Abstract
NAD is a vital redox carrier, and its degradation is a key element of important regulatory pathways. NAD-mediated functions are compartmentalized and have to be fueled by specific biosynthetic routes. However, little is known about the different pathways, their subcellular distribution, and regulation in human cells. In particular, the route(s) to generate mitochondrial NAD, the largest subcellular pool, is still unknown. To visualize organellar NAD changes in cells, we targeted poly(ADP-ribose) polymerase activity into the mitochondrial matrix. This activity synthesized immunodetectable poly(ADP-ribose) depending on mitochondrial NAD availability. Based on this novel detector system, detailed subcellular enzyme localizations, and pharmacological inhibitors, we identified extracellular NAD precursors, their cytosolic conversions, and the pathway of mitochondrial NAD generation. Our results demonstrate that, besides nicotinamide and nicotinic acid, only the corresponding nucleosides readily enter the cells. Nucleotides (e.g. NAD and NMN) undergo extracellular degradation resulting in the formation of permeable precursors. These precursors can all be converted to cytosolic and mitochondrial NAD. For mitochondrial NAD synthesis, precursors are converted to NMN in the cytosol. When taken up into the organelles, NMN (together with ATP) serves as substrate of NMNAT3 to form NAD. NMNAT3 was conclusively localized to the mitochondrial matrix and is the only known enzyme of NAD synthesis residing within these organelles. We thus present a comprehensive dissection of mammalian NAD biosynthesis, the groundwork to understand regulation of NAD-mediated processes, and the organismal homeostasis of this fundamental molecule.
Keywords: ADP-ribosylation, Cell Compartmentation, Cell Metabolism, Enzymes, Metabolism, Mitochondria, NAD, Nicotinamide, Nicotinic Acid, Nucleoside Nucleotide Biosynthesis
Introduction
NAD is an essential electron carrier and a key molecule of signaling pathways (1–4). In bioenergetic pathways, NAD is reversibly converted between its oxidized (NAD+) and reduced (NADH) states, which would not require continuous regeneration. Indeed, when the principal pathway of NAD+ synthesis from nicotinic acid (NA)2 had been established (5), the “case” was nearly closed, because neither additional roles of NAD nor a regulatory importance of its synthesis were suspected.
Meanwhile, discoveries of signaling processes in which NAD+ is degraded have dramatically changed this view. Signaling conversions of NAD+ include the cleavage to nicotinamide (Nam), which is recycled into NAD+ synthesis, and a concomitant reaction of the remaining ADP-ribose moiety. NAD+-dependent deacetylases (members of the sirtuin family) and mono-ADP-ribosyltransferases control life span, the biological clock, insulin secretion, and key metabolic enzymes (6–9). In addition, NAD+ represents the substrate for poly(ADP-ribosylation) to regulate DNA repair, transcription, telomerase activity, and chromatin dynamics (10–12). NAD+ is also the precursor of cyclic ADP-ribose and NAADP, potent agents to mobilize calcium from intracellular stores (13). This multitude of NAD+-degrading reactions clearly necessitates permanent regeneration of the nucleotide.
NAD+ biosynthesis itself likely holds a key position in the control of metabolism as it can provide the nucleotide in a regulated fashion with regard to both time, localization and nutrient availability. Indeed, NAD+ biosynthetic activity and NAD+ contents in liver vary depending on the circadian rhythm (8, 9). Moreover, at least in the nucleus and cytosol, NAD+ availability appears to be limited (14) and specific spatial distribution of enzymes may selectively supply NAD+-dependent processes, as has been suggested for poly(ADP-ribosylation) in the nucleus (15) and protection of nerve ends from degeneration (16).
In addition to the conversions of the vitamin precursors Nam and NA to NAD+ in yeast (2, 5), Brenner and co-workers (17) recently discovered the nucleoside derivatives of these precursors (the ribosides of Nam and NA, NR, and NAR) as precursors that are taken up and synthesized in yeast (18–20). These ribosides enter NAD+ biosynthesis via nicotinamide riboside kinases (NRKs) (Fig. 1A), which are also present in humans (17). Tryptophan can also serve as a NAD+ precursor when degraded to quinolinic acid in the kynurenine pathway (Fig. 1A). However, tryptophan alone is insufficient to support physiological NAD+ concentrations in mammals (21, 22). Thus, there are five different entry points into NAD+ biosynthesis in human cells as follows: NA, Nam, NR, NAR, and tryptophan (Fig. 1A). The biosynthetic pathways from all these precursors merge at the step of the dinucleotide formation catalyzed by NMN adenylyltransferases (NMNATs). In mammals, there are three isoforms of this activity (NMNAT1–3) that have been associated with the nucleus, the Golgi complex, and the mitochondria, respectively (23). Still, although the molecular identities of the human enzymes involved in NAD+ synthesis have been established, information regarding their subcellular distribution and regulation is scarce or inconclusive.
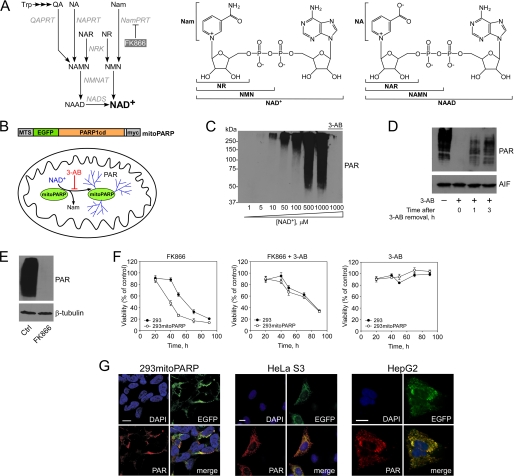
FIGURE 1.
Targeted expression of the catalytic domain of PARP1 as sensor for mitochondrial NAD+ levels. A, overview of NAD+ biosynthesis in humans. NAD+ can be synthesized from pyridine bases or nucleosides, which are initially converted to mononucleotides. The tryptophan (Trp) degradation product quinolinic acid (QA) is transformed to nicotinic acid mononucleotide (NAMN) by quinolinic acid phosphoribosyltransferase (QAPRT). Nicotinamide (Nam) and nicotinic acid (NA) are converted to the corresponding mononucleotides (NMN and NAMN) by nicotinamide phosphoribosyltransferase (NamPRT) and nicotinic acid phosphoribosyltransferase (NAPRT), respectively. FK866 inhibits NamPRT. NMN and NAMN are also generated through phosphorylation of nicotinamide riboside (NR) and nicotinic acid riboside (NAR), respectively, by nicotinamide riboside kinase (NRK) activity. NAMN and NMN are converted to the corresponding dinucleotide (NAAD or NAD+) by NMN adenylyltransferase (NMNAT). NAD synthetase (NADS) amidates NAAD to NAD+. The two right panels indicate the structures of NAD+ and its metabolites. B, experimental system to monitor mitochondrial NAD+ levels. mitoPARP automodifies itself, and its activity is inhibited by 3-AB. Generation and functional verification of the mitoPARP construct have been described (36). MTS, mitochondrial targeting sequence. C, PAR accumulation depends on the NAD+ concentration. The Western blot shows automodification of purified, recombinant PARP1cd (overexpressed in E. coli) after incubation with the indicated concentrations of NAD+. The smear reflects the heterogeneity of polymer lengths. D, 293mitoPARP cells constitutively contain PAR in mitochondria (left lane). Incubation with 3-AB for 24 h diminishes the PAR signal (2nd lane). Following washout of 3-AB, polymers accumulate over time. E, decrease of mitochondrial NAD+ by NamPRT inhibition. Relative NAD+ content in mitochondria was detected in lysates of 293mitoPARP cells treated or not (Ctrl) with FK866 for 24 h. F, mitoPARP activity increases the sensitivity toward NamPRT inhibition. Parental 293 and 293mitoPARP cells were treated with FK866 or 3-AB as indicated. G, mitoPARP (detected by its enhanced GFP(EGFP) portion) leads to mitochondrial PAR accumulation when stably (293mitoPARP) or transiently (HeLa S3 and HepG2) expressed. Bar, 10 μm.
Notably, the pathway of mitochondrial NAD+ generation in mammalian cells has remained an unresolved issue of fundamental importance. In mitochondria of tissues with high energy demand, up to 70% of cellular NAD+ are segregated within these organelles, presumably to provide maximal capacity for oxidative phosphorylation (24). Recent observations have reiterated the significance of mitochondrial NAD+ by demonstrating its critical role in signaling processes. Mitochondrial NAD+-dependent protein deacetylation and mono-ADP-ribosylation regulate key metabolic enzymes, including glutamate dehydrogenase (25, 26), acetyl-CoA synthetase 2 (27), and carbamoyl-phosphate synthetase 1 (28). The mitochondrial NAD+ pool is autonomous (29) and appears to be most critical for cell survival (30). In yeast and plants, cytosolic NAD+ is imported across the inner mitochondrial membrane (31, 32), whereas mammalian mitochondria need to take up a precursor from the cytosol, which can be converted into NAD+ within the organelles. So far, the precursor has not been identified.
NMNAT3 could be involved in mitochondrial NAD+ generation, as it has been associated with the organelles (23). It was also reported that rat Nam phosphoribosyltransferase (NamPRT) partially associated with mitochondria (30). The presence of both NamPRT and NMNAT activities in mitochondria would constitute a viable NAD+ synthetic route from Nam as precursor (cf. Fig. 1A). However, the presence of NamPRT in mitochondria was not confirmed in other studies (28, 29). Consequently, a conclusive assessment of the presence of NamPRT and NMNAT3 in mitochondria is required to understand NAD+ generation within these organelles.
Besides mitochondrial NAD+ synthesis, several general questions regarding this fundamental cellular process still remain unanswered. (i) Which extracellular NAD+ metabolites enter the cells, besides the vitamin B3 precursors Nam and NA? (ii) Which cytosolic conversions are required to supply subcellular NAD+ pools, in particular mitochondria? (iii) What are the rate-limiting steps in the different pathways? (iv) Is there an exchange of NAD+ metabolites across the borders of cells or do tissues rely exclusively on nutritional precursors?
A major challenge to address these questions has been the reliable detection of NAD+, a small molecule that is dynamically converted and distributed among different subcellular pools. So far, all methods to determine endogenous NAD+ contents require cell disintegration followed by nucleotide extraction, and it can only be assumed that organellar concentrations are not affected by these procedures.
In this study, we employed the catalytic domain of poly(ADP-ribose) polymerase-1 (PARP1cd) and targeted it to the mitochondrial matrix. The enzymatic activity of PARP1cd converts NAD+ into immunodetectable, protein-bound poly(ADP-ribose) (PAR) (10–12) and thereby provides a molecular tool for NAD+ detection. We show that the amount of polymers correlates with NAD+ availability and reports changes of mitochondrial NAD+ content brought about by modulation of NAD+ biosynthesis. Based on this detector system and the determination of the subcellular distribution of all known enzymes involved in human NAD+ synthesis, we identified the extracellular precursors of mitochondrial NAD+, their uptake into cells, and intracellular conversions. Collectively, our observations constitute the first comprehensive dissection of NAD+ metabolism in human cells.
EXPERIMENTAL PROCEDURES
Chemicals, Reagents, and Media
Unless otherwise specified, all chemicals and reagents were of analytical grade and were purchased from Sigma and Merck. Cell culture reagents were from Biochrom, Cambrex Corp., Nunc, or Invitrogen. FK866 was obtained from the NIMH Chemical Synthesis and Drug Supply Program. The following antibodies were used: mouse anti-FLAG and mouse anti-β-tubulin (Sigma), rabbit anti-PAR (96−10−04; Alexis Biochemicals), and rabbit anti-apoptosis-inducing factor (Santa Cruz Biotechnology). Mouse anti-Myc (9E10) and mouse anti-PAR (10H) antibodies were from hybridoma cell culture supernatants. Fluorescent-conjugated secondary antibodies were from Invitrogen. Enhanced chemiluminescence (ECL) reagents and HRP-conjugated goat anti-mouse/goat anti-rabbit antibodies were from Pierce or GE Healthcare. DNA-modifying and restriction enzymes were purchased from Fermentas, New England Biolabs, or TaKaRa, and oligonucleotide synthesis was done by Sigma.
Cell Culture
Cells were cultivated in Ham's F-12 medium for HeLa S3 and Dulbecco's modified Eagle's medium (DMEM) for 293 and HepG2 cells. Media were supplemented with 10% (v/v) FCS, 2 mm l-glutamine, and penicillin/streptomycin. Stably transfected 293mitoPARP cells were maintained in 293 medium supplemented with 100 μg/ml G418. Transient transfection of eukaryotic cells was performed using Effectene reagent (Qiagen).
Cloning and Generation of Eukaryotic Expression Vectors
For expression of tagged proteins, their corresponding open reading frames were inserted into pFLAG-CMV-5a (Sigma), pcDNA3.1(+)-PARP1cd (33), or pCMV/myc/mito (Invitrogen). All cloned DNA sequences were verified by DNA sequence analysis.
Pharmacological Treatments
Inhibitors (2 μm FK866, 2 mm 3-AB, 10 μm NBTI, 2 μm dipyridamole, 2 mm CMP, 25 μm pyridoxyl phosphate-6-azophenyl-2′,4′-disulfonic acid, and 1 mm Ap4A) or metabolites (100 μm NAD+, 100 μm NAAD, 100 μm NMN, 100 μm NAMN, 100 μm NA, and 100 μm NR) were added to the cell culture medium as indicated. NR was generated as described previously (34). Treatment with inhibitors and metabolites was carried out for 24–48 h prior to analyses of PAR formation and cell viability (72 h for 293mitoPARP cells, 96 h for HeLa S3 cells, and 120 h for HepG2 cells). Cell viability was determined using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay.
Identification of Mitochondrial Matrix Proteins by PARAPLAY
The poly(ADP-ribose)-assisted protein localization assay (PARAPLAY) establishes luminal protein localization (33). Here, PARAPLAY was used to identify mitochondrial matrix proteins. The analyte proteins (NMNAT3, NamPRT, and NAPRT) were expressed N-terminally fused to PARP1cd in HeLa S3 cells. Because of low [NAD+] and/or high PAR degrading activity in the cytosol and mitochondrial intermembrane space, no PAR was detected if the fusion protein resides within these compartments. In contrast, matrix localization was readily established by PAR accumulation. Even if the majority of the fusion protein resided in the cytosol, a possible luminal fraction of the protein would generate sufficient immunodetectable PAR. If no PAR was detected, the localization of the protein outside the matrix was verified by testing the functionality of the PARP1cd portion of the fusion protein. This was done by adding an N-terminal mitochondrial targeting sequence to the fusion protein, which will direct it into the matrix and result in PAR accumulation.
Protein Determination, SDS-PAGE, and Western Blotting
Protein concentration was determined using BCA reagent (Pierce). Cell lysates were prepared in lysis buffer (50 mm Tris/HCl, pH 6.8, 2% (w/v) SDS, 10% (v/v) glycerol) or directly in SDS sample buffer (50 mm Tris/HCl, pH 6.8, 2% (w/v) SDS, 10% (v/v) glycerol, 100 mm β-mercaptoethanol, and 0.01% (w/v) bromphenol blue). Gel electrophoresis and immunoblotting were carried out according to standard procedures. ECL was used for immunodetection. Equal protein loading was confirmed by β-tubulin or AIF immunodetection.
Immunocytochemistry
Cells were fixed with 4% (v/v) formaldehyde in PBS and permeabilized using 0.5% (v/v) Triton X-100 in PBS. Nuclei were stained with DAPI and mitochondria with MitoTracker Red CMXRos (Invitrogen). Images were taken using a Leica DMI6000B epifluorescence microscope (Leica Microsystems) equipped with ×10, ×40, and ×100 objectives.
Detection of Changes in NAD+ Content by PARP Activity within Mitochondria
293mitoPARP cells express a fusion protein (“mitoPARP”) consisting of enhanced GFP and PARP1cd targeted to the mitochondrial matrix. Using mitochondrial NAD+ as substrate, these cells constitutively generate protein-bound PAR, which is visualized immunochemically. Variations in the extent of detected PAR thus reflect changes of the mitochondrial NAD+ content. Likewise, transient expression of mitoPARP in HepG2 and HeLa S3 cells was used.
Enzyme Activity Measurements
The activity of FLAG-tagged NADS was measured following expression in 293 cells and immunoprecipitation from whole cell extracts using anti-FLAG M2 affinity gel matrix (Sigma). Amidation of NAAD to NAD+ by the enzyme was carried out overnight in 50 mm Tris/HCl, pH 8.0, 60 mm KCl, 2 mm ATP, 8 mm MgCl2, 20 mm l-glutamine, and 2 mm NAAD. Generated NAD+ was then converted to NADH by incubation with 1% EtOH and 2 units of alcohol dehydrogenase for 30 min. Proteins were removed by ultrafiltration (3 kDa), and samples were analyzed by HPLC.
To test formation of NMN by NADS, the amidation reaction was carried out in the presence of NAMN instead of NAAD. NADS was then removed by ultrafiltration (3 kDa). Because NMN and NAMN did not separate sufficiently well in the subsequent analysis, the mononucleotide was adenylated (to yield NAD+ or NAAD) by adding 1.5 μg of purified NMNAT1 and incubated for 1 h. Nucleotides were then analyzed by HPLC.
Automodification of PARP proteins was measured in extracts from 293mitoPARP cells or using bacterially expressed and purified His6-tagged PARP1cd (amino acids 652–1014, kindly provided by Dr. F. Koch-Nolte, Hamburg, Germany). Cell extracts (1 μg/μl of total protein) or purified PARP1cd (40 ng/μl) was incubated for 1 h at 30 °C in PBS supplemented with 10 mm MgCl2 and NAD+ or NAAD as substrate. Samples were then analyzed by Western blotting.
HPLC Analysis of Nucleotides
The HPLC system consisted of an LC-20AB solvent delivery module, an SPD-M20A photodiode array detector, and a SIL-20AC autosampler (Shimadzu). Nucleotides were separated on a Nucleodur C18 gravity column (Macherey & Nagel) as described (35).
RESULTS
In Situ Detection of Relative Mitochondrial NAD+ Levels by Poly(ADP-ribose) Generation in the Matrix
Targeted expression of PARP1cd in the mitochondrial matrix, here fused to enhanced GFP and termed mitoPARP, leads to the constitutive presence of PAR within these organelles (36). We reasoned that mitoPARP could be used to monitor alterations of the mitochondrial NAD+ content (Fig. 1B), i.e. a fraction of the mitochondrial NAD+ pool would be converted into protein-bound, immunodetectable PAR. Thus, changes of matrix NAD+ levels would be revealed using differences in PAR accumulation as readout (an [NAD+]-dependent steady state is established in concert with a slow endogenous polymer degrading activity (36)). Automodification of recombinant PARP1cd is indeed sensitive to the concentration of available NAD+ and precluded by the PARP inhibitor 3-AB (Fig. 1C). The smear of the PAR signal reflects the heterogeneity of the polymers, which may consist of up to 200 ADP-ribose units per acceptor site (11). Prolonged incubation of 293 cells stably expressing mitoPARP (293mitoPARP cells) with 3-AB reduced the amounts of PAR to nondetectable levels. After washout of 3-AB, polymers reappeared within 1 h, and their level increased steadily (Fig. 1D), verifying the constitutive activity of mitoPARP. Inhibition of NAD+ synthesis by the NamPRT inhibitor FK866 depleted mitochondrial NAD+ within 24 h (as indicated by the absence of detectable PAR, Fig. 1E). Cell viability was only slightly affected (Fig. 1F, left panel), but prolonged exposure to FK866 is lethal (see below). Thus, the PAR signal depends on both the catalytic activity of mitoPARP and the presence of NAD+. Consequently, mitoPARP expression enables detection of changes in the mitochondrial NAD+ content. mitoPARP activity contributes to mitochondrial NAD+ turnover (36). Moreover, under conditions of NamPRT inhibition, the viability of 293mitoPARP cells was lower than that of control 293 cells (Fig. 1F). Inhibition of PARP activity by 3-AB during FK866 treatment increased cell viability to control levels. 3-AB alone had no effect on cell viability. These observations underscore the importance of the mitochondrial NAD+ pool for cell survival.
Besides stably transfected 293mitoPARP cells, we also transiently expressed mitoPARP in HeLa S3 and HepG2 cells (Fig. 1G). Upon mitoPARP expression, PAR was similarly detectable in the mitochondria. PAR was undetectable in nuclei, the location of endogenous, unmodified PARP-1, in any of the experiments reported here.
Identification of Extracellular NAD+ Metabolites That Support Mitochondrial NAD+ Generation
We then explored which extracellular NAD+ precursors support mitochondrial NAD+ generation. Clearly, Nam serves this function, as it is usually the only NAD+ precursor in cell culture media. Although tryptophan, also present in the medium, can be converted to NAD+, it is insufficient to maintain cellular NAD+ levels (cf. supplemental Fig. S1A). Increasing the concentration of tryptophan in the medium up to 500 μm did also not improve cell viability in the presence of FK866 (supplemental Fig. S1A).
Thus, when NamPRT is inhibited, an alternative precursor needs to be present for NAD+ generation. Addition of NA, NAMN, or NMN to the medium restored mitochondrial NAD+ content and cell survival (Fig. 2, A and B). Cell viability correlated with mitochondrial NAD+ levels, as measured by PAR formation. Untransfected HeLa S3 cells displayed a similar sensitivity to FK866 and restoration of survival by NA, NAMN, and NMN (Fig. 2C). In contrast, in HepG2 cells, NA did not support the generation of NAD+ and cell survival (Fig. 2, D and E) confirming the lack of NA phosphoribosyltransferase (NAPRT) activity in this cell line (37). Following NAD+ depletion by FK866 treatment of 293mitoPARP cells, the mitochondrial NAD+ pool could be replenished by NA in a time-dependent manner (supplemental Fig. S1B).
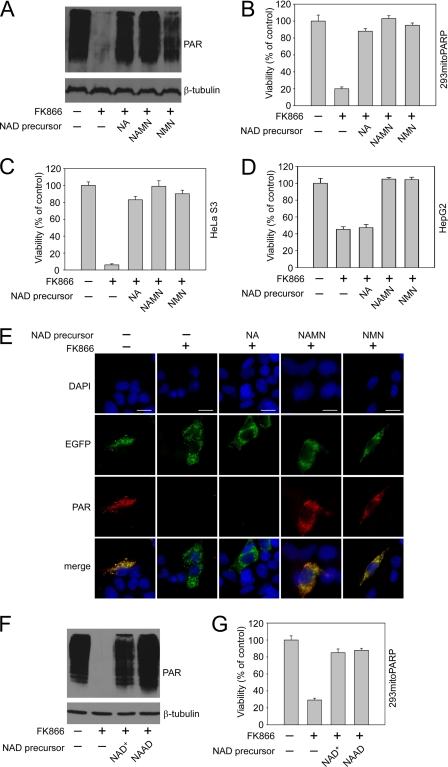
FIGURE 2.
Extracellular NAD+ derivatives supporting mitochondrial NAD+ generation and cell survival. Nam utilization was inhibited by FK866 addition, as indicated. Tryptophan is insufficient as NAD+ precursor to maintain cell viability (supplemental Fig. S1A). NA supports mitochondrial NAD+ synthesis in 293mitoPARP cells (supplemental Fig. S1B). “NAD precursor” designates the metabolite added to the medium. A, both NAMN and NMN support mitochondrial NAD+ formation. NAMN and NMN protect 293mitoPARP cells (B) and HeLa S3 cells (C) from cell death induced by NamPRT inhibition (FK866). D and E, mononucleotide precursors, but not NA, support generation of mitochondrial NAD+ and viability of HepG2 cells. Bar, 10 μm. NAD+ and NAAD support mitochondrial NAD+ (F) formation and cell viability (G) of 293mitoPARP cells.
We also tested NAD+ and NAAD as potential precursors of intracellular NAD+. Both dinucleotides were sufficient to preserve mitochondrial NAD+ and cell viability (Fig. 2, F and G). Thus, in addition to Nam and NA, their corresponding pyridine mono- and dinucleotides supported mitochondrial PAR formation when added to the cells. Although for some cells uptake of pyridine nucleotides has been proposed (38), the plasma membrane is generally considered impermeable to phosphorylated molecules. Therefore, we tested if maintenance of intracellular NAD+ by exogenous nucleotides required their extracellular degradation to intermediates that can then enter the cells.
Extracellular Nucleotides Are Degraded to Their Corresponding Nucleosides, which Then Enter Cells as NAD+ Precursors
The presence of nucleotide pyrophosphatase, also referred to as phosphodiesterase, on the external surface of the plasma membrane has been shown (39). This activity cleaves NAD+ to NMN and AMP (40). The nucleotide pyrophosphatase inhibitor pyridoxyl phosphate-6-azophenyl-2′,4′-disulfonic acid (41) prevented restoration of the mitochondrial NAD+ pool and cell survival when NAD+ was used as extracellular precursor (Fig. 3A). Likewise, addition of Ap4A as competitive nucleotide pyrophosphatase substrate (42) exerted a similar effect on cell viability both in 293mitoPARP and HeLa S3 cells (supplemental Fig. S2A). Consequently, to serve as precursor, extracellular NAD+ needs to be degraded to NMN.
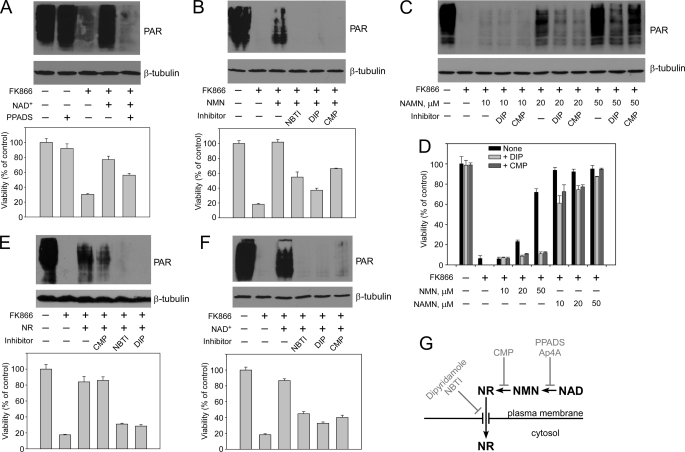
FIGURE 3.
Extracellular nucleotide precursors are degraded, and only the resultant ribosides enter the cell. Nam utilization by the 293mitoPARP cells was inhibited by FK866 addition, as indicated. Treatments with the indicated inhibitors were conducted as described under “Experimental Procedures.” The effects on mitochondrial NAD+ content (as detected by PAR formation) and cell viability are shown. A, extracellular degradation of NAD+ to NMN, which is inhibited by pyridoxyl phosphate-6-azophenyl-2′,4′-disulfonic acid (PPADS), is required to support mitochondrial NAD+ generation and cell viability. B, NMN must be dephosphorylated to NR to support mitochondrial NAD+ synthesis. Inhibitors of 5′-nucleotidase (CMP) or nucleoside transporters (dipyridamole (DIP), NBTI) were added as indicated. C, NAMN utilization requires dephosphorylation to NAR. Increasing NAMN concentrations in the medium were used to overcome the inhibition of NAD+ synthesis by DIP or CMP. D, when using NMN or NAMN as NAD+ precursors, cell viability depends on the extracellular degradation of the mononucleotides. Increasing concentrations of NMN or NAMN in the medium were used to overcome inhibition of NAD+ synthesis by DIP or CMP. The weaker inhibitory effect using NAMN as extracellular precursor may be due to the presence of NA (see text). E, extracellular NR enters cells via nucleoside transporters and supports mitochondrial NAD+ synthesis and cell viability. Nucleoside carrier inhibitors (NBTI, DIP), but not CMP, strongly reduce mitochondrial NAD+ generation when NR is used as extracellular precursor. F, extracellular NAD+ is degraded to NR to serve as precursor of intracellular NAD+. Both CMP and inhibition of nucleoside carriers strongly reduce the generation of mitochondrial NAD+ when NAD+ is added as extracellular precursor. G, schematic representation of the results. The effect of Ap4A is demonstrated in supplemental Fig. S2A.
We then studied the fate of NMN as extracellular NAD+ precursor. Addition of CMP, which competes with NMN as substrate for dephosphorylation by the external 5′-nucleotidase (39), notably reduced cell viability of 293mitoPARP cells (Fig. 3B) and HeLa S3 cells (supplemental Fig. S2B). As shown in Fig. 3B, this correlated with a reduced mitochondrial NAD+ content and was dose-dependent (supplemental Fig. S2D). NMN lost its capacity as extracellular NAD+ precursor also in the presence of dipyridamole and NBTI, inhibitors of plasma membrane nucleoside transporters (43). The decrease in immunodetectable PAR was again paralleled by reduced cell viability in 293mitoPARP cells (Fig. 3B) and HeLa S3 cells (supplemental Fig. S2B). Neither CMP nor the nucleoside transport inhibitors alone had any effect on the amount of detectable PAR or cell viability (supplemental Fig. S2C), when Nam could be used as NAD+ precursor.
Mitochondrial NAD+ generation from added NAMN was also sensitive to these inhibitors (Fig. 3, C and D), albeit less strongly than with NMN as precursor. Inhibition could be overcome by increasing the concentration of NAMN. A likely explanation for the lower inhibitor sensitivity using NAMN (and NAAD, see below) is the potential degradation into NA, an NAD+ precursor that bypasses inhibition of both NamPRT (FK866) and nucleoside transporters (DIP). Likewise, Nam is a degradation product of NMN and NAD+, but its utilization is prevented by the presence of FK866. Accordingly, NR supported mitochondrial PAR formation and cell viability in the presence of CMP, whereas both dipyridamole and NBTI inhibited NR utilization (Fig. 3E). Therefore, inhibition of nucleoside uptake or NMN dephosphorylation should also affect the use of NAD+ as an extracellular precursor. Indeed, as shown in Fig. 3F, NAD+ was considerably less efficient in maintaining mitochondrial PAR formation and cell viability in the presence of NBTI, dipyridamole, or CMP. Similar treatments of HeLaS3 cells had the same effect on cell viability (supplemental Fig. S2E). When using NAAD as extracellular NAD+ precursor, a similar tendency was found (supplemental Fig. S2F). However, the presence of NA (see above) may have provided an additional source of NAD synthesis and thus led to a less pronounced effect. Together, these results established that, besides NA and Nam, only the riboside precursors NR and NAR serve as extracellular precursors of intracellular NAD+, whereas mono- (NMN and NAMN) and dinucleotides (NAD+ and NAAD) need to be processed to the corresponding nucleosides (Fig. 3G). This interpretation assumes that entry of extracellular NAD+ precursors into cells could actually be monitored using mitoPARP, a probe that resides within mitochondria. As will be shown below, all known intermediates of NAD+ metabolism, including NAD+ itself, can be converted to a precursor of mitochondrial NAD+ in the cytosol. Consequently, all extracellular NAD+ precursors indeed contribute to mitochondrial NAD+ synthesis.
All NAD+ Biosynthetic Enzymes Localize to the Cytoplasm or the Nucleus, Except a Mitochondrial NMNAT Isoform
To understand the intracellular pathways of NAD+ biosynthesis, we next conducted comprehensive analyses of the subcellular distribution of the human enzymes. When expressed with a C-terminal FLAG epitope in HeLa S3 cells (Fig. 4A), all enzymes, except NMNAT3, localized to the cytoplasm or nucleus. NMNAT3 colocalized with Mitotracker, as expected (23). The two other NMNAT isoforms, not shown here, are known to be nuclear (23) or at the cytosolic face of the Golgi apparatus (23, 44).
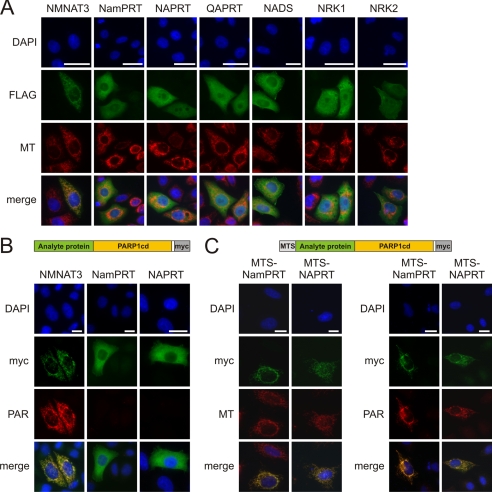
FIGURE 4.
Subcellular localization of NAD+ biosynthetic enzymes. A, all known NAD+ biosynthetic enzymes localize to the cytoplasm or the nucleus except for mitochondrial NMNAT3. HeLa S3 cells were transiently transfected with vectors encoding the indicated enzymes (C-terminally FLAG-tagged). The fluorescence micrographs show nuclei (DAPI), expressed recombinant proteins (FLAG), and mitochondria (MT). Bar, 20 μm. Mitochondrial localization of a potential variant of NAPRT was ruled out (supplemental Fig. S3A). B and C, NMNAT3, but not NamPRT or NAPRT, is a mitochondrial matrix protein. Submitochondrial protein localization was conducted following overexpression in HeLa S3 cells using PARAPLAY (33). B, analyte proteins (NMNAT3, NamPRT, and NAPRT) were overexpressed, endowed with a C-terminal PARP1cd and Myc tag (see scheme on top). If the fusion protein resides within the matrix, PAR accumulates and is detected by immunocytochemistry (as found for NMNAT3). Fusion proteins that localize to the intermembrane space or the cytosol do not support PAR accumulation (33). Bar, 20 μm. C, to verify the functionality of the PARAPLAY assay for the negative results (NamPRT and NAPRT), the corresponding constructs were additionally endowed with an established N-terminal mitochondrial targeting sequence (MTS, see scheme on top). Overexpression of these constructs resulted in mitochondrial PAR accumulation demonstrating that the NamPRT- or NAPRT-PARPcd fusion constructs are catalytically active. However, the authentic proteins (without added MTS) do not localize to the mitochondrial matrix, and therefore PAR accumulation was not observed (B).
Both NamPRT and NAPRT generate substrates for NMNATs (Fig. 1A). If they were present in the matrix, they could contribute to mitochondrial NAD+ synthesis provided NMNAT3 is a matrix protein. To address this issue, we applied PARAPLAY, a novel assay specifically suited to resolve suborganellar protein localization (33). This method includes overexpression of the analyte protein fused to PARP1cd. For mitochondrial proteins, PAR accumulation is only observed if the fusion protein resides within the matrix. We expressed NMNAT3, NamPRT, and NAPRT in fusion with PARP1cd. The localization of these constructs (Fig. 4B) was similar to that of the corresponding FLAG-tagged proteins (Fig. 4A). NMNAT3-PARP1cd protein supported PAR accumulation in mitochondria, demonstrating that NMNAT3 is indeed a matrix protein (45). Neither the NamPRT- nor the NAPRT-PARP1cd fusion proteins led to any detectable polymer formation verifying that these are not mitochondrial matrix proteins in human cells (Fig. 4B). To exclude that the catalytic activity of PARP1cd was impaired upon fusion to NamPRT or NAPRT, we added an established mitochondrial targeting sequence to the N termini of the fusion constructs to force them into the matrix. The resulting proteins now localized to the mitochondria and generated PAR (Fig. 4C). We also excluded the possibility that a truncated NAPRT, which could potentially originate from an alternative translational start, would lead to a mitochondrial isoform (supplemental Fig. S3A).
We thus established that NMNAT3 is the only enzyme of NAD+ synthesis that is present in mitochondria of human cells. All other enzymatic activities of human NAD+ biosynthesis reside in the nucleus and/or cytoplasm. Consequently, nuclear/cytosolic NAD+ can be synthesized from all identified extracellular precursors, Nam and NA as well as their corresponding ribosides.
NMN Is the Cytosolic Precursor of Mitochondrial NAD+ Synthesis
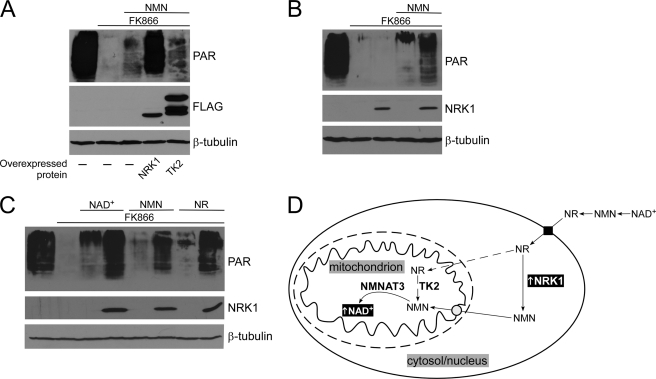
The presence of only NMNAT3 in the matrix strongly indicated NMN as the cytosolic precursor of mitochondrial NAD+. The absence of NADS from mitochondria ruled out the possibility of amidating NA precursors within the organelles (Fig. 4A). Moreover, localization of both NRK isoforms outside mitochondria (Fig. 4A) suggested that phosphorylation of NR does not occur within the organelles. Still, a possible side activity of thymidine kinase 2 (TK2), a mitochondrial matrix isoform, could potentially mediate phosphorylation of NR (Fig. 5D), because NRKs are homologous to pyrimidine nucleoside kinases (17). However, as shown in Fig. 5A, overexpression of TK2 had no detectable effect on mitochondrial PAR levels when NMN was the only extracellular NAD+ precursor (which is imported into cells after conversion to NR, see Fig. 3). The mitochondrial localization of the overexpressed TK2 protein was verified (supplemental Fig. S3B). Consequently, following entry into the cell, NR would have to be converted to NMN in the cytosol. Indeed, overexpression of NRK1 (which is cytoplasmic, see Fig. 4A) dramatically increased the amount of mitochondrial PAR (Fig. 5A), but only if an extracellular precursor (such as NMN) was available to provide NR (Fig. 5B). Thus, NR is a potent precursor of mitochondrial NAD+, if phosphorylated to NMN in the cytosol (Fig. 5D). Therefore, NMN has been functionally confirmed as the cytosolic precursor of mitochondrial NAD+. Overexpression of NRK1 considerably improved the use of extracellular NAD+ or NMN or NR itself for mitochondrial PAR formation (Fig. 5C). These observations strongly reinforce the notion that extracellular NMN and NAD+ are not taken up into the cells but rather have to be degraded to NR first.
FIGURE 5.
NMN is the cytosolic precursor of mitochondrial NAD+ synthesis in 293mitoPARP cells. A, overexpression of cytoplasmic NRK1, but not mitochondrial matrix TK2, increases mitochondrial NAD+ content. 293mitoPARP cells were transiently transfected with vectors encoding FLAG-tagged human NRK1 or TK2. The mitochondrial localization of TK2 was verified (supplemental Fig. S3B). B and C, increase of mitochondrial NAD+ levels by overexpressed NRK1 depends on a source of extracellular NR (NAD and NMN are extracellularly degraded to NR, Fig. 3). D, cytosolic NMN serves as precursor for mitochondrial NAD+. Increased NRK activity in the cytosol enhances mitochondrial NAD+ content when NR, NMN, or NAD+ are provided as extracellular NAD+ precursors. The conversion of NR to NMN by mitochondrial thymidine kinase 2 (TK2) activity was ruled out (dashed line).
Generation of Mitochondrial NAD+ from Nicotinic Acid Precursors Requires Synthesis and Subsequent Cleavage of Cytosolic NAD+
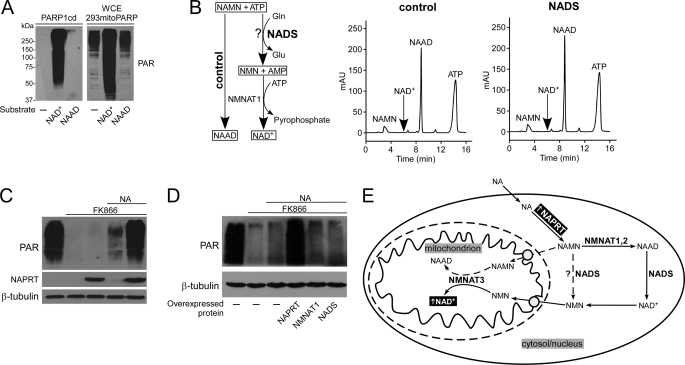
Given that NMN is the cytosolic precursor of mitochondrial NAD+, the question arises how extracellular NA precursors can serve to generate NAD+ within mitochondria. The scheme in Fig. 6E illustrates the possibilities to be considered when using NA as precursor (NAR would similarly lead to NAMN using NRK activity instead of NAPRT). First of all, we verified that the detector system was specific for NAD+. Purified PARP1cd did not use NAAD as substrate for PAR synthesis (Fig. 6A, left panel). Moreover, whole cell extracts from 293mitoPARP cells catalyzed the generation of PAR, in excess to the endogenously present polymers, only following addition of NAD+ but not NAAD (Fig. 6A, right panel). Generation of PAR in 293mitoPARP cells is thus not a result of NAAD turnover. Even if NAMN entered the mitochondria (where it could be converted to NAAD by NMNAT3), it would not support NAD+ generation and, therefore, not change the signal of PAR detection.
FIGURE 6.
Cytosolic nicotinic acid derivatives have to be converted to NAD+ before they can serve as mitochondrial NAD+ precursor. A, PARP1cd and mitoPARP do not synthesize PAR from NAAD. Recombinant PARP1cd or whole cell extracts (WCE) from 293mitoPARP cells were incubated with 1 mm NAD+ or NAAD. Endogenous PAR in whole cell extracts (WCE) of 293mitoPARP cells derives from PAR generated by the mitoPARP protein. As opposed to NAD+, addition of NAAD did not lead to further PAR generation. B, human NADS does not amidate NAMN. Overexpressed FLAG-tagged NADS was immunoprecipitated (supplemental Fig. S4A), and its activity to amidate NAAD to NAD+ was verified (supplemental Fig. S4B). After incubation with NAMN, ATP, and glutamine, NADS was removed by ultrafiltration, and purified NMNAT1 was added to convert NMN or NAMN to the respective dinucleotide. If NAMN had been amidated to NMN, NAD+ would be detected. The control sample did not contain NADS. C, NAPRT overexpression increases mitochondrial NAD+ formation from extracellular NA. FLAG-tagged NAPRT was transiently expressed in 293mitoPARP cells, as indicated. D, NAPRT is rate-limiting for mitochondrial NAD+ generation from extracellular NA. 293mitoPARP cells transiently expressed FLAG-tagged NAPRT, NMNAT1, or NADS, as indicated. E, NA-dependent mitochondrial NAD+ synthesis requires generation of cytosolic NAD+. Direct conversion of NAMN to NMN by NAD synthetase was excluded (dashed line). Import of NAMN into mitochondria and conversion by NMNAT3 to NAAD would not be detected (A). Therefore, NA undergoes several cytosolic conversions including NAAD amidation to NAD+ and degradation to NMN, which then enters the mitochondria. Note that use of extracellular NAR to generate mitochondrial NAD+ is similar, because this nucleoside is also first converted to NAMN by NRK (not shown).
A rather straightforward cytosolic conversion of NAMN to NMN would be direct amidation (Fig. 6E, dashed arrow). A bacterial NADS with this activity was recently identified (46). We therefore tested human NADS in this regard. The purified enzyme (supplemental Fig. S4A) exhibited its expected activity to convert NAAD to NAD+ (supplemental Fig. S4B). However, it did not amidate NAMN (Fig. 6B). Thus, cytosolic NAMN has to be converted to NAAD by NMNAT1 or -2 before amidation can take place. Consequently, to become a precursor for mitochondrial NAD+, NA derivatives have to be converted to cytosolic NAD+, which is then cleaved to NMN (Fig. 6E). Surprisingly, the use of NA as an extracellular precursor of mitochondrial NAD+ is efficient, even though four enzymatic activities (NAPRT, NMNAT, NADS, and NAD+ cleavage, Fig. 6E) are required, compared with a single one (e.g. NRK) when using amidated precursors (Fig. 5D). However, only overexpression of NAPRT, but not NMNAT1 or NADS, substantially increased mitochondrial PAR when NA was added as NAD+ precursor (Fig. 6, C and D) indicating that only NAPRT activity is rate-limiting.
DISCUSSION
This study provides the first comprehensive dissection of NAD+ biosynthesis in human cells. Based on mitoPARP expression, we developed a novel detector system, which permitted us to monitor changes in the mitochondrial NAD+ pool without cell disruption. We identified the extracellular NAD+ precursors that can enter cells to generate NAD+ for the nucleo-cytoplasm and NMN as precursor for mitochondrial NAD+. Moreover, our results establish the subcellular localization of all known enzymes involved in human NAD+ biosynthesis. Quite surprisingly, except for mitochondrial NAD+ generation by NMNAT3, the cellular pathways of NAD+ synthesis appear to be confined to the nucleo-cytoplasm, even though other subcellular NAD+ pools are likely to exist.
Generation of the Mitochondrial NAD+ Pool
A key result of this study is the identification of the pathway of mitochondrial NAD+ generation in mammalian cells. The utility of mitoPARP enabled monitoring of changes in mitochondrial NAD+ content in intact living cells and functionally established NMN as a cytosolic precursor. Moreover, using PARAPLAY (33), we demonstrated that NMNAT3, but not NamPRT, is located within the matrix of these organelles (Fig. 4B). Because ATP is readily available in mitochondria, the presence of NMNAT3 in the matrix and import of NMN are sufficient to maintain the organellar NAD+ pool. Several lines of evidence support the conclusion that NMN has to enter the matrix as substrate for mitochondrial NAD+ generation (Fig. 7). First, the absence of NAPRT and NamPRT from the mitochondrial matrix (Fig. 4, B and C) excludes NMN or NAMN generation within the organelles from the pyridine base precursors. Second, both NRK1 and -2 localize to the cytosol or nucleus (Fig. 4A), and the mitochondrial relative, TK2, does not support mitochondrial NAD+ generation from an extracellular source of NR (Fig. 5A). Therefore, mitochondrial generation of NMN (or NAMN) from the nucleoside precursors is also not possible. Third, the absence of NADS from mitochondria (Fig. 4A) rules out amidation within the organelles. Fourth, enhanced NMN generation in the cytosol by overexpression of NRK1 strongly increases mitochondrial NAD+ content (Fig. 5). Moreover, enhanced production of NMN by NamPRT (which is cytosolic, see Fig. 4) increases mitochondrial NAD+ content using Nam (28, 30). Mitochondrial NMN content rises, in fact, when the concentration of extracellular NAD+ precursors is elevated (47). Finally, only NMNAT3 resides within the mitochondrial matrix. From these observations, we conclude that NMN has to be imported into mitochondria to generate NAD+ in the matrix. So far, no NMN-specific mitochondrial transport system has been identified. In principle, the results presented here do not formally rule out the possibility of direct uptake of NAD form the cytosol. However, for mammalian mitochondria, this is considered rather unlikely under physiological conditions for a variety of reasons (48). Moreover, unlike their counterparts in yeast and plants (31, 32), mammalian mitochondria do not seem to have a mitochondrial NAD carrier (30).
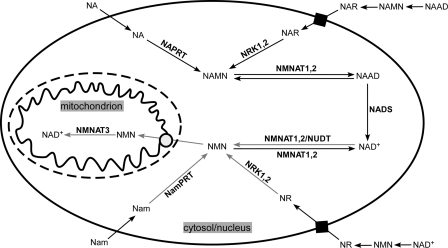
FIGURE 7.
Schematic representation of NAD+ biosynthesis in human cells. Nam and NA enter the cell and are converted to NMN and NAMN by NamPRT and NAPRT, respectively. The respective mononucleotides can also be generated from NR and NAR, which enter the cell via nucleoside transporters and are phosphorylated in the cytosol by NRK1 or NRK2. Extracellular NAAD, NAD+, NAMN, and NMN have to be degraded to the corresponding riboside. Cytosolic/nuclear NMN and NAMN are adenylated by NMNAT1 or -2 to NAD+ and NAAD, respectively. NAAD is amidated to NAD+ by NADS. NAD+ (or NAAD) can be cleaved to yield NMN (or NAMN) by Nudix hydrolases (NUDT) activity or in the reverse reaction of NMNATs. Therefore, all NA precursors can be converted into NMN within the nucleo-cytoplasm. NMN is the cytosolic precursor of NAD+ synthesis in mitochondria. NMNAT3 is located in the matrix and uses imported NMN (and mitochondrial ATP) to form NAD+.
Subcellular Distribution of NAD+ Biosynthesis and NAD+ Pools
The requirement for mitochondrial uptake of NMN, rather than exchange with cytosolic NAD+, as known for plants (31) and yeast (32), has important implications. It provides a rationale for the situation that, at least in some mammalian tissues, mitochondrial NAD+ is highly concentrated compared with the nuclear/cytosolic pool (24).
The conversion of NA precursors (NA, NAR, and NAMN) to NAD+ does not include NMN formation as an intermediate step (Fig. 1A, left panel). Nevertheless, they are efficiently used to generate mitochondrial NAD+. Consequently, there has to be an activity that mediates the cleavage of cytosolic NAD+ to NMN. Two principal possibilities can be considered (Fig. 7). First, the reverse reaction of NMNATs (NAD+ + PPi and ATP + NMN) leads to NMN generation. This possibility could involve either NMNAT1 or -2, both of which have access to cytosolic NAD+. However, it would require a source of pyrophosphate. Second, Nudix hydrolases would cleave NAD+ to NMN and AMP. In humans, there are several members in this family (49), and at least for NudT12, the activity toward NAD has been demonstrated (50).
The potential to generate NMN from all cytosolic intermediates provides human cells with a high flexibility to maintain the mitochondrial NAD+ pool. NamPRT has been suggested to be present within the organelles in rat liver and 293 cells (30). It was argued that NamPRT is essential for mitochondrial NAD+ generation in mammals and therefore plays a key role for mitochondrial function. However, our results rule out the absolute requirement for NamPRT to generate mitochondrial NAD+. Because all common cell culture media contain Nam as sole precursor of NAD+, it is obvious that under these conditions the activity of NamPRT is of critical importance. However, as we show, when other extracellular precursors are present, NamPRT inhibition is harmless, and the mitochondrial NAD+ pool is maintained. Moreover, the lack of NAPRT activity in HepG2 cells might indicate previously unrecognized tissue- or cell-specific differences in the utilization of NAD+ precursors.
Interestingly, NamPRT is the rate-limiting step in NAD+ generation from Nam (51). Our results suggest that this is true for the initial conversion of all extracellular precursors (Nam, NA, NR, and NAR). Overexpression of NAPRT or NRK activity led to considerable increase of mitochondrial NAD+, whereas overexpression of any subsequent activity in the pathway (NMNAT or NADS) had no effect, i.e. formation of the mononucleotide (NAMN or NMN), which is specific for the entry of the individual precursors into NAD+ synthesis (Figs. 1A and. 7), regulates the flux and thereby controls NAD+ levels. This notion explains why it is possible, for example, to increase cellular NAD+ content when NA is added to the medium in addition to Nam (37). The control of NAD+ synthesis by the initial steps thus suggests that combination of various NAD+ precursors could be beneficial to enhance cellular NAD+ levels.
Cellular Uptake of NAD+ Precursors
The demonstration of NR and NAR as intermediates in the NAD+ metabolome (17–20) has added an important aspect to cellular NAD+ homeostasis and extended the scope of potential extracellular NAD+ precursors. NAD+ itself has been proposed to be taken up by human cell lines, including those used in this study (38). Our results do not support this notion. Nevertheless, NAD+ uptake can be mediated, for example, by connexin 43 hemichannels, which are cell type-specific however (52). We demonstrated that inhibition of nucleotide degradation or nucleoside transporters markedly reduced the utilization of extracellular nucleotides as precursors (Fig. 3). Moreover, overexpression of cytosolic NRK1 increased NAD+ synthesis when extracellular NAD+ or NMN or NR was added as precursor (Fig. 5). These observations strongly support the conclusion that both NAD+ and NMN need to be degraded to NR outside the cell to serve as precursors of intracellular NAD+. Therefore, our data indicate that bases (NA and Nam) and nucleosides (NR and NAR) are taken up by human cells, but not nucleotides (NAD+, NMN, NAAD or NAMN), unless cell type-specific transport systems are present.
In conclusion, our study has provided fundamentally new insights into the metabolism of NAD+ in human cells. It has solved the long standing problem of mitochondrial NAD+ generation and identified pyridine ribosides as extracellular precursors of cellular NAD+ metabolism. Even though tryptophan is widely referred to as precursor of de novo NAD+ synthesis, its role to maintain cellular NAD+ levels in human cells seems negligible. The results have important implications for the understanding of compartment-specific bioenergetics and NAD+-dependent signaling processes as well as for organismal NAD+ homeostasis.
Supplementary Material
This work was supported by the Norwegian Research Council.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–4.
- NA
- nicotinic acid
- NRK
- nicotinamide riboside kinase
- NMNAT
- NMN adenylyltransferase
- NamPRT
- Nam phosphoribosyltransferase
- Ap4A
- diadenosine tetraphosphate
- 3-AB
- 3-aminobenzamide
- PARP
- poly(ADP-ribose) polymerase
- PAR
- poly(ADP-ribose)
- NADS
- NAD synthetase
- NAMN
- nicotinic acid mononucleotide
- Nam
- nicotinamide
- NAPRT
- NA phosphoribosyltransferase
- NAR
- nicotinic acid riboside
- DIP
- dipyridamole.
REFERENCES
- 1. Belenky P., Bogan K. L., Brenner C. (2007) Trends Biochem. Sci. 32, 12–19 [DOI] [PubMed] [Google Scholar]
- 2. Magni G., Orsomando G., Raffelli N., Ruggieri S. (2008) Front. Biosci. 13, 6135–6154 [DOI] [PubMed] [Google Scholar]
- 3. Berger F., Ramírez-Hernández M. H., Ziegler M. (2004) Trends Biochem. Sci. 29, 111–118 [DOI] [PubMed] [Google Scholar]
- 4. Houtkooper R. H., Cantó C., Wanders R. J., Auwerx J. (2010) Endocr. Rev. 31, 194–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Preiss J., Handler P. (1958) J. Biol. Chem. 233, 493–500 [PubMed] [Google Scholar]
- 6. Finkel T., Deng C. X., Mostoslavsky R. (2009) Nature 460, 587–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Liang F., Kume S., Koya D. (2009) Nat. Rev. Endocrinol. 5, 367–373 [DOI] [PubMed] [Google Scholar]
- 8. Nakahata Y., Sahar S., Astarita G., Kaluzova M., Sassone-Corsi P. (2009) Science 324, 654–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ramsey K. M., Yoshino J., Brace C. S., Abrassart D., Kobayashi Y., Marcheva B., Hong H. K., Chong J. L., Buhr E. D., Lee C., Takahashi J. S., Imai S., Bass J. (2009) Science 324, 651–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hassa P. O., Hottiger M. O. (2008) Front. Biosci. 13, 3046–3082 [DOI] [PubMed] [Google Scholar]
- 11. Bürkle A. (2005) FEBS J. 272, 4576–4589 [DOI] [PubMed] [Google Scholar]
- 12. Schreiber V., Dantzer F., Ame J. C., de Murcia G. (2006) Nat. Rev. Mol. Cell Biol. 7, 517–528 [DOI] [PubMed] [Google Scholar]
- 13. Fliegert R., Gasser A., Guse A. H. (2007) Biochem. Soc. Trans. 35, 109–114 [DOI] [PubMed] [Google Scholar]
- 14. Zhang Q., Piston D. W., Goodman R. H. (2002) Science 295, 1895–1897 [DOI] [PubMed] [Google Scholar]
- 15. Berger F., Lau C., Ziegler M. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 3765–3770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Coleman M. P., Freeman M. R. (2010) Annu. Rev. Neurosci. 33, 245–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bieganowski P., Brenner C. (2004) Cell 117, 495–502 [DOI] [PubMed] [Google Scholar]
- 18. Belenky P., Christensen K. C., Gazzaniga F., Pletnev A. A., Brenner C. (2009) J. Biol. Chem. 284, 158–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Belenky P., Racette F. G., Bogan K. L., McClure J. M., Smith J. S., Brenner C. (2007) Cell 129, 473–484 [DOI] [PubMed] [Google Scholar]
- 20. Bogan K. L., Evans C., Belenky P., Song P., Burant C. F., Kennedy R., Brenner C. (2009) J. Biol. Chem. 284, 34861–34869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Henderson L. M., Deodhar T., Krehl W. A., Elvehjem C. A. (1947) J. Biol. Chem. 170, 261–268 [Google Scholar]
- 22. Watson M., Roulston A., Bélec L., Billot X., Marcellus R., Bédard D., Bernier C., Branchaud S., Chan H., Dairi K., Gilbert K., Goulet D., Gratton M. O., Isakau H., Jang A., Khadir A., Koch E., Lavoie M., Lawless M., Nguyen M., Paquette D., Turcotte E., Berger A., Mitchell M., Shore G. C., Beauparlant P. (2009) Mol. Cell. Biol. 29, 5872–5888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Berger F., Lau C., Dahlmann M., Ziegler M. (2005) J. Biol. Chem. 280, 36334–36341 [DOI] [PubMed] [Google Scholar]
- 24. Di Lisa F., Ziegler M. (2001) FEBS Lett. 492, 4–8 [DOI] [PubMed] [Google Scholar]
- 25. Haigis M. C., Mostoslavsky R., Haigis K. M., Fahie K., Christodoulou D. C., Murphy A. J., Valenzuela D. M., Yancopoulos G. D., Karow M., Blander G., Wolberger C., Prolla T. A., Weindruch R., Alt F. W., Guarente L. (2006) Cell 126, 941–954 [DOI] [PubMed] [Google Scholar]
- 26. Schlicker C., Gertz M., Papatheodorou P., Kachholz B., Becker C. F., Steegborn C. (2008) J. Mol. Biol. 382, 790–801 [DOI] [PubMed] [Google Scholar]
- 27. Hallows W. C., Lee S., Denu J. M. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 10230–10235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nakagawa T., Lomb D. J., Haigis M. C., Guarente L. (2009) Cell 137, 560–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pittelli M., Formentini L., Faraco G., Lapucci A., Rapizzi E., Cialdai F., Romano G., Moneti G., Moroni F., Chiarugi A. (2010) J. Biol. Chem. 285, 34106–34114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yang H., Yang T., Baur J. A., Perez E., Matsui T., Carmona J. J., Lamming D. W., Souza-Pinto N. C., Bohr V. A., Rosenzweig A., de Cabo R., Sauve A. A., Sinclair D. A. (2007) Cell 130, 1095–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Palmieri F., Rieder B., Ventrella A., Blanco E., Do P. T., Nunes-Nesi A., Trauth A. U., Fiermonte G., Tjaden J., Agrimi G., Kirchberger S., Paradies E., Fernie A. R., Neuhaus H. E. (2009) J. Biol. Chem. 284, 31249–31259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Todisco S., Agrimi G., Castegna A., Palmieri F. (2006) J. Biol. Chem. 281, 1524–1531 [DOI] [PubMed] [Google Scholar]
- 33. Dölle C., Niere M., Lohndal E., Ziegler M. (2010) Cell. Mol. Life Sci. 67, 433–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dölle C., Ziegler M. (2009) Anal. Biochem. 385, 377–379 [DOI] [PubMed] [Google Scholar]
- 35. Pollak N., Niere M., Ziegler M. (2007) J. Biol. Chem. 282, 33562–33571 [DOI] [PubMed] [Google Scholar]
- 36. Niere M., Kernstock S., Koch-Nolte F., Ziegler M. (2008) Mol. Cell. Biol. 28, 814–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hara N., Yamada K., Shibata T., Osago H., Hashimoto T., Tsuchiya M. (2007) J. Biol. Chem. 282, 24574–24582 [DOI] [PubMed] [Google Scholar]
- 38. Billington R. A., Travelli C., Ercolano E., Galli U., Roman C. B., Grolla A. A., Canonico P. L., Condorelli F., Genazzani A. A. (2008) J. Biol. Chem. 283, 6367–6374 [DOI] [PubMed] [Google Scholar]
- 39. Zimmermann H. (2000) Naunyn-Schmiedebergs Arch. Pharmacol. 362, 299–309 [DOI] [PubMed] [Google Scholar]
- 40. Aleo M. F., Giudici M. L., Sestini S., Danesi P., Pompucci G., Preti A. (2001) J. Cell. Biochem. 80, 360–366 [DOI] [PubMed] [Google Scholar]
- 41. Grobben B., Anciaux K., Roymans D., Stefan C., Bollen M., Esmans E. L., Slegers H. (1999) J. Neurochem. 72, 826–834 [DOI] [PubMed] [Google Scholar]
- 42. Vollmayer P., Clair T., Goding J. W., Sano K., Servos J., Zimmermann H. (2003) Eur. J. Biochem. 270, 2971–2978 [DOI] [PubMed] [Google Scholar]
- 43. Griffith D. A., Jarvis S. M. (1996) Biochim. Biophys. Acta 1286, 153–181 [DOI] [PubMed] [Google Scholar]
- 44. Lau C., Dölle C., Gossmann T. I., Agledal L., Niere M., Ziegler M. (2010) J. Biol. Chem. 285, 18868–18876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yahata N., Yuasa S., Araki T. (2009) J. Neurosci. 29, 6276–6284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sorci L., Martynowski D., Rodionov D. A., Eyobo Y., Zogaj X., Klose K. E., Nikolaev E. V., Magni G., Zhang H., Osterman A. L. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 3083–3088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Formentini L., Moroni F., Chiarugi A. (2009) Biochem. Pharmacol. 77, 1612–1620 [DOI] [PubMed] [Google Scholar]
- 48. Tyler D. D. (1992) The Mitochondrion in Health and Disease, VCH Publishers, Inc., New York [Google Scholar]
- 49. McLennan A. G. (2006) Cell. Mol. Life Sci. 63, 123–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Abdelraheim S. R., Spiller D. G., McLennan A. G. (2003) Biochem. J. 374, 329–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Revollo J. R., Grimm A. A., Imai S. (2004) J. Biol. Chem. 279, 50754–50763 [DOI] [PubMed] [Google Scholar]
- 52. Bruzzone S., Guida L., Zocchi E., Franco L., De Flora A. (2001) FASEB J. 15, 10–12 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.