Abstract
The R7 family of regulators of G protein signaling (RGS) proteins, comprising RGS6, RGS7, RGS9, and RGS11, regulate neuronal G protein signaling pathways. All members of the R7 RGS form trimeric complexes with the atypical G protein β subunit, Gβ5, and membrane anchor R7BP or R9AP. Association with Gβ5 and membrane anchors has been shown to be critical for maintaining proteolytic stability of the R7 RGS proteins. However, despite its functional importance, the mechanism of how R7 RGS forms complexes with Gβ5 and membrane anchors remains poorly understood. Here, we used protein-protein interaction, co-localization, and protein stability assays to show that association of RGS9 with membrane anchors requires Gβ5. We further establish that the recruitment of R7BP to the complex requires an intact interface between the N-terminal lobe of RGS9 and protein interaction surface of Gβ5. Site-directed mutational analysis reveals that distinct molecular determinants in the interface between Gβ5 and N-terminal Dishevelled, EGL-10, Pleckstrin/DEP Helical Extension (DEP/DHEY) domains are differentially involved in R7BP binding and proteolytic stabilization. On the basis of these findings, we conclude that Gβ5 contributes to the formation of the binding site to the membrane anchors and thus is playing a central role in the assembly of the proteolytically stable trimeric complex and its correct localization in the cell.
Keywords: Adaptor Proteins, Cell Surface Receptor, G Protein-coupled Receptors (GPCR), G Proteins, Heterotrimeric G Proteins, Neuroscience, Protein Assembly, Protein Stability, Signal Transduction, RGS Proteins
Introduction
Regulators of G protein signaling (RGS)2 proteins are key components of G protein-coupled receptor (GPCR) signaling pathways (1). They limit the duration that heterotrimeric G proteins spend in their active state and thus serve as powerful negative regulators of GPCR signal transmission. Biochemically, RGS proteins function as GTPase-accelerating proteins that facilitate GTP hydrolysis on G protein α subunits (Gα) thereby promoting their inactivation (2, 3). There are more than 30 RGS proteins that are grouped into six families according to the homology and structural features (3, 4). Cumulatively, RGS proteins are thought to be ubiquitously expressed and have been demonstrated to regulate a range of the vital physiological processes (5–8).
The R7 family of RGS proteins comprises four members: RGS6, RGS7, RGS9, and RGS11 and plays prominent roles in the nervous and cardiovascular systems (9, 10). The best studied member of this family, RGS9, is involved in regulating diverse GPCR signaling cascades in mammals (11, 12). Its short splice isoform, RGS9-1, regulates phototransduction cascade in rod and cone photoreceptors of the retina and is essential for the high temporal resolution of our vision (13–15). The long splice isoform, RGS9-2, is predominantly expressed in the striatum, a region of the brain involved in movement control and reward behavior (16–18). RGS9-2 is thought to control G protein signaling downstream from the μ-opioid and D2 dopamine receptors and has been implicated in drug addiction and movement disorders (19–23).
Both RGS9 splice isoforms exist as constitutive heterotrimers with two other proteins. In the retina, RGS9-1 is found in the complex with type 5 G protein β subunit (Gβ5) (24) and RGS9 anchor protein (R9AP) (25). Similarly, in the striatum, RGS9-2 is complexed with Gβ5 and R9AP-like protein called R7-binding protein (R7BP) (26). Both R9AP and R7BP are small SNARE-like membrane proteins. Their binding dictates localization of both RGS9 isoforms in photoreceptors and striatal neurons, respectively (27–31). In addition, both R7BP/R9AP and Gβ5 are critical for regulating the expression level of RGS9. Knock out of Gβ5 (32), R7BP (29), or R9AP (33) leads to a dramatic reduction in RGS9 protein levels by increasing its susceptibility to proteolytic degradation. Conversely, overexpression of R7BP (31, 34) or R9AP (35) results in an increase in RGS9 levels. Interestingly, reciprocal effects on the stability of RGS9-containing complexes have been also described: elimination of RGS9 in photoreceptors leads to down-regulation in the levels of Gβ5 (14), and knock-out of Gβ5 compromises the stability of R7BP (29, 36). Furthermore, association of RGS9 with the binding partners and consequently its expression level is modulated by oxygen concentration and neuronal activity, suggesting that reorganization in RGS9 complexes could contribute to the neuronal plasticity mechanisms (37). These observations lead to a general view that RGS9 forms an obligatory trimeric complex with its bona fide subunits R9AP/R7BP and Gβ5 and that the interactions between the subunits are responsible for setting an appropriate expression level of the complex. However, the mechanisms governing complex assembly and intrasubunit interactions are far from being understood.
The recently solved crystal structure of the RGS9-Gβ5 complex reveals tight integration of the Gβ5 subunit, which interacts with multiple domains of RGS9 (38). One critical focal contact point is established between the so-called “protein interaction interface” of Gβ5 and the N-terminal segment of RGS9 containing the DEP domain. Interestingly, the same DEP domain of the molecule is also indispensable for the recruitment of R7BP and R9AP membrane anchors to the RGS9-Gβ5 complex (26, 28, 30, 34). However, the molecular determinants mediating this interaction and mechanisms by which proteolytic stabilization of the trimeric complex is achieved are unknown.
In this study we examined assembly of the RGS9-Gβ5-R7BP complex and mechanisms by which interactions between individual subunits regulate its expression and localization. We report an unexpected observation that Gβ5 is required for the interaction of RGS9 with its membrane anchors R7BP and R9AP, which was previously thought to be mediated exclusively by the DEP domain of RGS9. We further established that the binding site for the membrane anchors is formed by the interface between the protein interaction surface of Gβ5 and the N-terminal lobe of RGS9. Based on our site-directed mutagenesis results we propose a model where R7BP acts to remodel the interface that is essential for the stability of the complexes and their membrane recruitment.
EXPERIMENTAL PROCEDURES
Plasmid Constructs
Cloning of RGS9-2, RGS9-1-HA, Gβ5S, Gβ5L, R7BP, R9AP, and RGS9NT-myc (amino acids 1–209) in pcDNA3.1/V5-His-TOPO was described previously (26, 30, 39). To generate linker and DEP mutants, D202A/Y203A/G204A/L205A/R207A/V208A/T209A and F16E/F76E substitutions, respectively, were introduced into RGS9-1-HA in pcDNA3.1/V5-His-TOPO vector (Invitrogen) by using the QuikChange Multi-site-directed Mutagenesis kit (Stratagene). The coding sequence of mouse R7BP was cloned in pcDNA4/HisMax-TOPO (Invitrogen).
Antibodies
Generation of anti-R9AP (against amino acids 144–223) (40), sheep anti-RGS9-1 (24), and sheep anti-RGS9-2 (26) antibodies was described previously. Rabbit anti-Gβ5 and rabbit anti-R7BP were gifts from Dr. William Simonds (NIDDK, National Institutes of Health, Bethesda, MD). Mouse anti-HA (12CA5) (Roche Applied Science), rat anti-HA high affinity (3F10) (Roche Applied Science), mouse anti-β-actin (AC-15) (Sigma), rabbit anti-Gαo (K-20), and rabbit anti-c-myc (GenScript) were purchased.
Cell Culture and Transfection
HEK293T/17 cells were chosen because of their high transfectability (41). The cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), MEM nonessential amino acids, 1 mm sodium pyruvate, and antibiotics (100 units/ml penicillin and 100 μg/ml streptomycin) at 37 °C in a humidified incubator containing 5% CO2. For transfection, cells were seeded into 6-cm dishes at a density of 1 × 106 cells/dish. After 24 h, expression constructs (0.83 μg/construct, total 2.5 μg/dish) were transfected into the cells using Lipofectamine LTX (8 μl/dish) and PLUS (2.5 μl/dish) reagents. Empty vector was used to normalize the amount of transfected DNA. The cells were used for experiments at 48 h after transfection.
Selection of Stable Cell Lines
R7BP stable cell lines were generated by transfecting linearized R7BP in pcDNA4/HisMax-TOPO into HEK293T/17 cells. Cells were selected in 100 μg/ml zeocin (Invitrogen). Resistant colonies were expanded, and the expression of R7BP protein was evaluated by Western blotting and immunocytochemistry.
Immunoprecipitation Assay
HEK293T/17 cells in 6-cm plates were transfected with the indicated constructs. Forty-eight hours after transfection, cells were washed once with ice-cold PBS and lysed with 0.5 ml of lysis buffer (50 mm Tris, pH 7.4, 0.5% n-dodecanoylsucrose, 300 mm NaCl, and Complete Mini Protease Inhibitor Mixture (Roche Applied Science)) by sonication on ice. The resultant whole cell lysates were incubated for 30 min at 4 °C with rotary agitation to solubilize membrane proteins. After lysis, cell lysates were centrifuged at 14,000 rcf for 15 min at 4 °C. A 5 μg/sample of the indicated antibody and 20 μl of Dynabeads Protein G (Invitrogen) were added, and the supernatants were tumbled for 1 h at 4 °C. After three washes with 1 ml of ice-cold wash buffer (50 mm Tris, pH 7.4, 0.5% n-dodecanoylsucrose, 300 mm NaCl, and 0.5 mm phenylmethanesulfonyl fluoride), proteins bound to the beads were eluted with SDS-sample buffer (50 mm Tris, pH 6.8, 1% SDS, 143 mm β-mercaptoethanol, 0.08 mg/ml bromphenol blue, 10% glycerol). Immunoprecipitation from the mouse brain extracts was performed as described previously (42). Immunoprecipitates were subjected to SDS-PAGE, transferred to polyvinylidene difluoride membranes, and probed with the indicated antibodies.
Quantitative Western Blot Analysis
Fluorescent Western blot analysis was performed using the Odyssey Infrared Imaging System according to the manufacturer's instructions (LI-COR Biosciences), which is reported to have a 16–250-fold wider quantifiable linear range with increased reproducibility than a chemiluminescence Western blot detection system. For detection, blots were blocked with Odyssey Blocking Buffer for 1 h at room temperature followed by a 1.5-h incubation with specific antibodies diluted in Odyssey Blocking Buffer containing 0.1% Tween 20. Blots were washed in PBS containing 0.1% Tween 20 and incubated for 45 min with a 1:5,000 dilution of secondary antibodies labeled with IRDye 800CW in Odyssey buffer containing 0.1% Tween 20 and 0.01% SDS. Proteins were visualized using the Odyssey Infrared Imager, and molecular analysis of band intensities was quantified using the Odyssey Application software by measuring the integrated intensity. The integrated intensity is proportional to the amount of dye-labeled antibodies bound to the blot.
Immunocytochemistry
HEK293T/17 cells were seeded onto poly-l-lysine-coated coverslips. After 24 h, expression constructs (total, 5 μg/well) were transfected into the cells using Lipofectamine LTX (μl/well) and PLUS (0.5 μl/well) reagents. The cells were used for immunocytochemistry at 48 h after transfection. Cells were fixed with 4% paraformaldehyde in PBS for 20 min and permeabilized by incubation in 0.5% Triton X-100 in PBS for 5 min. Cells were blocked with 10% goat serum in PBS containing 0.1% Triton X-100 (0.1% PBS-T) for 1 h and were then incubated with primary antibody in 2% goat serum in 0.1% PBS-T for 90 min. The cells were washed with 0.1% PBS-T, and the antibody was visualized with fluorophore-conjugated secondary antibody in 2% goat serum in 0.1% PBS-T for 45 min. The coverslips were washed three times with 0.1% PBS-T and twice with PBS and mounted on glass slides with Fluoromount (Sigma). Microscopy was performed with an Olympus Fluoview 1000 confocal microscope. The following concentrations of the antibodies were used: 2.5 μg/ml anti-HA (12CA5), 1.2 μg/ml anti-R7BP, and 1.4 μg/ml anti-Gβ5 antibodies.
RESULTS
Proteolytic Stability of RGS9-Gβ5-R7BP Complex Is Achieved by Mutually Reciprocal Contribution of Individual Subunits
To begin examining the relationship between subunits in RGS9-Gβ5-R7BP we compared their individual contributions toward establishing a proteolytically stable complex. We have previously determined that expression levels of RGS9 in the cells are limited by its proteolytic degradation rate as measured by the pulse-chase method (34, 39). Therefore, proteolytic stability of the complex could be approximated by monitoring changes in protein expression levels. To facilitate these studies we chose RGS9-2 as a model isoform due to its relatively short lifetime in transfected cells and large degree of stabilization by R7BP (34). Mammalian HEK293 cells were transfected with RGS9-2, Gβ5, and R7BP subunits of the complex in all possible combinations, and the effects of co-expression on modulating protein levels were evaluated by quantitative Western blotting. We found no change in RGS9-2 protein levels when Gβ5 or R7BP was added separately (Fig. 1, A and B). In contrast, co-expression of RGS9-2 with both Gβ5 and R7BP produced a substantial increase in the amount of RGS9-2 (Fig. 1, A and B). We further found that the levels of Gβ5 increase only upon co-expression with both RGS9-2 and R7BP, and neither of the proteins alone affects Gβ5 expression (Fig. 1, C and D). Similarly, R7BP levels are elevated only when it is expressed together with both Gβ5 and R7BP (Fig. 1, E and F). The degree of the effect on the protein expression level by co-expressing all components was found to be similar for all three subunits. These results indicate that stabilizing effects of the individual proteins in the RGS9-Gβ5-R7BP complex are not additive but rather require synergistic contributions from all three subunits to achieve proteolytic stabilization.
FIGURE 1.
Co-expression of all three components: RGS9, Gβ5, and R7BP, is essential for achieving proteolytic stabilization of the complex. A, C, and E, Western blot analysis of protein expression in HEK293 cells. RGS9-2, Gβ5, and R7BP were expressed alone or in different combinations. The proteins extracted from the cells were subjected to Western blot analysis using the indicated specific antibodies. B, D, and F, quantification of the Western blotting data presented in A, C, and E, respectively. Band intensities of indicated proteins were determined and normalized to levels of β-actin present in the same sample. All values were expressed as -fold increase relative to control cells expressing a single subunit of RGS9-Gβ5-R7BP heterotrimer. Values shown are the mean ± S.E. (error bars) from a representative experiment performed in triplicate, which was repeated several times and yielded similar results. Asterisks indicate statistical significance of the differences in protein expression: **, p < 0.01; ***, p < 0.001, Student's t test.
Gβ5 Is Required for Interaction of RGS9 with Membrane Anchors
RGS9 binds to Gβ5 directly via the Gγ-like domain, and this association was shown not to require contributions from membrane anchors R7BP/R9AP (29, 38). On the other hand, the complex formation of RGS9 with R7BP/R9AP has not been fully characterized. Therefore, we have next analyzed the impact of Gβ5 on the association between RGS9 and R7BP (Fig. 2). In these studies, we used the short splice isoform of RGS9, RGS9-1, as the absence of intrinsically disordered C-terminal domain unique to RGS9-2 makes this isoform more proteolytically stable and therefore less dependent on Gβ5 and R7BP for achieving high expression levels in transfected cells (42). Indeed, we observed robust expression of RGS9-1 in HEK293 cells line stably transfected with R7BP alone (Fig. 2A). Co-expression with Gβ5 in this cell line had only minimal effect on the stability of RGS9-1 (Fig. 2A). Strikingly, co-immunoprecipitation assays showed that RGS9-1 can form complexes with R7BP only when Gβ5 is present (Fig. 2A). Similar observations were made when cells were transfected with R9AP instead of R7BP (Fig. 2B). Again, R9AP failed to co-precipitate with RGS9-1 in the absence of Gβ5 (Fig. 2B). Nevertheless, RGS9-1 expressed without Gβ5 retained its ability to interact with the AlF4−-activated Gαo, arguing that monomeric RGS9-1 retains the ability to associate with its functional partners (Fig. 2C).
FIGURE 2.
RGS9 interacts with its membrane anchors R7BP and R9AP in a Gβ5-dependent manner. A and B, RGS9-1 forms a complex with membrane anchors only in the presence of Gβ5 in vitro. The R7BP-stable cell line (A) or HEK293 cells transiently expressing R9AP (B) were transfected with indicated constructs. Cells were lysed and subjected to immunoprecipitation (IP) with anti-HA antibody. Expression levels of the transfected constructs (left) and their presence in the eluates after IP (right) were determined by Western blotting with their specific antibodies. Transfected cells without RGS9-1-HA served as a control for nonspecific binding. C, RGS9-1 interacts with activated Gαo both with and without Gβ5. Indicated constructs were co-transfected into HEK293 cells, cells were lysed in the buffer supplemented with GDP and AlF4−, and RGS9-1 complexes were immunoprecipitated with anti-HA antibodies. Protein content in the lysate and eluate fractions was analyzed with indicated specific antibodies. D, in native tissues, RGS9-2 co-immunoprecipitates with R7BP only in the presence of Gβ5. Striatal protein extracts were prepared from wild-type (WT) or knock-out mice lacking RGS9 (RGS9−/−), Gβ5 (Gβ5−/−), R7BP (R7BP−/−), and used for co-IP studies with anti-RGS9-2 antibody. Lysates and IP eluates were probed with indicated specific antibodies.
We further extended these observations and analyzed binding of RGS9 with its subunits in the native striatal tissues derived from mouse knock-out strains lacking individual proteins of the complex (Fig. 2D). We observed that knock-out of R7BP or Gβ5 resulted in the similar reduction in RGS9-2 levels. However, although RGS9-2 effectively co-precipitated with Gβ5 in the absence of R7BP, it failed to pull down R7BP in the absence of Gβ5 (Fig. 2D). Collectively, these findings demonstrate that the interaction between RGS9 and R7BP is dependent on Gβ5, both in transfected cells and in native striatal neurons.
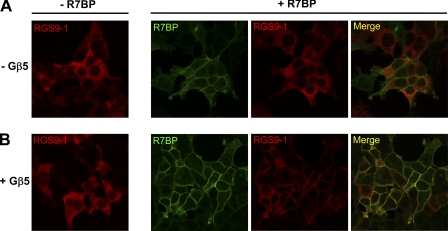
Plasma Membrane Recruitment of RGS9 by R7BP Is Gβ5-dependent
At the cellular level, one of the central effects elicited by the association with R7BP is the recruitment of RGS proteins to the plasma membrane which substantially augments their ability to regulate GPCR signaling (27, 31). Therefore, we next examined the effect of R7BP on the subcellular localization of RGS9 in the absence or presence of Gβ5. In the absence of its binding partners, monomeric RGS9-1 was found to be evenly distributed through the cytoplasm (Fig. 3A) as indicated by the presence of a nuclear shadow and the decrease in immunofluorescence toward the cell periphery. Co-expression with Gβ5 did not change this localization pattern, and the dimeric RGS9-1-Gβ5 complex remained mainly cytoplasmic (Fig. 3B). Co-transfection of RGS9-1 with Gβ5 into a R7BP-stable cell line resulted in the marked recruitment of the complex to the plasma membrane, as visualized by the sharp staining in the cell periphery and co-localization with R7BP immunofluorescence (Fig. 3B), as noted before (28, 43). In contrast, in the absence of Gβ5, RGS9-1 failed to be recruited to the plasma membrane by R7BP and remained mostly cytosolic (Fig. 3A). We therefore conclude that R7BP-driven recruitment of RGS9-1 to the plasma membrane requires Gβ5. These observations support a critical role of Gβ5 in the interaction of RGS9 isoforms with membrane anchors.
FIGURE 3.
Expression of Gβ5 induces R7BP-dependent plasma membrane localization of RGS9. Effect of co-expression with R7BP on RGS9-1 localization was studied in the absence (A) or presence (B) of Gβ5. HEK293 or an R7BP-stable cell line was transiently transfected with the indicated constructs encoding RGS9-1-HA or Gβ5. At 48 h after transfection, cells were fixed and subjected to immunostaining for RGS9-1 (red) and R7BP (green) followed by the image acquisition on a confocal microscope. Green and red images were merged to determine co-localization, and overlapping pixels appear as yellow pseudofluorescence. Representative images of the intracellular distribution for each condition are shown.
Gβ5 and an N-terminal DEP/DHEX Module Are Not Sufficient for Binding to R7BP
The observation that RGS9 can bind to R7BP only in the presence of Gβ5 could be explained if Gβ5 binds directly to R7BP. Because this possibility has not been addressed before, we have next investigated potential protein-protein interaction between Gβ5 and R7BP. Using a co-immunoprecipitation approach we did not detect any appreciable interaction of Gβ5 with R7BP in the absence of RGS9-1 (Fig. 4A). Accordingly, when protein localization was studied in the intact cells by immunostaining, Gβ5 failed to be recruited to the plasma membranes by R7BP when RGS9-1 was not present in the cells (Fig. 4B). In contrast, addition of RGS9-1 induced Gβ5 binding to R7BP (Fig. 4A) and resulted in its translocation to the plasma membrane (Fig. 4B). These data indicate that Gβ5 by itself does not have any significant R7BP binding affinity.
FIGURE 4.
Gβ5, N-terminal DEP-DHEX module of RGS9, or their complex does not interact with R7BP. A, co-IP analysis of interaction between R7BP and Gβ5. Cell lysates were prepared from HEK293 cells transiently transfected with indicated expression plasmids and used for co-immunoprecipitation studies with anti-R7BP antibody. B, R7BP recruits Gβ5 to the plasma membrane only in the presence of RGS9. HEK293 cells and/or an R7BP-stable cell line was transiently transfected with Gβ5 with or without RGS9-1-HA. At 48 h after transfection, cells were subjected to immunofluorescence staining with anti-Gβ5 antibody. C, R7BP fails to bind to DEP/DHEX module of RGS9 in the absence or presence of Gβ5. Cell lysates were prepared from an R7BP-stable cell line transiently transfected with indicated expression plasmids and used for co-immunoprecipitation with anti-myc antibody.
The association of RGS9-Gβ5 with R7BP has been also shown to require the presence of the intact DEP/DHEX domain of RGS9 (30, 34) that forms direct contacts with Gβ5 (38, 44). Therefore, we have next tested whether R7BP could bind to the complex between DEP/DHEX domains and Gβ5 reconstituted in trans. Although the DEP/DHEX domain was able to pull down Gβ5 efficiently upon co-expression in the cells, it failed to recruit R7BP to the complex as evidenced by the immunoprecipitation data (Fig. 4C). Neither did we find evidence for an appreciable interaction between R7BP and the DEP/DHEX domain in the absence of Gβ5. These data indicate that neither Gβ5 nor the N-terminal DEP/DHEX module alone or in combination is sufficient to enable binding of R7BP to the complex.
Interaction of R7BP with RGS9-Gβ5 Complex Requires Intact Conformation of the Interface between Gβ5 and N Terminus of RGS9
The inability to reconstitute R7BP binding to the complex between the DEP/DHEX domains and Gβ5 suggests that recruitment of R7BP is sensitive to the conformational orientation of the elements involved in the establishment of interface between Gβ5 and DEP/DHEX module. Analysis of the crystal structure of RGS9-1-Gβ5 complex reveals that contacts with Gβ5 mainly mediated by either the DEP domain with helixes α2–α3 contributing the most, and the linker loop connecting α4 helix and short β1 in the DHEX domain (Fig. 5A,B) (38). To study the role of these regions in mediating the interaction with R7BP, we have introduced point mutations predicted to disrupt the conformation of the interface between Gβ5 and the N-terminal lobe of RGS9. The F16E/F76E double mutation was introduced to the DEP domain of RGS9 (DEP mutant), and the amino acids 202DYGL205 and 207RVT209 were replaced to alanines in the DHEX linker loop (Linker mutant). We have next examined the effects of these mutations on the proteolytic stability and Gβ5-induced interaction with R7BP. When transfected in cells without Gβ5, both mutants showed expression levels similar to wild-type RGS9-1, indicating that their basal stability is uncompromised (Fig. 5, C and D). However, the ability of Gβ5 to increase the levels of RGS9-1 mutants in R7BP-dependent fashion was markedly compromised (Fig. 5, C and E). Although the R7BP/Gβ5-induced stabilization of the Linker mutant was reduced by ∼70%, the protective effect of Gβ5-R7BP was completely lost in case of the DEP mutant.
FIGURE 5.
Mutations in DHEX linker and DEP domain of RGS9 differentially affect its proteolytic stability and R7BP binding. A, mapping of regions in Gβ5 interface with the N-terminal DEP/DHEX domains on the structure of the RGS9-Gβ5 complex. Residues in DHEX linker (red) and DEP domain (blue) involved in interaction with Gβ5 are highlighted by spheres. B, mapping of regions in Gβ5 involved in contacts with the DHEX linker (red) and DEP domain (blue). The crystal structure described in Ref. 38 was used to generate information presented in A and B. C, effects of mutations in DHEX linker and DEP domain on expression level of RGS9- and R7BP-dependent stabilization by Gβ5. The wild-type, Linker, and DEP mutants of HA-tagged RGS9-1 were expressed in R7BP-stable cell line with or without Gβ5. Expression levels of the transfected constructs were determined by quantitative Western blotting with specific antibodies. D and E, quantification of RGS9 expression levels from C. Band intensities of RGS9-1 were determined and normalized to those of β-actin present in the same samples. Data are plotted relative to wild-type RGS9-1. F, effects of the mutations on R7BP-binding property of RGS9. Protein content in immunoprecipitates was determined by quantitative Western blotting with specific antibodies. G and H, quantification of interaction relative to wild-type RGS9-1. Band intensities of indicated proteins were determined and normalized to those of RGS9-1-HA present in the same immunoprecipitates. Values shown are the mean ± S.E. (error bars) from a representative experiment performed in quadruplicate, which was repeated with similar results. Values in E are obtained from two independent experiments performed in quadruplicate. ##, p < 0.01; ***, p < 0.001; ns, no significance, Student's t test.
We next studied the ability of the mutants to interact with Gβ5 and R7BP by co-immunoprecipitation (Fig. 5, F–H). Both DEP and Linker mutants showed normal interaction with Gβ5. In contrast, their association with R7BP was substantially compromised. Although both mutants retained some ability to recruit R7BP, the strength of this association was much lower than observed for the wild-type RGS9-1. Interestingly, the DEP mutant was affected half as much as the Linker mutant. These differences did not correlate with the differences seen in the stabilizing effects of R7BP on the expression level of RGS9 mutants. These observations indicate that interaction interface between Gβ5 and the N-terminal lobe of RGS9 controls association of the RGS9-Gβ5 complex with R7BP. Our results also suggest that R7BP recruitment is separate from the proteolytic stabilization effects that likely require additional conformational changes.
DISCUSSION
The main observation of this study is that the formation of the RGS9-Gβ5-R7BP (R9AP) complex occurs sequentially and there is a distinct hierarchy of RGS9 association with its subunits. First, RGS9 forms a complex with Gβ5, and then this dimer recruits membrane-targeting subunit R7BP or R9AP. Interestingly, we find that neither RGS9 nor Gβ5 alone can bind to R7BP(R9AP). Thus, our study for the first time establishes the assembly relationship between the subunits of the trimeric complex and demonstrates that the Gβ5 subunit is required for the interaction of RGS9-containing complex with R7BP or R9AP.
Previous studies have shown that the N-terminal DEP domain of RGS9 is necessary for the interaction of the RGS9-Gβ5 complex with both R9AP and R7BP (26, 30) and that it acts synergistically with the DHEX domain to establish the selectivity of R7BP/R9AP binding (34). Furthermore, it was reported that the N termini of RGS9 homologues, RGS7 and Egl-10, can interact directly with Gβ5 (44, 45). The crystal structure of the RGS9-Gβ5 complex confirmed that DEP and DHEX domains form a structurally integrated module that makes direct contacts with the protein interaction interface of Gβ5 (38). Therefore, to account for the dual requirement for both Gβ5 and the DEP domain we suggest that the binding site for R7BP and R9AP is formed by the interface between the DEP/DHEX module and Gβ5. This model is consistent with our site-directed mutagenesis data showing that substitution of amino acids in RGS9 involved in direct contacts with Gβ5 substantially reduces R7BP recruitment to the complex (39).
Unexpectedly, we found that there was no correlation between the ability of the “interface” mutations to disrupt R7BP binding and affect the stability of RGS9-Gβ5 complexes. For example, mutating the linker loop in DHEX domain severely compromises R7BP binding but does not eliminate its effects on the proteolytic stability of the complexes. Conversely, mutations of the DEP domain amino acids involved in contacts with Gβ5 completely eliminate proteolytic stabilization by R7BP but exert less severe effects on R7BP binding compared with the DHEX linker mutant. The latter observation is in line with the report that reciprocal mutations in Gβ5 that disrupt interaction with the DEP domain also completely abolish stabilizing effects of R7BP while preserving its recruitment to the complex (39). Taken together, these observations indicate that even though binding of R7BP leads to the proteolytic stabilization of the complex, these are two mechanistically separate events that are not firmly hardwired together. Furthermore, our data pinpoint the underlying molecular determinants: the contacts of Gβ5 with the DEP domain appear to regulate the proteolytic stability of the complexes, whereas coordination of the DHEX linker region seems to be primarily engaged in R7BP (R9AP) binding. Interestingly, Gβ5 uses distinct clusters on its surface for establishing contacts with the DEP and DHEX linker regions (38) (Fig. 5B). Although understanding precise details of R7BP interaction with RGS9-Gβ5 would certainly require determination of the RGS9-Gβ5-R7BP complex structure at the atomic level, it is tempting to speculate that R7BP binds to the interdomain cleft between the DHEX and the Gβ5 in the close vicinity to the DHEX loop region. Supporting this model is the observation that RGS7, a close homologue of RGS9 that also binds Gβ5, has a markedly different composition of the DHEX linker region (containing ∼25 additional unique amino acids). This correlates with drastically lower affinity of RGS7 for R7BP binding (37). Furthermore, the RGS7-Gβ5 complex does not require R7BP binding to achieve proteolytic stabilization (29, 34).
The recruitment of Gβ5 to the R7 RGS proteins is primarily mediated by the Gγ-like domain of the RGS proteins. This interaction resembles classical stable pairing of the G protein βγ subunits (38) and is mediated by the specialized CCT chaperonin complex that permanently locks the subunits together (46). In contrast, the association of the Gβ5 at the opposite interface with the DEP/DHEX module of the RGS proteins appears to be dynamic. Narayanan et al. (44) reported that Gβ5 can bind to the DEP domain of RGS7 and that this interaction is dynamic and is modulated by the R7BP. In addition, the same group has shown that the DEP domain of RGS7 can also bind to the third intracellular loop of M3 receptor and that this interaction is negatively modulated by R7BP (47). On the basis of these observations it was proposed that the interaction of Gβ5 with the DEP/DHEX module is dynamic. Our findings suggest that a similar mechanism modulating the interface between DEP/DHEX and Gβ5 is involved in controlling the proteolytic stability of the R7 RGS complexes. We propose that the opening of the interface at the DEP/Gβ5 site exposes degradation determinants located in the DEP/DHEX module and facilitates proteolysis of the complex. Binding of R7BP in the vicinity of the DHEX linker located on the other side of the interface serves as a “prop” to restrict the dynamics of the interactions and to promote closed conformation, thus resulting in shielding of the degradation determinants and proteolytic stabilization (Fig. 6). In summary, our results indicate that intramolecular interactions between Gβ5 and DEP/DHEX module of RGS9 determine protein expression of the complex and control its localization in cells, revealing a powerful regulatory hot-spot in the R7 RGS proteins.
FIGURE 6.
Model of the heterotrimeric RGS9-Gβ5-R7BP complex formation. RGS9 forms tight complex with Gβ5 via its Gγ-like (GGL) domain. In addition, the N-terminal DEP/DHEX module transiently interacts with the opposite surface of Gβ5 through two points of contact involving DHEX linker (red circles) and DEP domain (blue circles). The association of membrane anchors R7BP or R9AP with the complex is mediated by the interface between Gβ5 and the DEP/DHEX module, primarily involving DHEX linker region. Binding of R7BP/R9AP changes the conformation of the interface stabilizing the DEP contacts that contain instability determinants, thus providing proteolytic protection.
Acknowledgments
We thank Dr. William Simonds (NIDDK, National Institutes of Health) for the anti-Gβ5 and anti-R7BP antibodies.
This work was supported, in whole or in part, by National Institutes of Health Grants EY018139 (to K. A. M.), DA026405 (to K. A. M.), and DA021743 (to K. A. M.).
- RGS
- regulator of G protein signaling
- Gβ5
- type 5 G protein β subunit
- GPCR
- G protein-coupled receptor
- R7BP
- R7-binding protein
- R9AP
- RGS9 anchor protein
- DEP
- Dishevelled, EGL-10, Pleckstrin
- DHEX
- DEP Helical Extension.
REFERENCES
- 1. Siderovski D. P., Willard F. S. (2005) Int. J. Biol. Sci. 1, 51–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hollinger S., Hepler J. R. (2002) Pharmacol. Rev. 54, 527–559 [DOI] [PubMed] [Google Scholar]
- 3. Ross E. M., Wilkie T. M. (2000) Annu. Rev. Biochem. 69, 795–827 [DOI] [PubMed] [Google Scholar]
- 4. De Vries L., Zheng B., Fischer T., Elenko E., Farquhar M. G. (2000) Annu. Rev. Pharmacol. Toxicol. 40, 235–271 [DOI] [PubMed] [Google Scholar]
- 5. Hooks S. B., Martemyanov K., Zachariou V. (2008) Biochem. Pharmacol. 75, 76–84 [DOI] [PubMed] [Google Scholar]
- 6. Kehrl J. H. (2006) Immunol. Res. 34, 211–227 [DOI] [PubMed] [Google Scholar]
- 7. Traynor J. R., Neubig R. R. (2005) Mol. Interv. 5, 30–41 [DOI] [PubMed] [Google Scholar]
- 8. Wieland T., Lutz S., Chidiac P. (2007) Curr. Opin. Pharmacol. 7, 201–207 [DOI] [PubMed] [Google Scholar]
- 9. Anderson G. R., Posokhova E., Martemyanov K. A. (2009) Cell Biochem. Biophys. 54, 33–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Slepak V. Z. (2009) Prog. Mol. Biol. Transl. Sci. 86, 157–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Martemyanov K. A., Arshavsky V. Y. (2009) in Progress in Molecular Biology and Translation Science (Fisher R. ed) pp. 205–227, Academic Press, Orlando FL: [DOI] [PubMed] [Google Scholar]
- 12. Traynor J. R., Terzi D., Caldarone B. J., Zachariou V. (2009) Trends Pharmacol. Sci. 30, 105–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. He W., Cowan C. W., Wensel T. G. (1998) Neuron 20, 95–102 [DOI] [PubMed] [Google Scholar]
- 14. Chen C. K., Burns M. E., He W., Wensel T. G., Baylor D. A., Simon M. I. (2000) Nature 403, 557–560 [DOI] [PubMed] [Google Scholar]
- 15. Nishiguchi K. M., Sandberg M. A., Kooijman A. C., Martemyanov K. A., Pott J. W., Hagstrom S. A., Arshavsky V. Y., Berson E. L., Dryja T. P. (2004) Nature 427, 75–78 [DOI] [PubMed] [Google Scholar]
- 16. Thomas E. A., Danielson P. E., Sutcliffe J. G. (1998) J. Neurosci. Res. 52, 118–124 [DOI] [PubMed] [Google Scholar]
- 17. Gold S. J., Ni Y. G., Dohlman H. G., Nestler E. J. (1997) J. Neurosci. 17, 8024–8037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang K., Howes K. A., He W., Bronson J. D., Pettenati M. J., Chen C., Palczewski K., Wensel T. G., Baehr W. (1999) Gene 240, 23–34 [DOI] [PubMed] [Google Scholar]
- 19. Cabrera-Vera T. M., Hernandez S., Earls L. R., Medkova M., Sundgren-Andersson A. K., Surmeier D. J., Hamm H. E. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 16339–16344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gold S. J., Hoang C. V., Potts B. W., Porras G., Pioli E., Kim K. W., Nadjar A., Qin C., LaHoste G. J., Li Q., Bioulac B. H., Waugh J. L., Gurevich E., Neve R. L., Bezard E. (2007) J. Neurosci. 27, 14338–14348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kovoor A., Seyffarth P., Ebert J., Barghshoon S., Chen C. K., Schwarz S., Axelrod J. D., Cheyette B. N., Simon M. I., Lester H. A., Schwarz J. (2005) J. Neurosci. 25, 2157–2165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rahman Z., Schwarz J., Gold S. J., Zachariou V., Wein M. N., Choi K. H., Kovoor A., Chen C. K., DiLeone R. J., Schwarz S. C., Selley D. E., Sim-Selley L. J., Barrot M., Luedtke R. R., Self D., Neve R. L., Lester H. A., Simon M. I., Nestler E. J. (2003) Neuron 38, 941–952 [DOI] [PubMed] [Google Scholar]
- 23. Zachariou V., Georgescu D., Sanchez N., Rahman Z., DiLeone R., Berton O., Neve R. L., Sim-Selley L. J., Selley D. E., Gold S. J., Nestler E. J. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 13656–13661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Makino E. R., Handy J. W., Li T., Arshavsky V. Y. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 1947–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hu G., Wensel T. G. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 9755–9760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Martemyanov K. A., Yoo P. J., Skiba N. P., Arshavsky V. Y. (2005) J. Biol. Chem. 280, 5133–5136 [DOI] [PubMed] [Google Scholar]
- 27. Drenan R. M., Doupnik C. A., Jayaraman M., Buchwalter A. L., Kaltenbronn K. M., Huettner J. E., Linder M. E., Blumer K. J. (2006) J. Biol. Chem. 281, 28222–28231 [DOI] [PubMed] [Google Scholar]
- 28. Drenan R. M., Doupnik C. A., Boyle M. P., Muglia L. J., Huettner J. E., Linder M. E., Blumer K. J. (2005) J. Cell Biol. 169, 623–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Anderson G. R., Lujan R., Semenov A., Pravetoni M., Posokhova E. N., Song J. H., Uversky V., Chen C. K., Wickman K., Martemyanov K. A. (2007) J. Neurosci. 27, 14117–14127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Martemyanov K. A., Lishko P. V., Calero N., Keresztes G., Sokolov M., Strissel K. J., Leskov I. B., Hopp J. A., Kolesnikov A. V., Chen C. K., Lem J., Heller S., Burns M. E., Arshavsky V. Y. (2003) J. Neurosci. 23, 10175–10181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cao Y., Kolesnikov A. V., Masuho I., Kefalov V. J., Martemyanov K. A. (2010) J. Neurosci. 30, 13784–13793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chen C. K., Eversole-Cire P., Zhang H., Mancino V., Chen Y. J., He W., Wensel T. G., Simon M. I. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 6604–6609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Keresztes G., Martemyanov K. A., Krispel C. M., Mutai H., Yoo P. J., Maison S. F., Burns M. E., Arshavsky V. Y., Heller S. (2004) J. Biol. Chem. 279, 1581–1584 [DOI] [PubMed] [Google Scholar]
- 34. Anderson G. R., Semenov A., Song J. H., Martemyanov K. A. (2007) J. Biol. Chem. 282, 4772–4781 [DOI] [PubMed] [Google Scholar]
- 35. Krispel C. M., Chen D., Melling N., Chen Y. J., Martemyanov K. A., Quillinan N., Arshavsky V. Y., Wensel T. G., Chen C. K., Burns M. E. (2006) Neuron 51, 409–416 [DOI] [PubMed] [Google Scholar]
- 36. Grabowska D., Jayaraman M., Kaltenbronn K. M., Sandiford S. L., Wang Q., Jenkins S., Slepak V. Z., Smith Y., Blumer K. J. (2008) Neuroscience 151, 969–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Anderson G. R., Lujan R., Martemyanov K. A. (2009) Mol. Cell. Biol. 29, 3033–3044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cheever M. L., Snyder J. T., Gershburg S., Siderovski D. P., Harden T. K., Sondek J. (2008) Nat. Struct. Mol. Biol. 15, 155–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Porter M. Y., Xie K., Pozharski E., Koelle M. R., Martemyanov K. A. (2010) J. Biol. Chem. 285, 41100–41112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Keresztes G., Mutai H., Hibino H., Hudspeth A. J., Heller S. (2003) Mol. Cell. Neurosci. 24, 687–695 [DOI] [PubMed] [Google Scholar]
- 41. Pear W. S., Nolan G. P., Scott M. L., Baltimore D. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 8392–8396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Posokhova E., Uversky V., Martemyanov K. A. (2010) J. Proteome Res. 9, 1510–1521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Song J. H., Waataja J. J., Martemyanov K. A. (2006) J. Biol. Chem. 281, 15361–15369 [DOI] [PubMed] [Google Scholar]
- 44. Narayanan V., Sandiford S. L., Wang Q., Keren-Raifman T., Levay K., Slepak V. Z. (2007) Biochemistry 46, 6859–6870 [DOI] [PubMed] [Google Scholar]
- 45. Patikoglou G. A., Koelle M. R. (2002) J. Biol. Chem. 277, 47004–47013 [DOI] [PubMed] [Google Scholar]
- 46. Howlett A. C., Gray A. J., Hunter J. M., Willardson B. M. (2009) J. Biol. Chem. 284, 16386–16399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sandiford S. L., Slepak V. Z. (2009) Biochemistry 48, 2282–2289 [DOI] [PMC free article] [PubMed] [Google Scholar]