshetnikova L., Reiser C. O., Schirmer N. K., Sprinzl M., Hilgenfeld R. (1993) Nature 365, 126–132 [DOI] [PubMed] [Google Scholar]
- 31. Voorhees R. M., Schmeing T. M., Kelley A. C., Ramakrishnan V. (2010) Science 330, 835–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nissen P., Kjeldgaard M., Thirup S., Polekhina G., Reshetnikova L., Clark B. F., Nyborg J. (1995) Science 270, 1464–1472 [DOI] [PubMed] [Google Scholar]
- 33. Schmeing T. M., Voorhees R. M., Kelley A. C., Gao Y. G., Murphy F. V., 4th, Weir J. R., Ramakrishnan V. (2009) Science 326, 688–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Martemyanov K. A., Gudkov A. T. (2000) J. Biol. Chem. 275, 35820–35824 [DOI] [PubMed] [Google Scholar]
- 35. Mohr D., Wintermeyer W., Rodnina M. V. (2002) Biochemistry 41, 12520–12528 [DOI] [PubMed] [Google Scholar]
- 36. Valle M., Zavialov A., Sengupta J., Rawat U., Ehrenberg M., Frank J. (2003) Cell 114, 123–134 [DOI] [PubMed] [Google Scholar]
- 37. Frank J., Gao H., Sengupta J., Gao N., Taylor D. J. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 19671–19678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wolf H., Chinali G., Parmeggiani A. (1974) Proc. Natl. Acad. Sci. U.S.A. 71, 4910–4914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Vogeley L., Palm G. J., Mesters J. R., Hilgenfeld R. (2001) J. Biol. Chem. 276, 17149–17155 [DOI] [PubMed] [Google Scholar]
- 40. Hansson S., Singh R., Gudkov A. T., Liljas A., Logan D. T. (2005) FEBS Lett. 579, 4492–4497 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.